Promising New Methods Based on the SOD Enzyme and SAUR36 Gene to Screen for Canola Materials with Heavy Metal Resistance
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Experimental Method
2.2.1. Treatment Methods for Heavy Metal Stress
2.2.2. Physiological Indexes under Heavy Metal Stress
2.2.3. Omics Analysis
2.2.4. Quantitative Real-Time PCR (RT-qPCR) Detection
2.2.5. Field Experiment
2.3. Data Analysis
3. Results
3.1. Performance of Canola NILs at Different Heavy Metal Concentrations
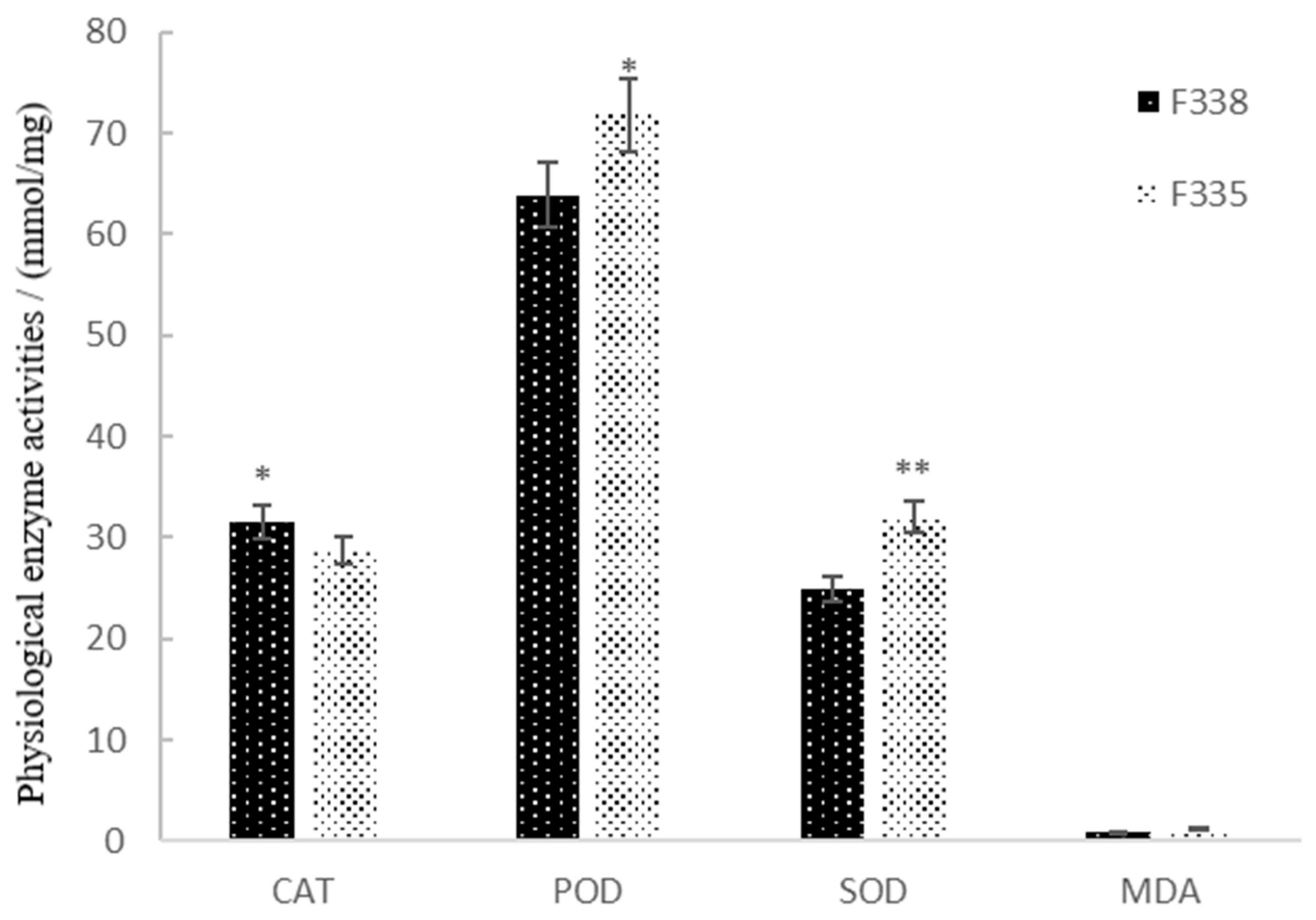
3.2. Physiological Performance of Canola NILs in Resisting Heavy Metal Stress
3.3. Omics Association Analyses of the Canola NILs in Response to Heavy Metals
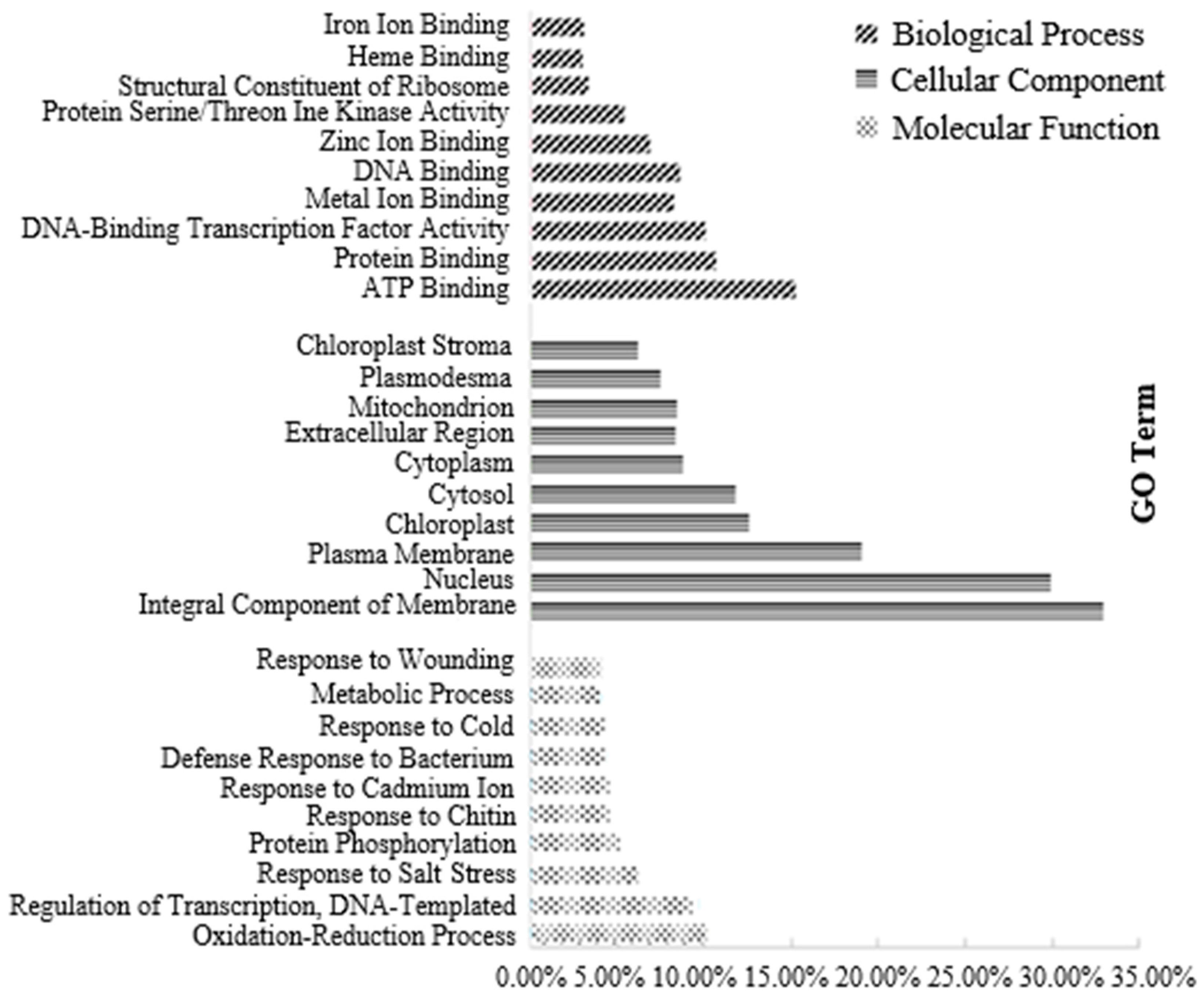
3.3.1. Transcriptome Analysis of Canola NILs under 100× Heavy Metal Stress
3.3.2. iTRAQ Analysis of Canola NILs under 100× Heavy Metal Stress
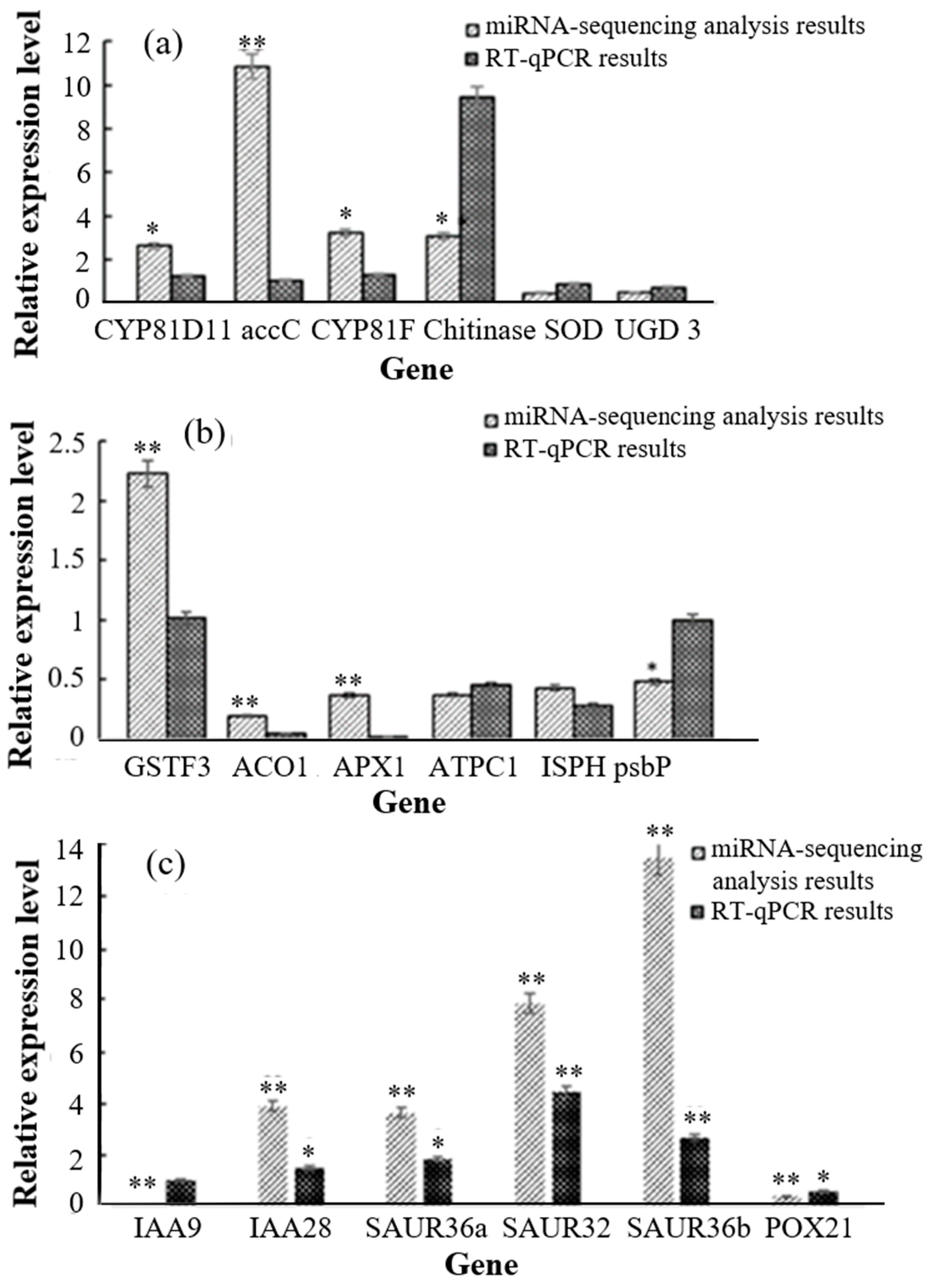
3.4. Validation of Transcription and Proteomics Analyses Using Real-Time Quantitative PCR (RT-qPCR)
3.5. The Functional Verification of SAUR Genes
4. Discussion
4.1. Effects of Heavy Metal Stress on Physiological Performance of Canola
4.2. The Key Genes for Heavy Metal Stress Tolerance in Canola
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meilke, K.D.; Griffith, G.R. An application of the Market Share Approach to the demand for soyabean and rapeseed oil. Eur. Rev. Agric. Econ. 1981, 8, 85–97. [Google Scholar] [CrossRef]
- Shahid, M.; Cai, G.; Zu, F.; Zhao, Q.; Qasim, M.U.; Hong, Y.; Fan, C.; Zhou, Y. Comparative Transcriptome Analysis of Developing Seeds and Silique Wall Reveals Dynamic Transcription Networks for Effective Oil Production in Brassica napus L. Int. J. Mol. Sci. 2019, 20, 1982. [Google Scholar] [CrossRef]
- Yan, L.; Shah, T.; Cheng, Y.; LÜ, Y.; Zhang, X.K.; Zou, X.L. Physiological and molecular responses to cold stress in rapeseed (Brassica napus L.). J. Integr. Agric. 2019, 18, 2742–2752. [Google Scholar] [CrossRef]
- Deng, J.; Li, W.; Xu, W.; He, Z.; Tan, X. Correlation and the concentrations of Pb, Cd, Hg and As in vegetables and soils of Chongqing, China. Environ. Geochem. Health 2021, 43, 2357–2376. [Google Scholar] [CrossRef]
- Guo, Z.H.; Song, J.; Xiao, X.Y.; Ming, H.; Miao, X.F.; Wang, F.Y. Spatial distribution and environmental characterization of sediment-associated metals from middle-downstream of Xiangjiang River, southern China. J. Cent. South Univ. Technol. 2010, 17, 68–78. [Google Scholar] [CrossRef]
- Asad, S.A.; Farooq, M.; Afzal, A.; West, H. Integrated phytobial heavy metal remediation strategies for a sustainable clean environment-A review. Chemosphere 2019, 217, 925–941. [Google Scholar] [CrossRef]
- Munir, O.; Ersin, Y.; Salih, G.; Serdal Aksoy, A. Plants as Biomonitors of Trace Elements Pollution in Soil. In Trace Elements as Contaminants and Nutrients: Consequences in Ecosystems and Human Health; John Wiley & Sons: Hoboken, NJ, USA, 2008; pp. 723–744. [Google Scholar] [CrossRef]
- Liu, J.G.; Liang, J.S.; Li, K.Q.; Zhang, Z.J.; Yu, B.Y.; Lu, X.L.; Yang, J.C.; Zhu, Q.S. Correlations between cadmium and mineral nutrients in absorption and accumulation in various genotypes of rice under cadmium stress. Chemosphere 2003, 52, 1467–1473. [Google Scholar] [CrossRef]
- Chandra, R.; Kang, H. Mixed heavy metal stress on photosynthesis, transpiration rate, and chlorophyll content in poplar hybrids. For. Sci. Technol. 2015, 12, 55–61. [Google Scholar] [CrossRef]
- Iannelli, M.A.; Pietrini, F.; Fiore, L.; Petrilli, L.; Massacci, A. Antioxidant response to cadmium in Phragmites australis plants. Plant Physiol. Biochem. 2002, 40, 977–982. [Google Scholar] [CrossRef]
- Yan, H.; Filardo, F.; Hu, X.; Zhao, X.; Fu, D. Cadmium stress alters the redox reaction and hormone balance in oilseed rape (Brassica napus L.) leaves. Environ. Sci. Pollut. Res. Int. 2016, 23, 3758–3769. [Google Scholar] [CrossRef]
- Dalyan, E.; Yüzbaşıoğlu, E.; Akpınar, I. Effect of 24-Epibrassinolide on Antioxidative Defence System Against Lead-Induced Oxidative Stress in The Roots of Brassica juncea L. seedlings. Russ. J. Plant Physiol. 2018, 65, 570–578. [Google Scholar] [CrossRef]
- Kohli, S.K.; Handa, N.; Sharma, A.; Gautam, V.; Arora, S.; Bhardwaj, R.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P. Combined effect of 24-epibrassinolide and salicylic acid mitigates lead (Pb) toxicity by modulating various metabolites in Brassica juncea L. seedlings. Protoplasma 2018, 255, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.X.; Wang, G.Z.; Zhang, Y.X.; Zhao, H.J. Hydroxyapatite nanoparticles in root cells: Reducing the mobility and toxicity of Pb in rice. Environ. Sci. Nano 2018, 5, 398–407. [Google Scholar] [CrossRef]
- Meharg, A.A.; Hartley, W.J. Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytol. 2002, 154, 29–43. [Google Scholar] [CrossRef]
- Wei, B.; Yang, L. A review of heavy metal contaminations in urban soils, urban road dusts and agricultural soils from China. Microchem. J. 2010, 94, 99–107. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, C.; Chen, H.; Wang, P.; Wang, P.; Song, C.; Zhang, X.; Wang, D. Multi-gene co-expression can improve comprehensive resistance to multiple abiotic stresses in Brassica napus L. Plant Sci. 2018, 274, 410–419. [Google Scholar] [CrossRef]
- Hossain, M.A.; Piyatida, P.; Silva, J.A.T.; Fujita, M. Molecular Mechanism of Heavy Metal Toxicity and Tolerance in Plants: Central Role of Glutathione in Detoxification of Reactive Oxygen Species and Methylglyoxal and in Heavy Metal Chelation. J. Bot. 2012, 2012, 1–37. [Google Scholar] [CrossRef]
- Shahid, M.; Dumat, C.; Khalid, S.; Schreck, E.; Xiong, T.; Niazi, N.K. Foliar heavy metal uptake, toxicity and detoxification in plants: A comparison of foliar and root metal uptake. J. Hazard. Mater. 2017, 325, 36–58. [Google Scholar] [CrossRef]
- Yu, X.Z.; Lin, Y.J.; Zhang, Q. Metallothioneins enhance chromium detoxification through scavenging ROS and stimulating metal chelation in Oryza sativa. Chemosphere 2019, 220, 300–313. [Google Scholar] [CrossRef]
- Natasha, S.M.; Khalid, S.; Bibi, I.; Bundschuh, J.; Khan, N.N.; Dumat, C. A critical review of mercury speciation, bioavailability, toxicity and detoxification in soil-plant environment: Ecotoxicology and health risk assessment. Sci. Total Environ. 2020, 711, 134749. [Google Scholar] [CrossRef]
- Wang, S.; Sun, J.; Li, S.; Lu, K.; Meng, H.; Xiao, Z.; Zhang, Z.; Li, J.; Luo, F.; Li, N. Physiological, genomic and transcriptomic comparison of two Brassica napus cultivars with contrasting cadmium tolerance. Plant Soil. 2019, 441, 71–87. [Google Scholar] [CrossRef]
- Guo, J.; Dai, X.; Xu, W.; Ma, M. Overexpressing GSH1 and AsPCS1 simultaneously increases the tolerance and accumulation of cadmium and arsenic in Arabidopsis thaliana. Chemosphere 2008, 72, 1020–1026. [Google Scholar] [CrossRef]
- Xie, T.; Yang, W.; Chen, X.; Rong, H.; Wang, Y.; Jiang, J. Genome-Wide Identification and Expressional Profiling of the Metal Tolerance Protein Gene Family in Brassica napus. Genes 2022, 13, 761. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Jian, H.; Wang, T.; Di, F.; Wang, J.; Li, J.; Liu, L. Screening of candidate gene responses to cadmium stress by RNA sequencing in oilseed rape (Brassica napus L.). Environ. Sci. Pollut. Res. Int. 2018, 25, 32433–32446. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Parihar, P.; Singh, R.; Singh, V.P.; Prasad, S.M. Heavy Metal Tolerance in Plants: Role of Transcriptomics, Proteomics, Metabolomics, and Ionomics. Front. Plant Sci. 2015, 6, 1143. [Google Scholar] [CrossRef]
- Adejumo, S.A.; Tiwari, S.; Thul, S.; Sarangi, B.K. Evaluation of lead and chromium tolerance and accumulation level in Gomphrena celosoides: A novel metal accumulator from lead acid battery waste contaminated site in Nigeria. Int. J. Phytoremediation 2019, 21, 1341–1355. [Google Scholar] [CrossRef]
- Sruthi, P.; Puthur, J.T. Characterization of physiochemical and anatomical features associated with enhanced phytostabilization of copper in Bruguiera cylindrica (L.) Blume. Int. J. Phytoremediation 2019, 21, 1423–1441. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Zhang, R.; Hu, X.; Song, J.; Li, B.; Ou, D.; Hu, X.; Zhao, Y. Exogenous spermidine elevating cadmium tolerance in Salix matsudana involves cadmium detoxification and antioxidant defense. Int. J. Phytoremediation 2019, 21, 305–315. [Google Scholar] [CrossRef]
- Soares, T.; Dias, D.; Oliveira, A.M.S.; Ribeiro, D.M.; Dias, L. Exogenous brassinosteroids increase lead stress tolerance in seed germination and seedling growth of Brassica juncea L. Ecotoxicol. Environ. Saf. 2020, 193, 110296. [Google Scholar] [CrossRef]
- Kania, J.; Krawczyk, T.; Gillner, D.M. Oilseed rape (Brassica napus): The importance of aminopeptidases in germination under normal and heavy metals stress conditions. J. Sci. Food Agric. 2021, 101, 6533–6541. [Google Scholar] [CrossRef]
- Yang, P.M.; Huang, Q.C.; Qin, G.Y.; Zhao, S.P.; Zhou, J.G. Different drought-stress responses in photosynthesis and reactive oxygen metabolism between autotetraploid and diploid rice. Photosynthetica 2014, 52, 193–202. [Google Scholar] [CrossRef]
- Shi, J.; Fu, X.Z.; Peng, T.; Huang, X.S.; Fan, Q.J.; Liu, J.H. Spermine pretreatment confers dehydration tolerance of citrus in vitro plants via modulation of antioxidative capacity and stomatal response. Tree Physiol. 2010, 30, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Draper, H.H.; Squires, E.J.; Mahmoodi, H.; Wu, J.; Agarwal, S.; Hadley, M. A comparative evaluation of thiobarbituric acid methods for the determination of malondialdehyde in biological materials. Free Radic. Biol. Med. 1993, 15, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Yan, L.; Ma, X.; Chen, Y.; Wu, L.; Ma, T.; Zhao, L.; Yi, B.; Ma, C.; Tu, J. Combined BSA-Seq Based Mapping and RNA-Seq Profiling Reveal Candidate Genes Associated with Plant Architecture in Brassica napus. Int. J. Mol. Sci. 2022, 23, 2472. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Goloubinoff, P.; Christen, P. Heavy metal ions are potent inhibitors of protein folding. Biochem. Biophys. Res. Commun. 2008, 372, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Tamas, M.J.; Sharma, S.K.; Ibstedt, S.; Jacobson, T.; Christen, P. Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 2014, 4, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.L.; Rizwan, M.; Li, M.; Song, F.; Zhou, S.J.; He, X.; Ding, R.; Dai, Z.H.; Yuan, Y.; Cao, M.H.; et al. Comparative efficacy of organic and inorganic silicon fertilizers on antioxidant response, Cd/Pb accumulation and health risk assessment in wheat (Triticum aestivum L.). Environ. Pollut. 2019, 255, 113146. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.A.; Jiao, Y.Y.; Chen, C.; Shireen, F.; Zheng, Z.H.; Imtiaz, M.; Bie, Z.L.; Huang, Y. Melatonin pretreatment improves vanadium stress tolerance of watermelon seedlings by reducing vanadium concentration in the leaves and regulating melatonin biosynthesis and antioxidant-related gene expression. J. Plant Physiol. 2018, 220, 115–127. [Google Scholar] [CrossRef]
- Rahman, S.U.; Li, Y.L.; Hussain, S.; Hussain, B.; Khan, W.D.; Riaz, L.; Ashraf, M.N.; Khaliq, M.A.; Du, Z.J.; Cheng, H.F. Role of phytohormones in heavy metal tolerance in plants: A review. Ecol. Indic. 2023, 146, 109844. [Google Scholar] [CrossRef]
- Adrees, M.; Ali, S.; Rizwan, M.; Ibrahim, M.; Abbas, F.; Farid, M.; Zia-Ur-Rehman, M.; Irshad, M.K.; Bharwana, S.A. The effect of excess copper on growth and physiology of important food crops: A review. Environ. Sci. Pollut. Res. Int. 2015, 22, 8148–8162. [Google Scholar] [CrossRef]
- Barceló, J.; Poschenrieder, C. Plant water relations as affected by heavy metal stress: A review. J. Plant Nutr. 1990, 13, 1–37. [Google Scholar] [CrossRef]
- Laetitia, P.B.; Nathalie, L.; Alain, V.; Cyrille, F. Heavy metal toxicity cadmium permeates through calcium channels and disturbs the plant water status. Plant J. 2002, 32, 539–548. [Google Scholar]
- Karmous, I.; Bellani, L.M.; Chaoui, A.; Ferjani, E.; Muccifora, S. Effects of copper on reserve mobilization in embryo of Phaseolus vulgaris L. Environ. Sci. Pollut. Res. Int. 2015, 22, 10159–10165. [Google Scholar] [CrossRef] [PubMed]
- Baszyński, T. Interference of Cd2+ in functioning of the photosynthetic apparatus of higher plants. Acta Soc. Bot. Pol. 2014, 55, 291–304. [Google Scholar] [CrossRef]
- Luo, Z.B.; Jiali He, J.L.; Polle, A.; Rennenberg, H. Heavy metal accumulation and signal transduction in herbaceous and woody plants: Paving the way for enhancing phytoremediation efficiency. Biotechnol. Adv. 2016, 34, 1131–1148. [Google Scholar] [CrossRef] [PubMed]
- Jalmi, S.K.; Bhagat, P.K.; Verma, D.; Noryang, S.; Tayyeba, S.; Singh, K.; Sharma, D.; Sinha, A.K. Traversing the Links between Heavy Metal Stress and Plant Signaling. Front. Plant Sci. 2018, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Li, S.C.; Han, X.J.; Lu, Z.C.; Qiu, W.M.; Yu, M.; Li, H.Y.; He, Z.Q.; Zhuo, R.Y. MAPK Cascades and Transcriptional Factors: Regulation of Heavy Metal Tolerance in Plants. Int. J. Mol. Sci. 2022, 23, 4463. [Google Scholar] [CrossRef]
- Seneviratne, M.; Rajakaruna, N.; Rizwan, M.; Madawala, H.M.S.P.; Yong, S.O.; Vithanage, M. Heavy metal-induced oxidative stress on seed germination and seedling development: A critical review. Environ. Geochem. Health 2017, 41, 1813–1831. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Kudla, J.; Batistic, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef]
- Thao, N.P.; Khan, M.I.; Thu, N.B.; Hoang, X.L.; Asgher, M.; Khan, N.A.; Tran, L.S. Role of Ethylene and Its Cross Talk with Other Signaling Molecules in Plant Responses to Heavy Metal Stress. Plant Physiol. 2015, 169, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Lee, H.S.; Kwak, S.S. Differential responses of sweetpotato peroxidases to heavy metals. Chemosphere 2010, 81, 79–85. [Google Scholar] [CrossRef]
- Nazli, F.; Wang, X.; Ahmad, M.; Hussain, A.; Bushra Dar, A.; Nasim, M.; Jamil, M.; Panpluem, N.; Mustafa, A. Efficacy of Indole Acetic Acid and Exopolysaccharides-Producing Bacillus safensis Strain FN13 for Inducing Cd-Stress Tolerance and Plant Growth Promotion in Brassica juncea (L.). Appl. Sci. 2021, 11, 4160. [Google Scholar] [CrossRef]
- Stortenbeker, N.; Bemer, M. The SAUR gene family: The plant′s toolbox for adaptation of growth and development. J. Exp. Bot. 2019, 70, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G.; Guilfoyle, T.J.; Gray, W.M. Auxin signal transduction. In Plant Hormones; Springer: Berlin/Heidelberg, Germany, 2010; pp. 282–307. [Google Scholar] [CrossRef]
- Bemer, M.; Van, M.H.; Muino, J.M.; Ferrandiz, C.; Kaufmann, K.; Angenent, G.C. FRUITFULL controls SAUR10 expression and regulates Arabidopsis growth and architecture. J. Exp. Bot. 2017, 68, 3391–3403. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, W.; Gan, S.S. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013, 161, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Wang, J.; Gao, Z.; Dong, J.; He, H.; Terzaghi, W.; Wei, N.; Deng, X.W.; Chen, H. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc. Natl. Acad. Sci. USA 2016, 113, 6071–6076. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Verma, P.K.; Chakrabarty, D. Arsenic Bio-volatilization by Engineered Yeast Promotes Rice Growth and Reduces Arsenic Accumulation in Grains. Int. J. Environ. Res. 2019, 13, 475–485. [Google Scholar] [CrossRef]
- Rui, H.; Chen, C.; Zhang, X.; Shen, Z.; Zhang, F. Cd-induced oxidative stress and lignification in the roots of two Vicia sativa L. varieties with different Cd tolerances. J. Hazard. Mater. 2016, 301, 304–313. [Google Scholar] [CrossRef]
- Farooq, M.A.; Gill, R.A.; Ali, B.; Wang, J.; Islam, F.; Ali, S.; Zhou, W. Subcellular distribution, modulation of antioxidant and stress-related genes response to arsenic in Brassica napus L. Ecotoxicology 2016, 25, 350–366. [Google Scholar] [CrossRef]
- Garg, N.; Singla, P. Arsenic toxicity in crop plants: Physiological effects and tolerance mechanisms. Environ. Chem. Lett. 2011, 9, 303–321. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Mittler, R. The water-water cycle is essential for chloroplast protection in the absence of stress. J. Biol. Chem. 2003, 278, 38921–38925. [Google Scholar] [CrossRef] [PubMed]
- Myouga, F.; Hosoda, C.; Umezawa, T.; Iizumi, H.; Kuromori, T.; Motohashi, R.; Shono, Y.; Nagata, N.; Ikeuchi, M.; Shinozaki, K. A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 2008, 20, 3148–3162. [Google Scholar] [CrossRef] [PubMed]
- Basu, U.; Good, A.G.; Taylor, G.J. Transgenic Brassica napus plants overexpressing aluminium-induced mitochondrial manganese superoxide dismutase cDNA are resistant to aluminium. Plant Cell Environ. 2001, 24, 1269–1278. [Google Scholar] [CrossRef]
- Imtiaz, M.; Tu, S.; Xie, Z.; Han, D.; Ashraf, M.; Rizwan, M.S. Growth, V uptake, and antioxidant enzymes responses of chickpea (Cicer arietinum L.) genotypes under vanadium stress. Plant Soil. 2015, 390, 17–27. [Google Scholar] [CrossRef]
- Gokul, A.; Cyster, L.F.; Keyster, M. Efficient superoxide scavenging and metal immobilization in roots determines the level of tolerance to Vanadium stress in two contrasting Brassica napus genotypes. S. Afr. J. Bot. 2018, 119, 17–27. [Google Scholar] [CrossRef]
- Yu, X.Z.; Yang, L.; Feng, Y.X. Comparative response of SOD in different plants against cadmium and drought stress at the molecular level. Appl. Environ. Biotechnol. 2020, 5, 2. [Google Scholar] [CrossRef]
- Su, W.; Raza, A.; Gao, A.; Jiao, Z.Q.; Zhang, Y.; Hussain, M.A.; Mehmood, S.S.; Cheng, Y.; Lv, Y.; Zou, X.L. Genome-Wide Analysis and Expression Profile of Superoxide Dismutase (SOD) Gene Family in Rapeseed (Brassica napus L.) under Different Hormones and Abiotic Stress Conditions. Antioxidants 2021, 10, 1182. [Google Scholar] [CrossRef]
- Lee, S.Y.; Cheon, K.S.; Kim, S.Y.; Kim, J.H.; Kim, W.H. Expression of sod2 enhances tolerance to drought stress in roses. Hortic. Environ. Biotechnol. 2020, 61, 569–576. [Google Scholar] [CrossRef]
- Khare, S.; Singh, N.B.; Niharika Singh, A.; Amist, N.; Azim, Z.; Yadav, R.K. Phytochemicals mitigation of Brassica napus by IAA grown under Cd and Pb toxicity and its impact on growth responses of Anagallis arvensis. J. Biotechnol. 2022, 343, 83–95. [Google Scholar] [CrossRef]
- Ran, J.; Zheng, W.; Wang, H.; Wang, H.; Li, Q. Indole-3-acetic acid promotes cadmium (Cd) accumulation in a Cd hyperaccumulator and a non-hyperaccumulator by different physiological responses. Ecotoxicol. Environ. Saf. 2020, 191, 110213. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Y.; Prakash, V.; Yadav, V.; Chauhan, D.K.; Prasad, S.M.; Ramawat, N.; Singh, V.P.; Tripathi, D.K.; Sharma, S. Regulation of cadmium toxicity in roots of tomato by indole acetic acid with special emphasis on reactive oxygen species production and their scavenging. Plant Physiol. Biochem. 2019, 142, 193–201. [Google Scholar] [CrossRef]
- Fendrych, M.; Leung, J.; Friml, J. TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 2016, 5, e19048. [Google Scholar] [CrossRef] [PubMed]
- Spartz, A.K.; Lor, V.S.; Ren, H.; Olszewski, N.E.; Miller, N.D.; Wu, G.; Spalding, E.P.; Gray, W.M. Constitutive Expression of Arabidopsis SMALL AUXIN UP RNA19 (SAUR19) in Tomato Confers Auxin-Independent Hypocotyl Elongation. Plant Physiol. 2017, 173, 1453–1462. [Google Scholar] [CrossRef]
- Wu, J.; Liu, S.; He, Y.; Guan, X.; Zhu, X.; Cheng, L.; Wang, J.; Lu, G. Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 2012, 509, 38–50. [Google Scholar] [CrossRef]
- Kodaira, K.S.; Qin, F.; Tran, L.S.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi, S.K. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 2011, 157, 742–756. [Google Scholar] [CrossRef]
- He, Y.; Liu, Y.; Li, M.; Lamin-Samu, A.T.; Yang, D.; Yu, X.; Izhar, M.; Jan, I.; Ali, M.; Lu, G. The Arabidopsis SMALL AUXIN UP RNA32 Protein Regulates ABA-Mediated Responses to Drought Stress. Front. Plant Sci. 2021, 12, 625493. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Qi, M.Y.; Ding, X.H.; Zheng, Y.Y.; Zhou, T.J.; Chen, Y.; Han, N.; Zhu, M.Y.; Bian, H.W.; Wang, J.H. The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings. Ann. Bot. 2020, 125, 805–819. [Google Scholar] [CrossRef]
- Ma, X.Q.; Dai, S.T.; Qin, N.; Zhu, C.C.; Qin, J.F.; Li, J.X. Genome-wide identification and expression analysis of the SAUR gene family in foxtail millet (Setaria italica L.). BMC Plant Biol. 2023, 23, 31. [Google Scholar] [CrossRef]
- Huang, C.K.; Lo, P.C.; Huang, L.F.; Wu, S.J.; Yeh, C.H.; Lu, C.A. A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant Mol. Biol. 2015, 88, 269–286. [Google Scholar] [CrossRef]
- Stamm, P.; Kumar, P.P. Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep. 2013, 32, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wen, S.S.; Sun, T.T.; Wang, R.; Zuo, W.T.; Yang, T.; Wang, C.; Hu, J.J.; Lu, M.Z.; Wang, L.Q. PagWOX11/12a positively regulates the PagSAUR36 gene that enhances adventitious root development in poplar. J. Exp. Bot. 2022, 73, 7298–7311. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liang, W.; Li, Y.X.; Li, M.J.; Ma, B.Q.; Liu, C.H.; Ma, F.W.; Li, C.Y. Transcriptome analysis reveals the effects of alkali stress on root system architecture and endogenous hormones in apple rootstocks. J. Integr. Agric. 2019, 18, 2264–2271. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Yang, R.; Wang, F.; Fu, J.; Yang, W.; Bai, T.; Wang, S.; Yin, H. Effects of gibberellin priming on seedling emergence and transcripts involved in mesocotyl elongation in rice under deep direct-seeding conditions. J. Zhejiang Univ. Sci. B 2021, 22, 1002–1021. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, K.; Kereamy, A.E.; Kim, S.H.; Nambara, E.; Rothstein, S.J. ANAC032 Positively Regulates Age-Dependent and Stress-Induced Senescence in Arabidopsis thaliana. Kashif Mahmood Plant Cell Physiol. 2016, 57, 2029–2046. [Google Scholar] [CrossRef] [PubMed]
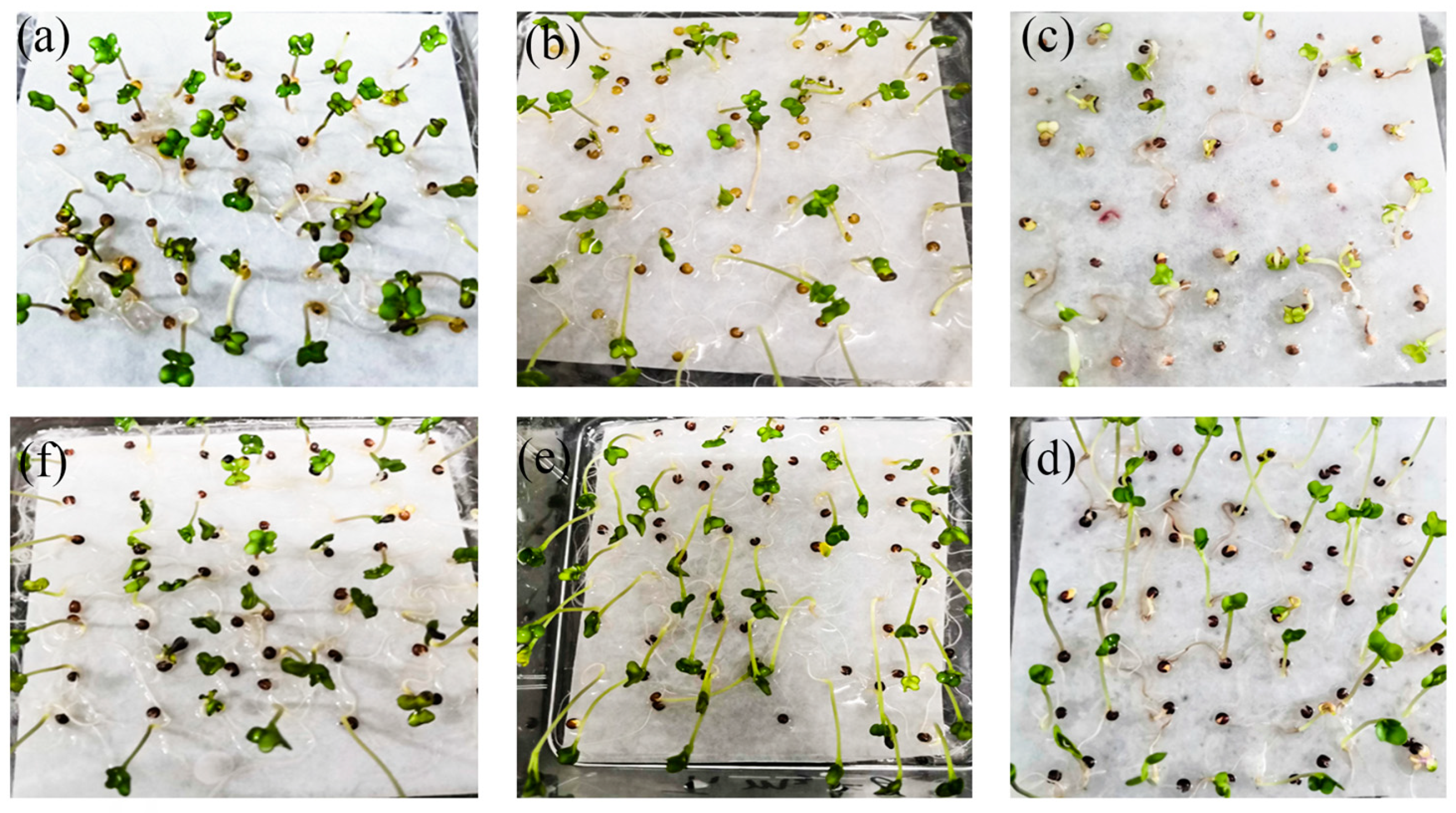
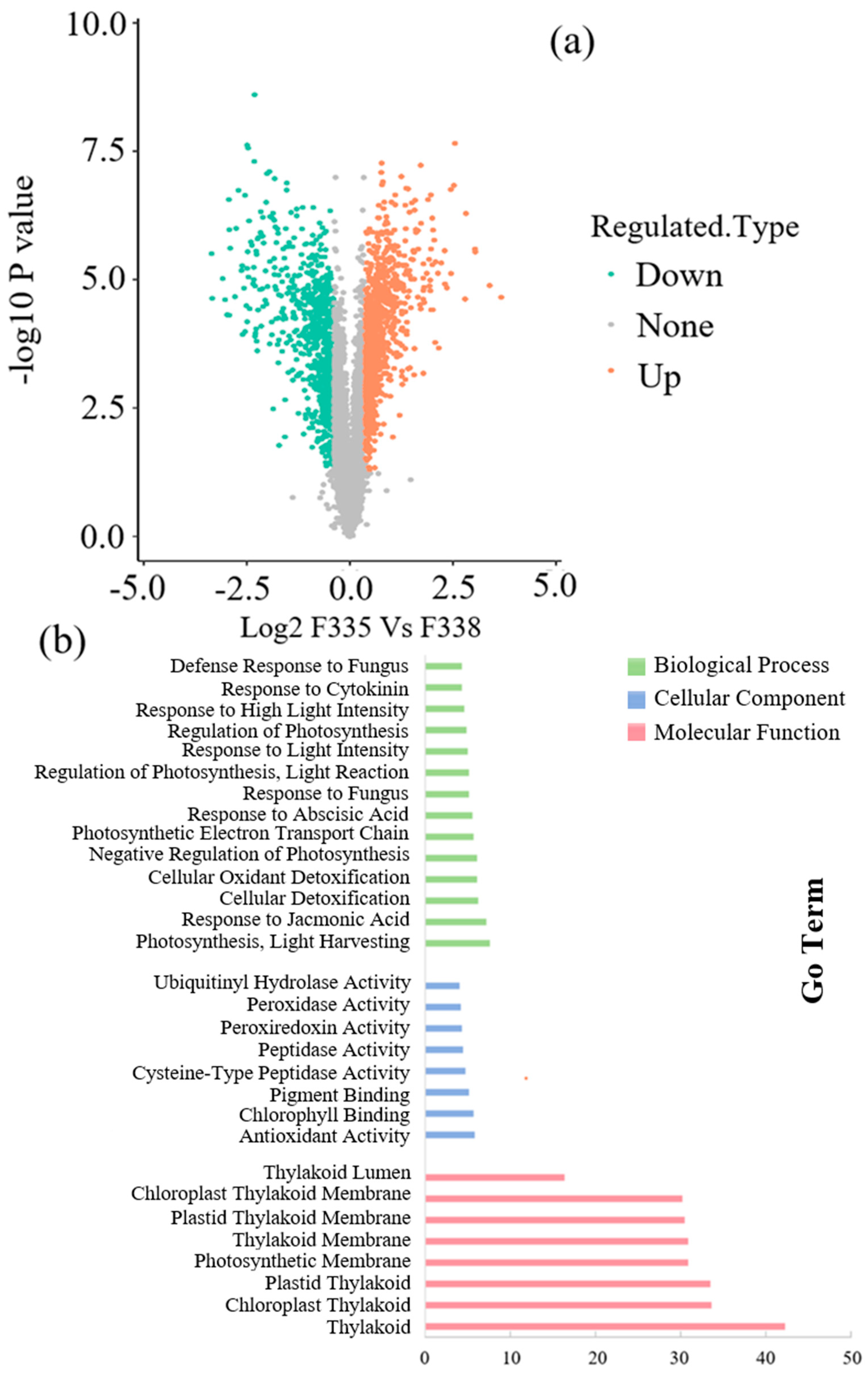
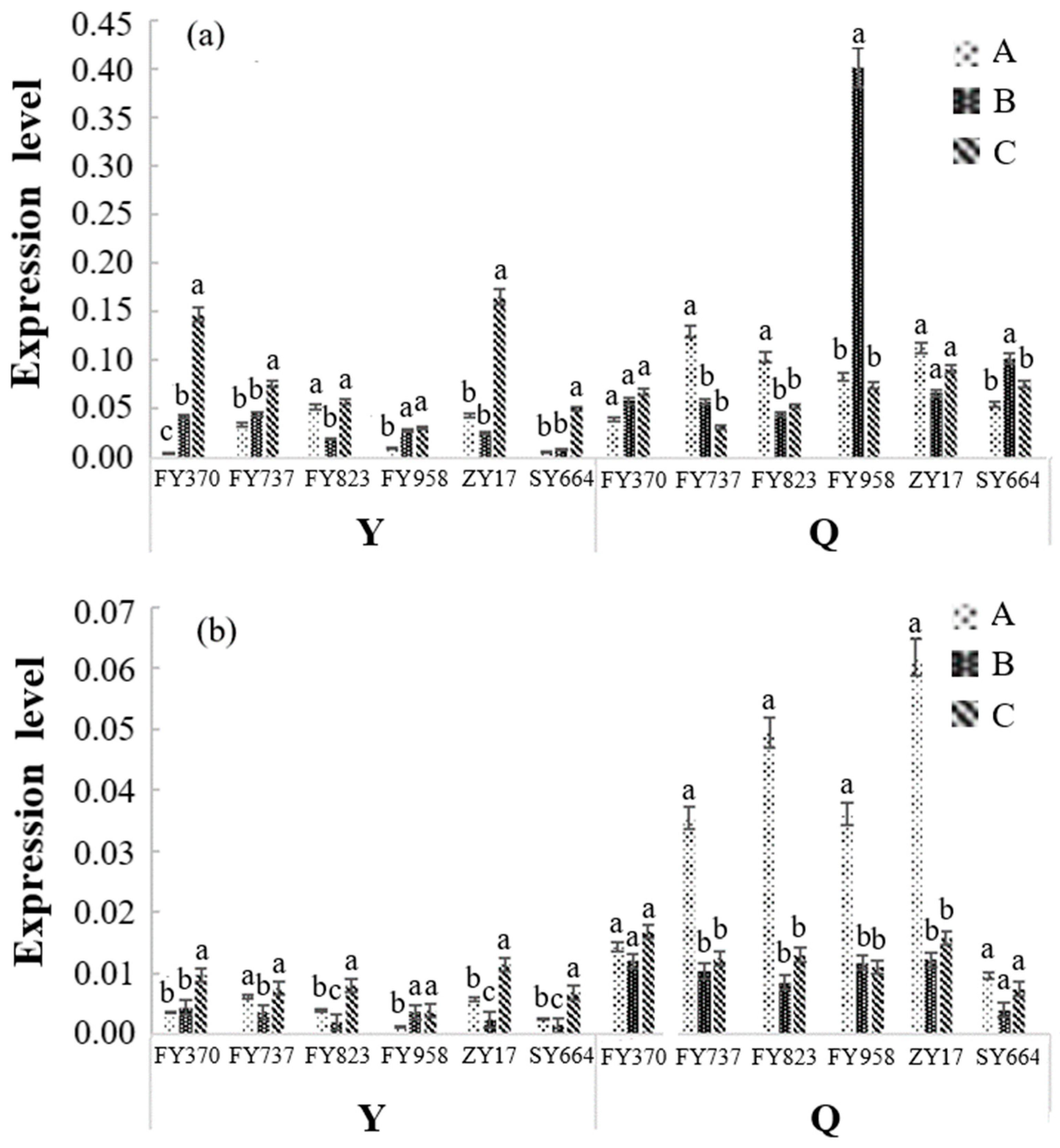
| Heavy Metal | Safety Standard (µg/kg) | 10× (µg/kg) | 50× (µg/kg) | 100× (µg/kg) |
|---|---|---|---|---|
| Cd | 5.00 | 50.00 | 250.00 | 500.00 |
| Pb | 20.00 | 200.00 | 1000.00 | 2000.00 |
| As | 10.00 | 100.00 | 500.00 | 1000.00 |
| Materials | 10× | 50× | 100× | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A (%) | B (%) | C (%) | D (g) | A (%) | B (%) | C (%) | D (g) | A (%) | B (%) | C (%) | D (g) | |
| F335 | 92.00 | 100.00 | 22.00 | 1.76 | 98.00 | 100.00 | 10.00 | 2.00 | 52.00 | 52.00 | 0.00 | 0.59 |
| F338 | 90.00 | 100.00 | 20.00 | 2.38 | 96.00 | 98.00 | 0.00 | 2.43 | 92.00 | 88.00 | 0.00 | 1.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Y.; Chen, H.; Li, Y.; Hui, R.; Zhang, Z. Promising New Methods Based on the SOD Enzyme and SAUR36 Gene to Screen for Canola Materials with Heavy Metal Resistance. Biology 2024, 13, 441. https://doi.org/10.3390/biology13060441
Dai Y, Chen H, Li Y, Hui R, Zhang Z. Promising New Methods Based on the SOD Enzyme and SAUR36 Gene to Screen for Canola Materials with Heavy Metal Resistance. Biology. 2024; 13(6):441. https://doi.org/10.3390/biology13060441
Chicago/Turabian StyleDai, Yue, Hao Chen, Yufang Li, Rongkui Hui, and Zhenqian Zhang. 2024. "Promising New Methods Based on the SOD Enzyme and SAUR36 Gene to Screen for Canola Materials with Heavy Metal Resistance" Biology 13, no. 6: 441. https://doi.org/10.3390/biology13060441
APA StyleDai, Y., Chen, H., Li, Y., Hui, R., & Zhang, Z. (2024). Promising New Methods Based on the SOD Enzyme and SAUR36 Gene to Screen for Canola Materials with Heavy Metal Resistance. Biology, 13(6), 441. https://doi.org/10.3390/biology13060441






