Transcriptomic Analysis of Hub Genes Reveals Associated Inflammatory Pathways in Estrogen-Dependent Gynecological Diseases
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Acquisition and Pre-Processing
2.2. Weighted Gene Co-Expression Network Analysis (WGCNA)
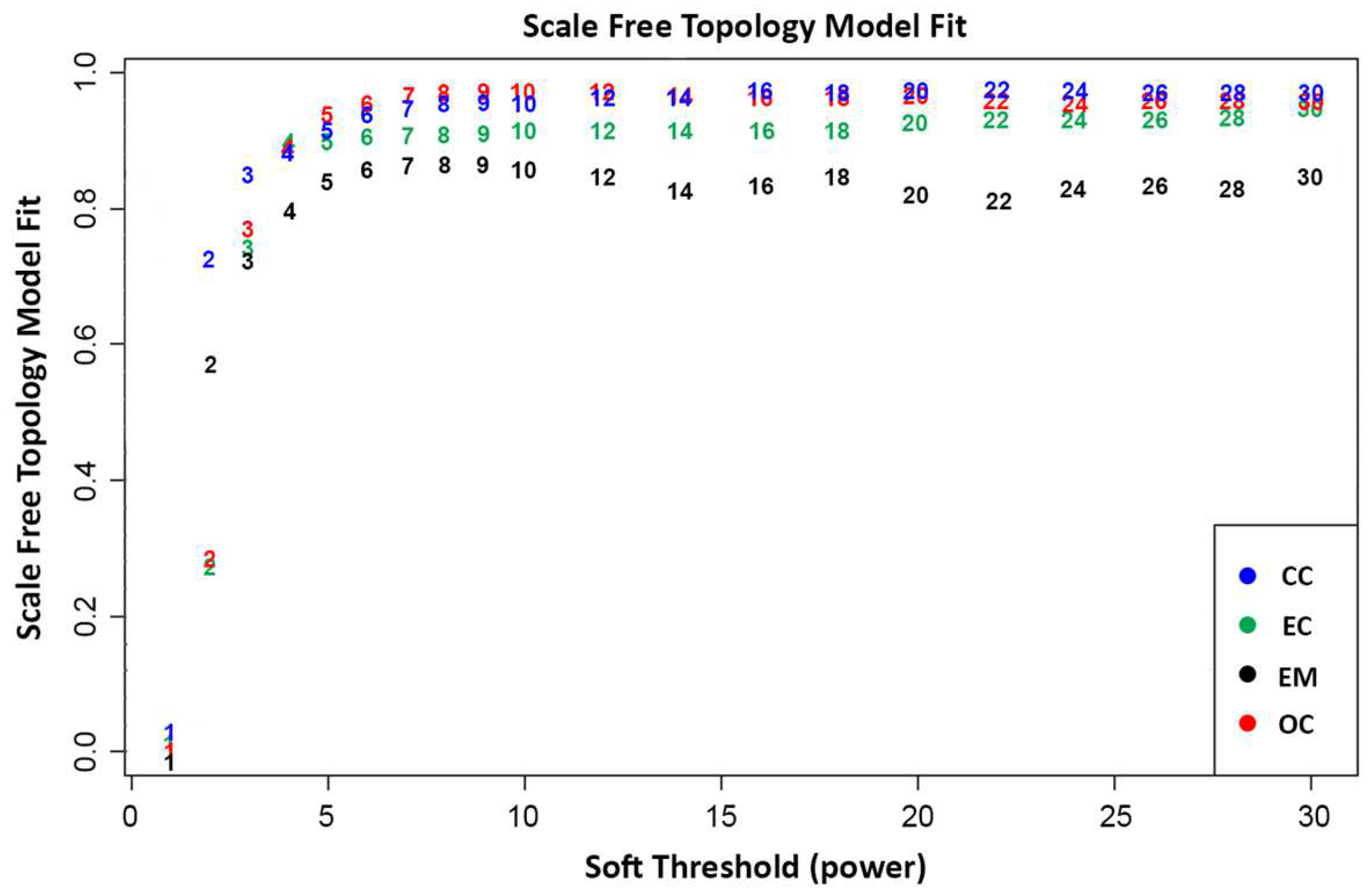
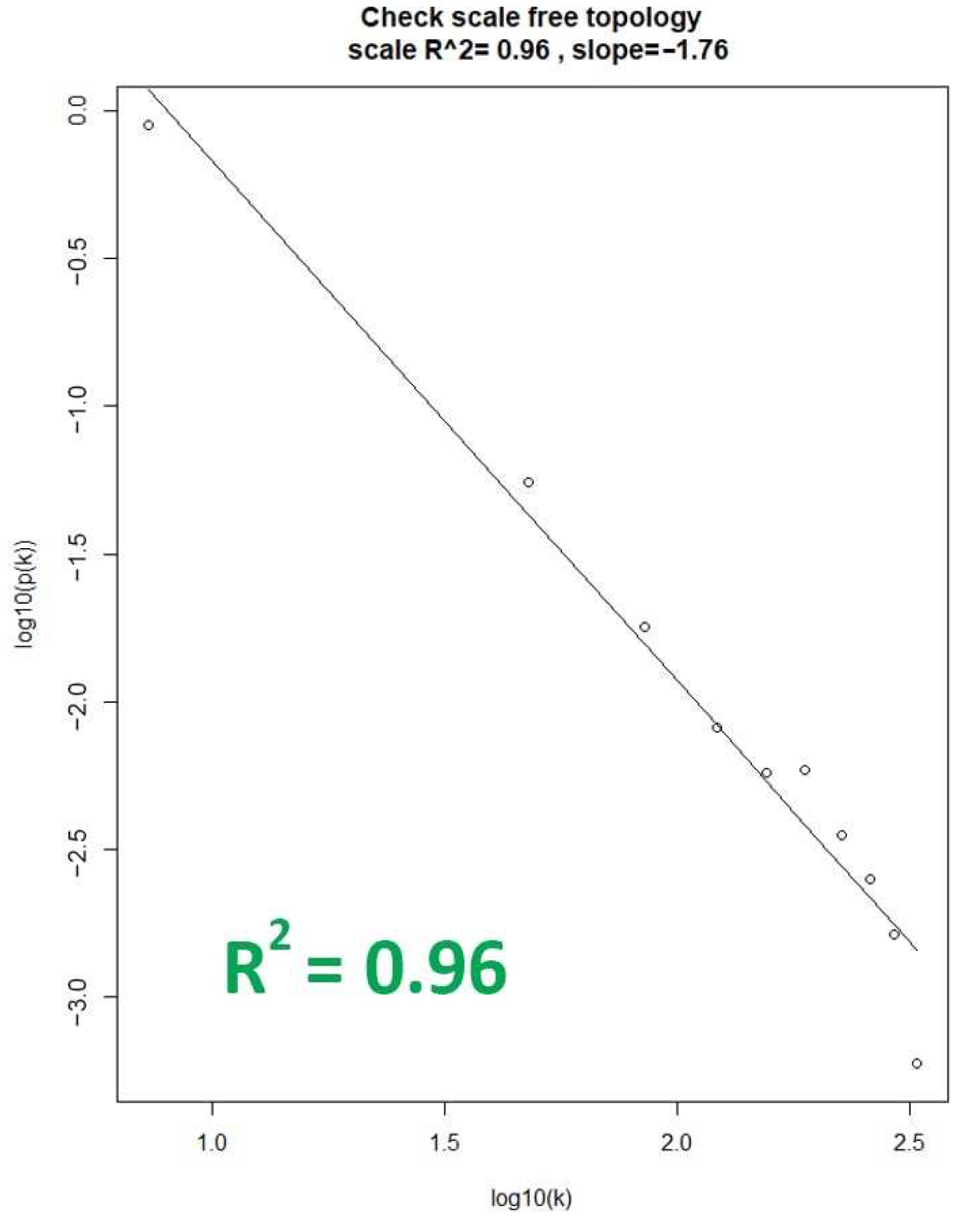
2.2.1. Scale-Free Network
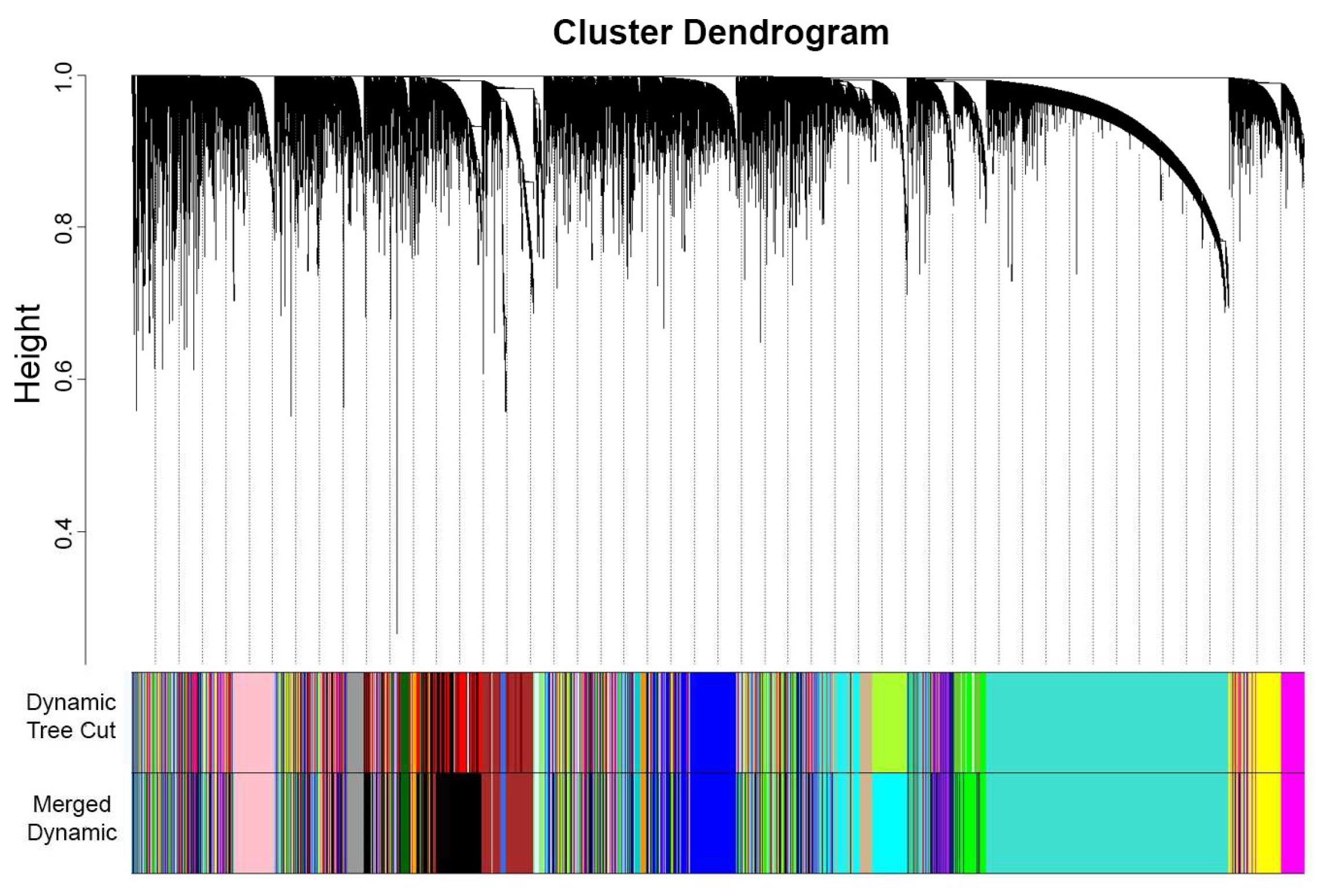
2.2.2. Network Construction and Module Identification
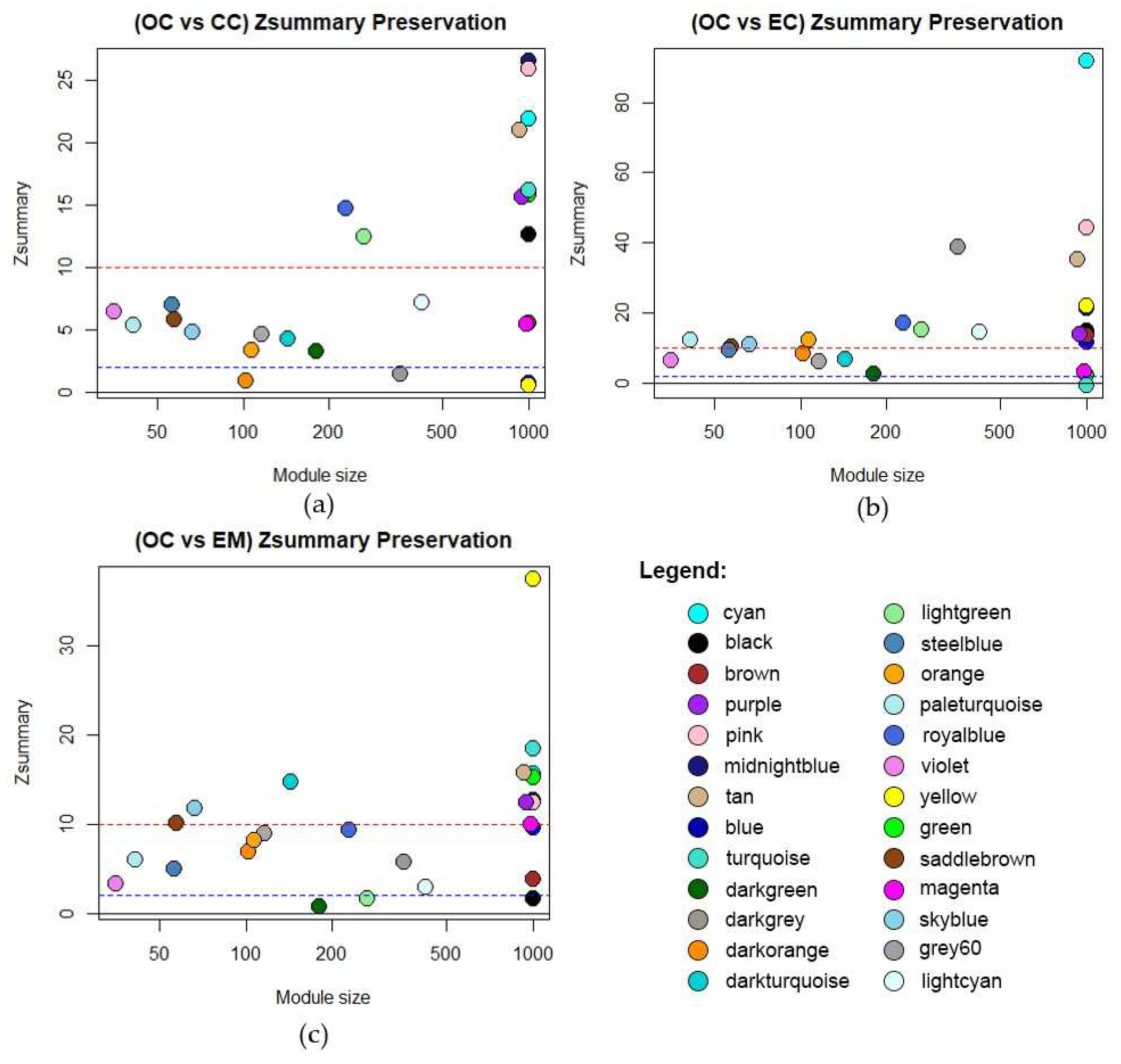
2.2.3. Module Preservation Analysis
2.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways
2.4. Protein-Protein Interaction (PPI) and Hub Genes
2.5. Signature-Based Approach for Drug Repurposing
3. Results
3.1. Weighted Gene Co-Expression Network Analysis (WGCNA)
3.1.1. Data Preparation and Scale-Free Networks
3.1.2. Network Construction and Module Identification
3.2. Module Preservation Analysis
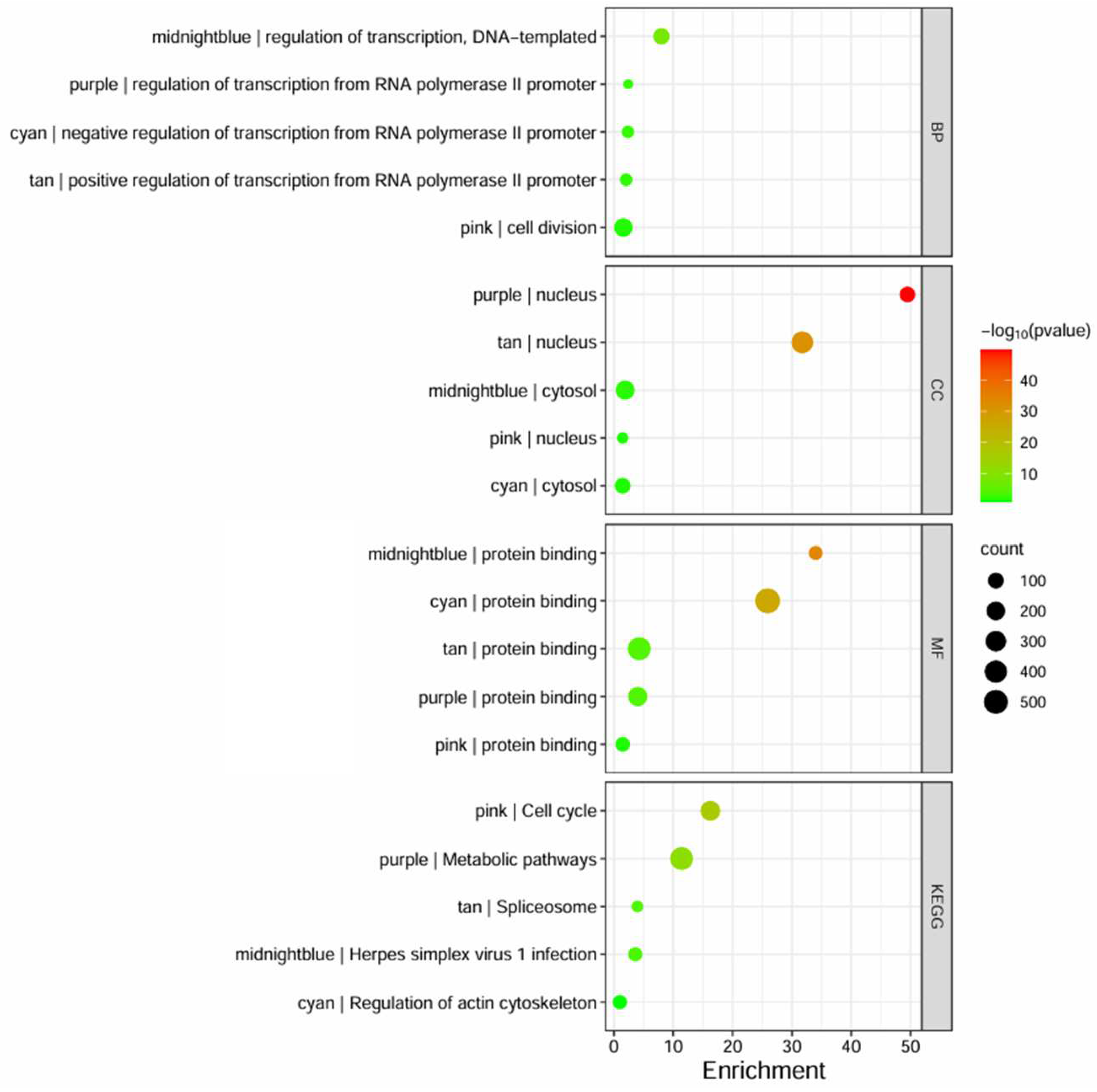
3.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways
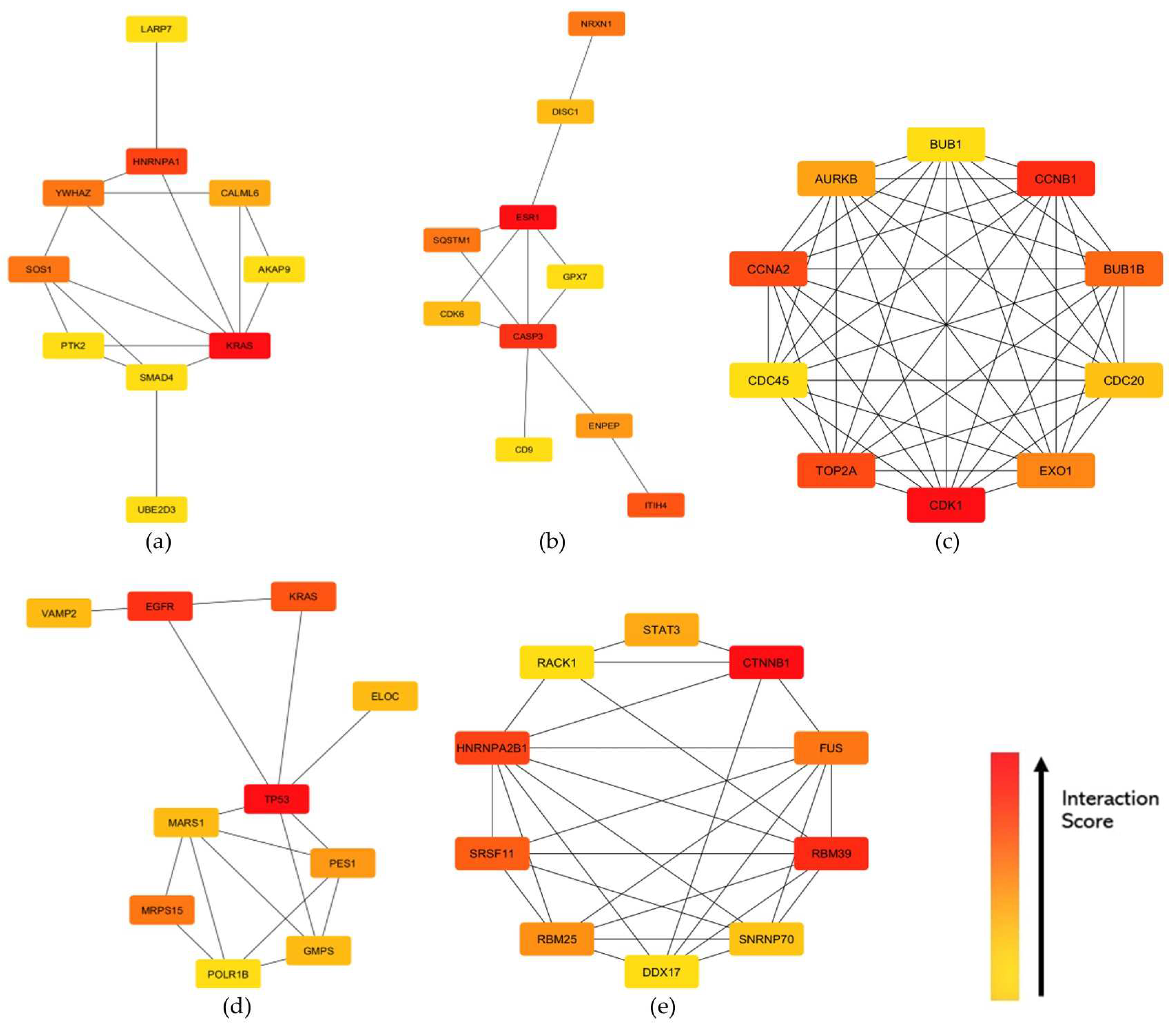
3.4. Protein–Protein Interaction (PPI) Networks and Hub Genes
3.5. Signature-Based Approach for Drug Repurposing
4. Discussion
4.1. Gene Co-Expression Modules across the Datasets
4.2. Module Hub Genes and Their Protein Functions
4.2.1. Inflammatory Pathways Affecting Transcription Dysfunction
4.2.2. Viral Infections Triggering Inflammatory Pathways
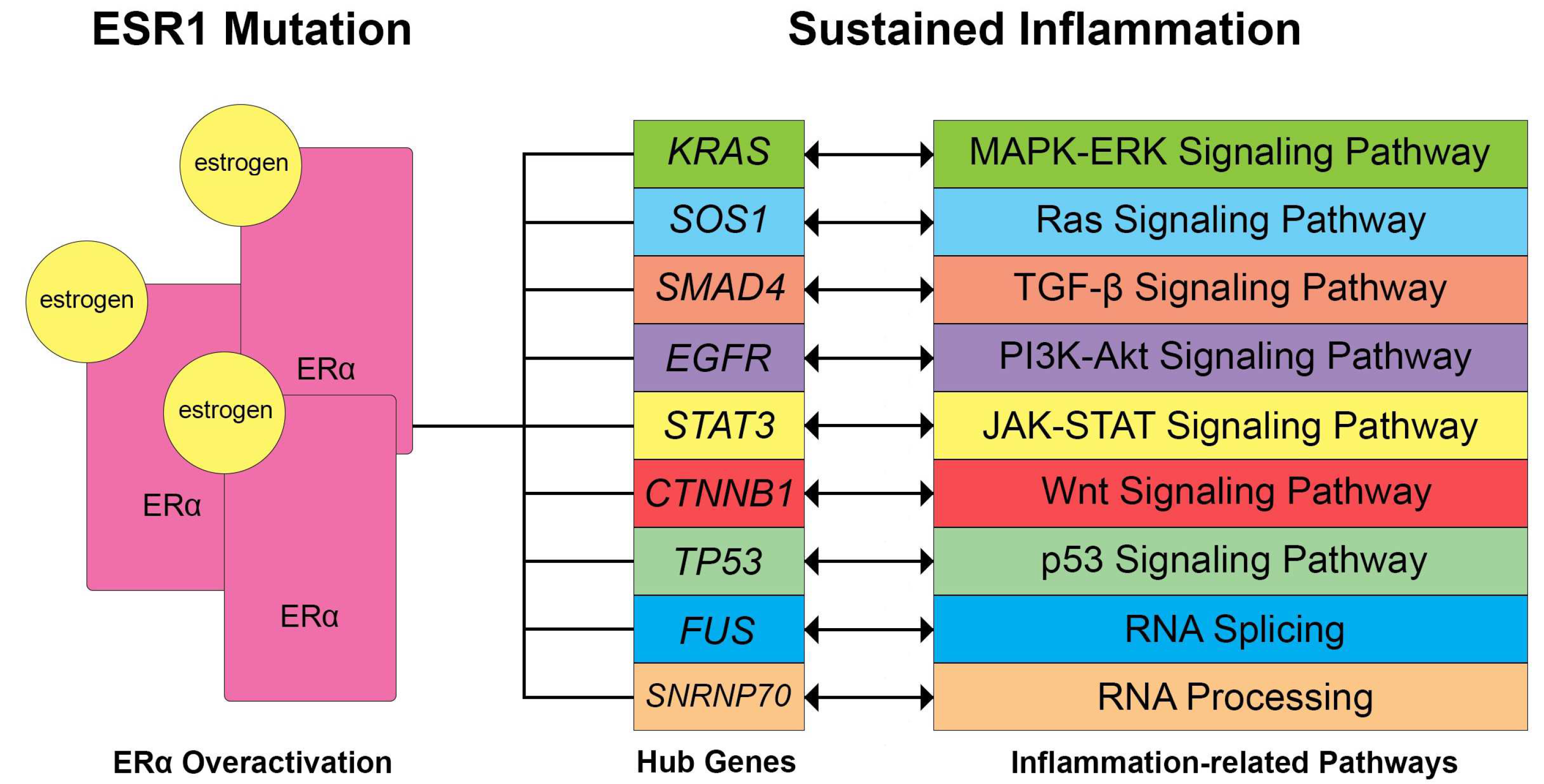
4.3. Hormonal Imbalance and Inflammation
4.4. Signature-Based Approach for Drug Repurposing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
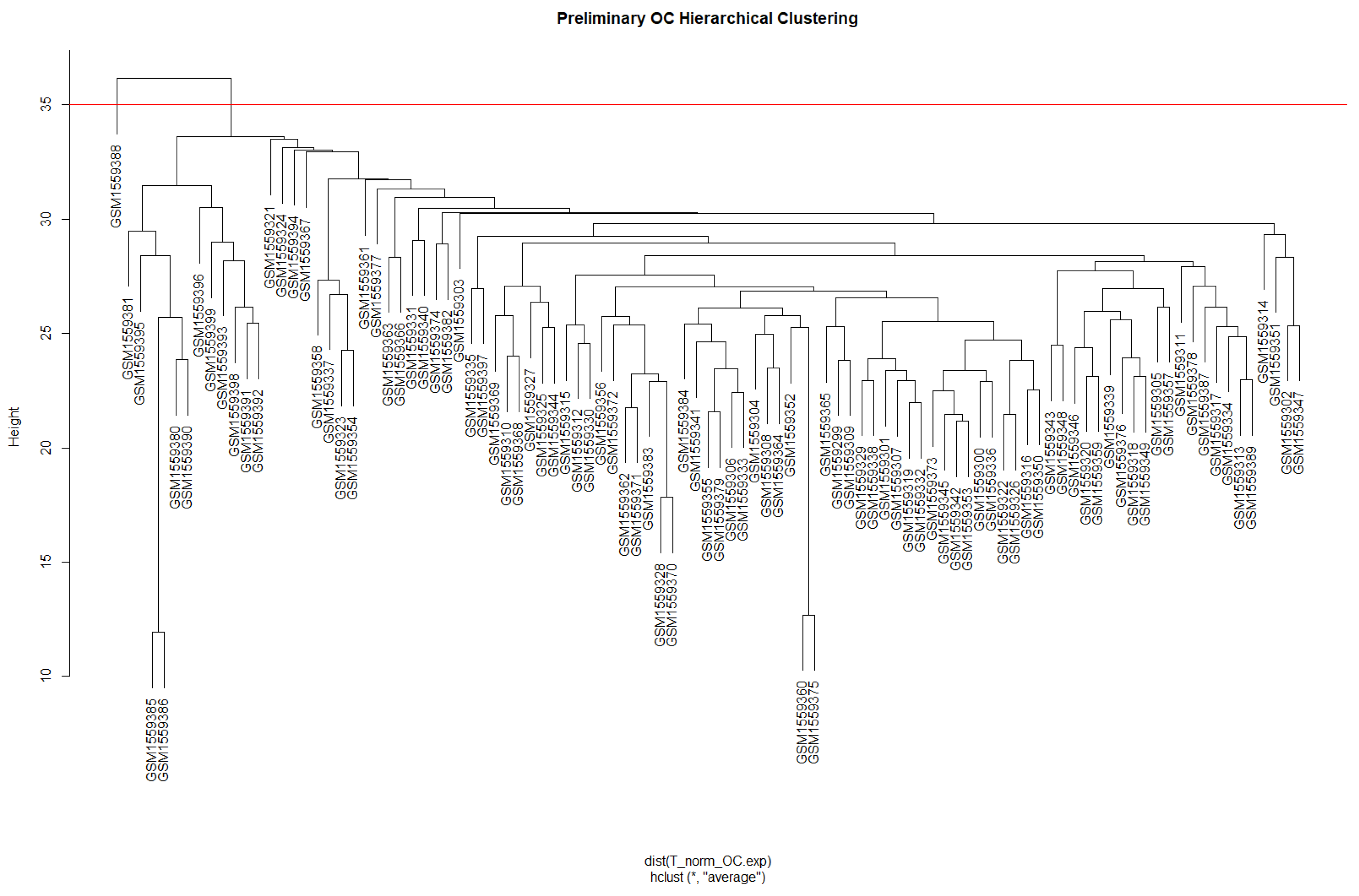
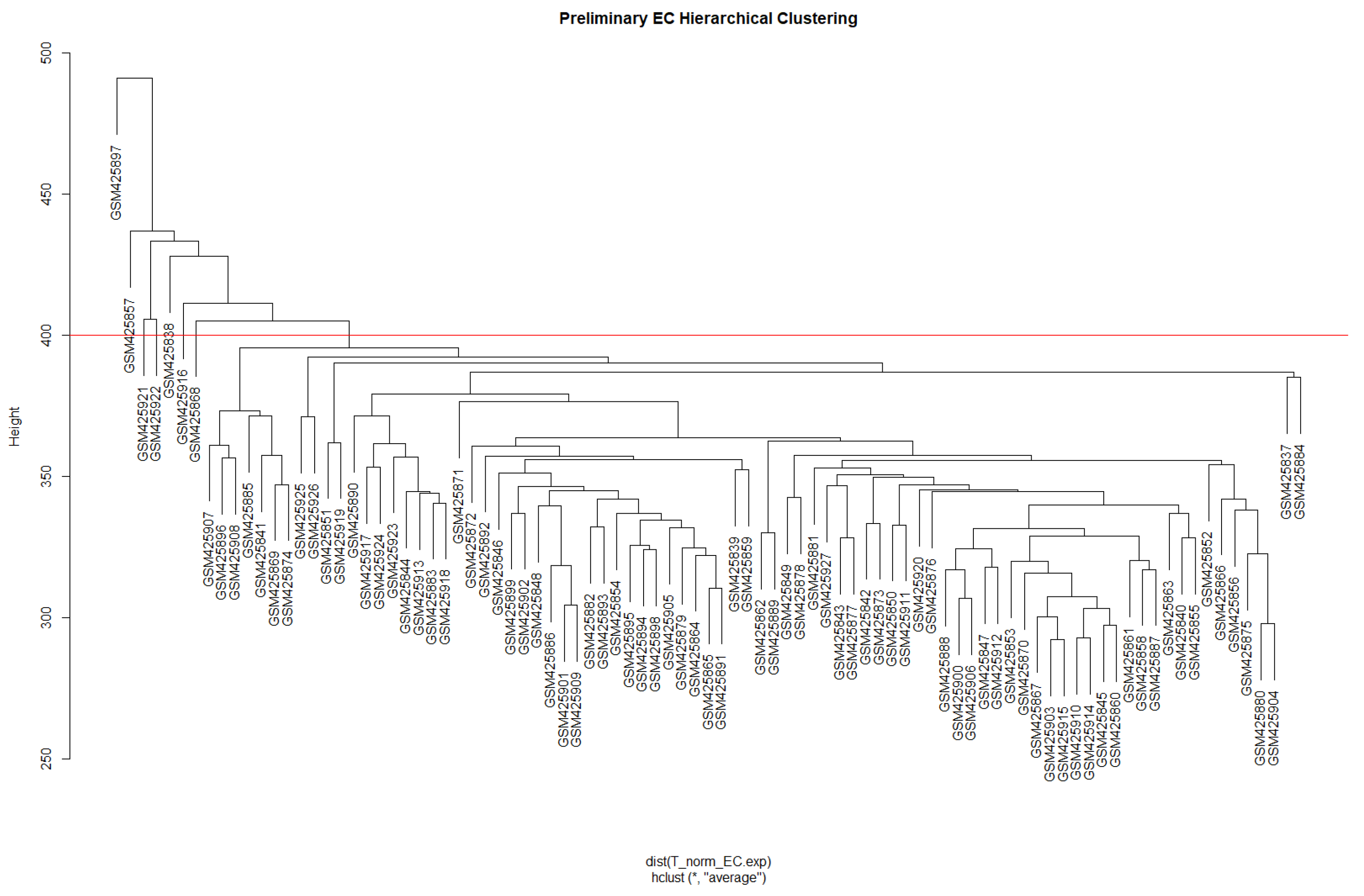
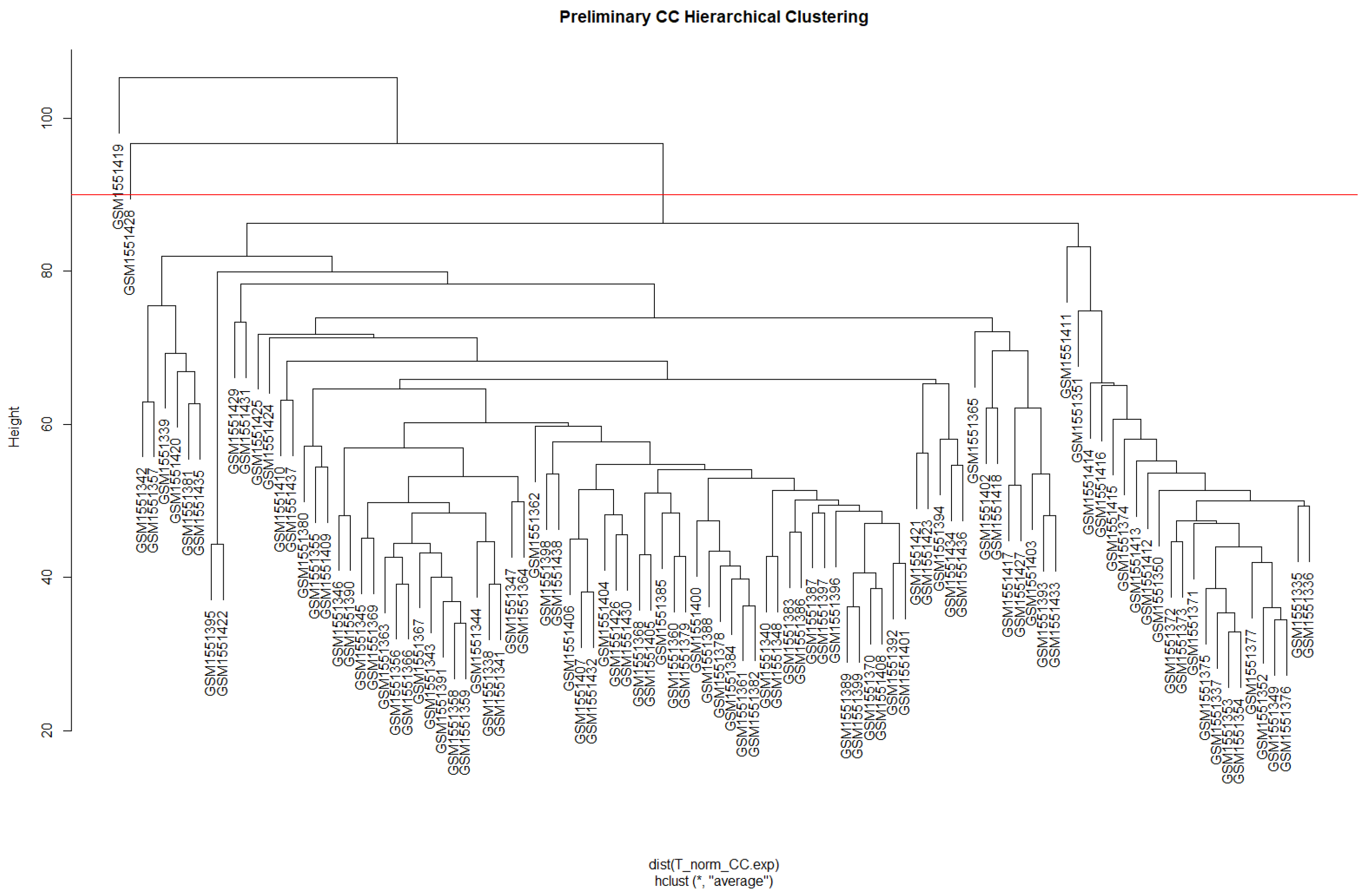
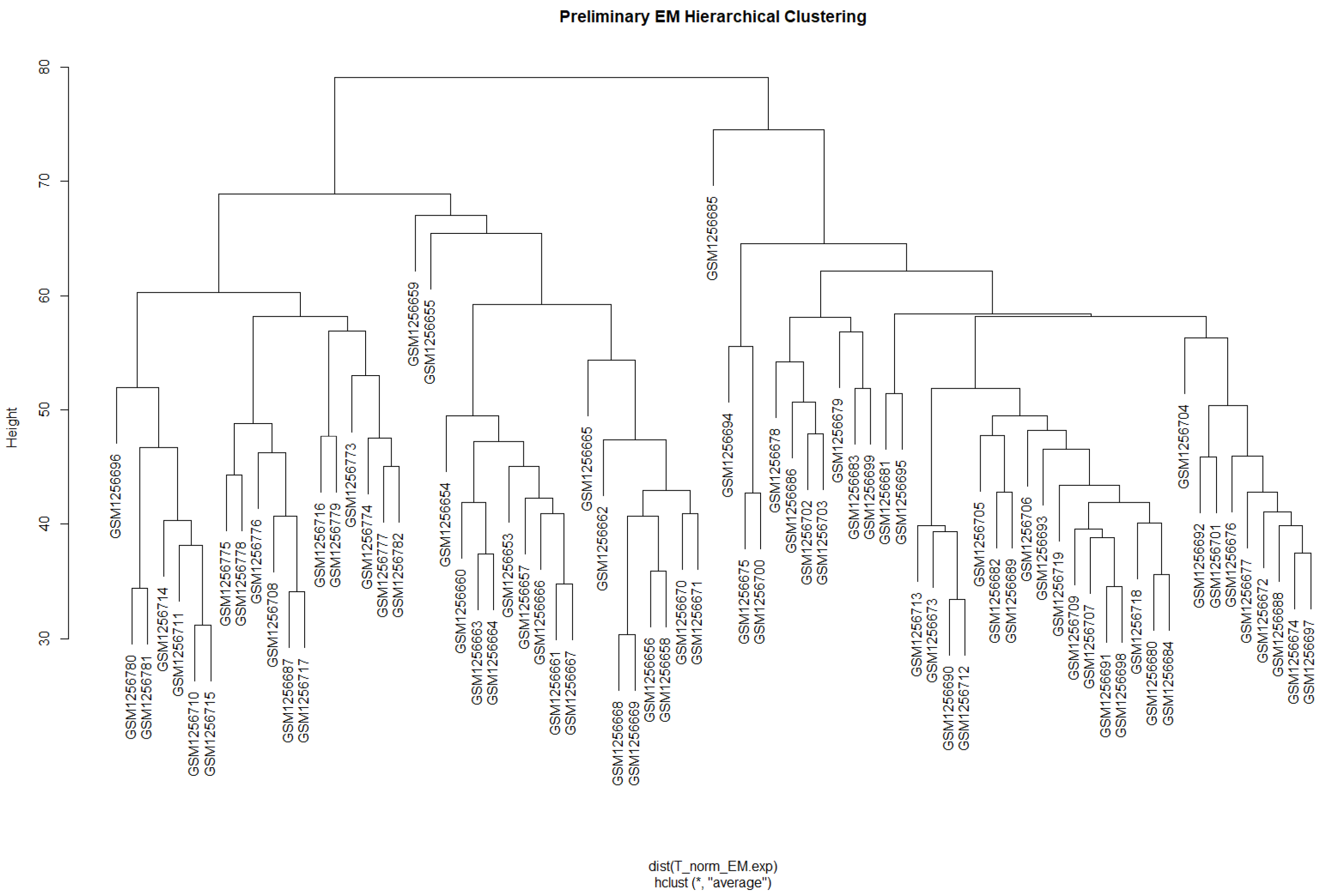
| Module | Dynamic Tree Cut (Genes) | Merge Modules (Genes) |
|---|---|---|
| Cyan | 653 | 1594 |
| Purple | 947 | 947 |
| Pink | 1053 | 1053 |
| Midnightblue | 524 | 1232 |
| Tan | 935 | 935 |
| Blue | 1952 | 1952 |
| Brown | 1938 | 2164 |
| Darkgreen | 181 | 181 |
| Darkgrey | 116 | 116 |
| Darkorange | 102 | 102 |
| Darkturquoise | 143 | 143 |
| Magenta | 984 | 984 |
| Orange | 107 | 107 |
| Paleturquoise | 41 | 41 |
| Royalblue | 228 | 228 |
| Violet | 35 | 35 |
| Yellow | 1424 | 1424 |
| Green | 1387 | 1630 |
| Saddlebrown | 57 | 57 |
| Grey | 327 | 327 |
| Skyblue | 66 | 66 |
| Grey60 | 354 | 354 |
| Lightcyan | 425 | 425 |
| Black | 1162 | |
| Lightgreen | 265 | |
| Lightyellow | 243 | |
| Salmon | 708 | |
| Steelblue | 56 | |
| Turquoise | 6116 | |
| Greenyellow | 941 | |
| Red | 1341 | |
| Darkred | 226 | |
| White | 101 |
| Module | Size | Zsummary Preservation | |
|---|---|---|---|
| Common High Preserve Modules | cyan | 1594 | 21.86149527 |
| midnightblue | 1232 | 26.52630433 | |
| pink | 1053 | 25.86352918 | |
| purple | 947 | 15.59037483 | |
| tan | 935 | 20.96990849 | |
| High Preserve Modules in OC VS CC only | black | 2604 | 12.59707559 |
| green | 1630 | 15.81619288 | |
| lightgreen | 265 | 12.46321267 | |
| royalblue | 228 | 14.71890028 | |
| turquoise | 6116 | 16.18415255 |
| Module | Size | Zsummary Preservation | |
|---|---|---|---|
| Common High Preserve Modules | cyan | 1594 | 91.87686929 |
| midnightblue | 1232 | 21.42695751 | |
| pink | 1053 | 44.18140552 | |
| purple | 947 | 13.8939192 | |
| tan | 935 | 35.24655182 | |
| High Preserve Modules in (OC VS EC) only | black | 2604 | 14.84965178 |
| blue | 1952 | 11.75575271 | |
| brown | 2164 | 13.40832857 | |
| grey60 | 354 | 38.92112853 | |
| lightcyan | 425 | 14.51331585 | |
| lightgreen | 265 | 15.06853233 | |
| orange | 107 | 12.18371176 | |
| paleturquoise | 41 | 12.3852009 | |
| royalblue | 228 | 17.13450004 | |
| saddlebrown | 57 | 10.25197434 | |
| skyblue | 66 | 10.88052848 | |
| yellow | 1424 | 22.08498616 |
| Module | Size | Zsummary Preservation | |
|---|---|---|---|
| Common High Preserve Modules | cyan | 1594 | 15.67053707 |
| midnightblue | 1232 | 12.67510038 | |
| pink | 1053 | 12.42696169 | |
| purple | 947 | 12.43896986 | |
| tan | 935 | 15.75233647 | |
| High Preserve Modules in (OC VS EM) only | darkturquoise | 143 | 14.73601775 |
| green | 1630 | 15.24494125 | |
| magenta | 984 | 10.04291952 | |
| saddlebrown | 57 | 10.13071697 | |
| skyblue | 66 | 11.86774714 | |
| turquoise | 6116 | 18.54903814 | |
| yellow | 1424 | 37.50425118 |
| Category | Term | Count | p-Value | |
|---|---|---|---|---|
| BP | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 25 | 4.2 × 10−3 |
| GO:0006355 | regulation of transcription, DNA-templated | 21 | 3.4 × 10−2 | |
| GO:0016310 | phosphorylation | 15 | 3.8 × 10−2 | |
| GO:0006338 | chromatin remodeling | 13 | 1.8 × 10−3 | |
| GO:0006468 | protein phosphorylation | 12 | 1.3 × 10−2 | |
| CC | GO:0005829 | cytosol | 116 | 1.5 × 10−8 |
| GO:0005634 | nucleus | 111 | 3.1 × 10−5 | |
| GO:0005737 | cytoplasm | 105 | 4.0 × 10−5 | |
| GO:0005654 | nucleoplasm | 77 | 2.5 × 10−4 | |
| GO:0005783 | endoplasmic reticulum | 28 | 2.4 × 10−3 | |
| MF | GO:0005515 | protein binding | 193 | 2.6 × 10−2 |
| GO:0046872 | metal ion binding | 60 | 4.2 × 10−4 | |
| GO:0003723 | RNA binding | 33 | 8.3 × 10−3 | |
| GO:0005524 | ATP binding | 33 | 1.5 × 10−2 | |
| GO:0005085 | guanyl-nucleotide exchange factor activity | 11 | 1.6 × 10−3 | |
| KEGG | hsa04810 | Regulation of actin cytoskeleton | 10 | 3.7 × 10−3 |
| hsa05208 | Chemical carcinogenesis—reactive oxygen species | 8 | 3.1 × 10−2 | |
| hsa04660 | T cell receptor signaling pathway | 6 | 2.4 × 10−2 | |
| hsa05100 | Bacterial invasion of epithelial cells | 5 | 2.0 × 10−2 | |
| hsa04012 | ErbB signaling pathway | 5 | 2.8 × 10−2 | |
| Category | Term | Count | p-Value | |
|---|---|---|---|---|
| BP | GO:0006355 | regulation of transcription, DNA-templated | 26 | 6.0 × 10−3 |
| GO:1905515 | non-motile cilium assembly | 6 | 2.3 × 10−3 | |
| GO:0070588 | calcium ion transmembrane transport | 6 | 3.5 × 10−2 | |
| GO:0030212 | hyaluronan metabolic process | 3 | 5.9 × 10−3 | |
| GO:1905606 | regulation of presynapse assembly | 3 | 8.7 × 10−2 | |
| CC | GO:0005829 | cytosol | 98 | 4.7 × 10−2 |
| GO:0005576 | extracellular region | 46 | 1.7 × 10−2 | |
| GO:0005739 | mitochondrion | 29 | 9.1 × 10−2 | |
| GO:0098978 | glutamatergic synapse | 13 | 2.4 × 10−2 | |
| GO:0005764 | lysosome | 10 | 3.5 × 10−2 | |
| MF | GO:0005515 | protein binding | 218 | 1.4 × 10−2 |
| GO:0004222 | metalloendopeptidase activity | 6 | 3.0 × 10−2 | |
| GO:0019838 | growth factor binding | 4 | 2.3 × 10−2 | |
| GO:0004709 | MAP kinase kinase kinase activity | 3 | 4.2 × 10−2 | |
| KEGG | hsa05168 | Herpes simplex virus 1 infection | 15 | 3.3 × 10−2 |
| hsa05010 | Alzheimer disease | 11 | 8.6 × 10−2 | |
| hsa05160 | Hepatitis C | 7 | 4.0 × 10−2 | |
| hsa04614 | Renin-angiotensin system | 4 | 5.5 × 10−2 | |
| Category | Term | Count | p-Value | |
|---|---|---|---|---|
| BP | GO:0051301 | cell division | 92 | 1.6 × 10−49 |
| GO:0007049 | cell cycle | 58 | 1.7 × 10−22 | |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 53 | 3.7 × 10−3 | |
| GO:0006281 | DNA repair | 52 | 5.2 × 10−21 | |
| GO:0006260 | DNA replication | 39 | 4.5 × 10−26 | |
| CC | GO:0005634 | nucleus | 352 | 1.6 × 10−30 |
| GO:0005829 | cytosol | 321 | 4.0 × 10−26 | |
| GO:0005654 | nucleoplasm | 296 | 1.1 × 10−43 | |
| GO:0005737 | cytoplasm | 268 | 1.7 × 10−9 | |
| GO:0005813 | centrosome | 77 | 7.1 × 10−21 | |
| MF | GO:0005515 | protein binding | 578 | 3.4 × 10−26 |
| GO:0003677 | DNA binding | 96 | 2.0 × 10−10 | |
| GO:0005524 | ATP binding | 94 | 5.2 × 10−7 | |
| GO:0003682 | chromatin binding | 47 | 2.7 × 10−9 | |
| GO:0016887 | ATPase activity | 46 | 1.4 × 10−10 | |
| KEGG | hsa04110 | Cell cycle | 52 | 8.7 × 10−35 |
| hsa04218 | Cellular senescence | 20 | 5.7 × 10−6 | |
| hsa05166 | Human T-cell leukemia virus 1 infection | 20 | 6.9 × 10−4 | |
| hsa05203 | Viral carcinogenesis | 15 | 2.2 × 10−2 | |
| hsa04115 | p53 signaling pathway | 12 | 9.1 × 10−5 | |
| Category | Term | Count | p-Value | |
|---|---|---|---|---|
| BP | GO:0006357 | regulation of transcription from RNA polymerase II promoter | 58 | 7.9 × 10−2 |
| GO:0007165 | signal transduction | 50 | 2.5 × 10−2 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 46 | 7.8 × 10−2 | |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 39 | 7.4 × 10−2 | |
| GO:0016310 | phosphorylation | 30 | 1.1 × 10−2 | |
| CC | GO:0005634 | nucleus | 220 | 7.5 × 10−5 |
| GO:0005829 | cytosol | 204 | 1.0 × 10−4 | |
| GO:0005737 | cytoplasm | 203 | 4.2 × 10−4 | |
| GO:0005654 | nucleoplasm | 154 | 1.1 × 10−4 | |
| GO:0005739 | mitochondrion | 61 | 2.8 × 10−3 | |
| MF | GO:0005515 | protein binding | 437 | 6.7 × 10−5 |
| GO:0046872 | metal ion binding | 106 | 1.4 × 10−2 | |
| GO:0003723 | RNA binding | 60 | 2.5 × 10−2 | |
| GO:0003700 | transcription factor activity, sequence-specific DNA binding | 32 | 4.9 × 10−3 | |
| GO:0003676 | nucleic acid binding | 18 | 1.7 × 10−2 | |
| KEGG | hsa04371 | Apelin signaling pathway | 10 | 3.0 × 10−2 |
| hsa04926 | Relaxin signaling pathway | 9 | 4.9 × 10−2 | |
| hsa03083 | Polycomb repressive complex | 8 | 1.6 × 10−2 | |
| hsa00520 | Amino sugar and nucleotide sugar metabolism | 6 | 1.8 × 10−2 | |
| hsa05219 | Bladder cancer | 5 | 3.8 × 10−2 | |
| Category | Term | Count | p-Value | |
|---|---|---|---|---|
| BP | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 51 | 2.1 × 10−3 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 43 | 3.2 × 10−3 | |
| GO:0045893 | positive regulation of transcription, DNA-templated | 38 | 1.1 × 10−4 | |
| GO:0006355 | regulation of transcription, DNA-templated | 38 | 2.0 × 10−2 | |
| GO:0016310 | phosphorylation | 33 | 3.5 × 10−4 | |
| CC | GO:0005634 | nucleus | 252 | 2.0 × 10−17 |
| GO:0005829 | cytosol | 219 | 1.3 × 10−11 | |
| GO:0005654 | nucleoplasm | 214 | 1.2 × 10−27 | |
| GO:0005737 | cytoplasm | 199 | 2.6 × 10−6 | |
| GO:0005739 | mitochondrion | 51 | 2.7 × 10−2 | |
| MF | GO:0005515 | protein binding | 434 | 3.5 × 10−12 |
| GO:0003723 | RNA binding | 99 | 1.5 × 10−15 | |
| GO:0046872 | metal ion binding | 95 | 3.6 × 10−2 | |
| GO:0005524 | ATP binding | 62 | 5.4 × 10−3 | |
| GO:0003677 | DNA binding | 60 | 6.8 × 10−4 | |
| KEGG | hsa03040 | Spliceosome | 18 | 1.2 × 10−4 |
| hsa05014 | Amyotrophic lateral sclerosis | 18 | 2.6 × 10−2 | |
| hsa05202 | Transcriptional misregulation in cancer | 12 | 1.9 × 10−2 | |
| hsa03015 | mRNA surveillance pathway | 9 | 5.6 × 10−3 | |
| hsa00310 | Lysine degradation | 6 | 3.1 × 10−2 | |
References
- Wijeratne, D.; Fiander, A. Gynaecological Disease in the Developing World: A Silent Pandemic. Obstet. Gynaecol. 2018, 20, 237–244. [Google Scholar] [CrossRef]
- Ye, J.; Peng, H.; Huang, X.; Qi, X. The Association between Endometriosis and Risk of Endometrial Cancer and Breast Cancer: A Meta-Analysis. BMC Womens Health 2022, 22, 455. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, A.E.W. Gynecologic Cancers: Cervical, Uterine, and Ovarian Cancers. In Mayo Clinic Internal Medicine Board Review; Wittich, C.M., Beckman, T.J., Bonnes, S.L., Schwenk, N.M., Szostek, J.H., Collins, N.M., Stephenson, C.R., Wittich, C.M., Eds.; Oxford University Press: Oxford, UK, 2019; ISBN 978-0-19-093836-9. [Google Scholar]
- Kitson, S.J.; Evans, D.G.; Crosbie, E.J. Identifying High-Risk Women for Endometrial Cancer Prevention Strategies: Proposal of an Endometrial Cancer Risk Prediction Model. Cancer Prev. Res. 2017, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Obesity and Endometrial Cancer. SpringerLink. Available online: https://link.springer.com/chapter/10.1007/978-3-319-42542-9_7 (accessed on 19 April 2024).
- Obesity and Ovarian Cancer. SpringerLink. Available online: https://link.springer.com/chapter/10.1007/978-3-319-42542-9_9 (accessed on 19 April 2024).
- Fernandes, J.V.; Fernandes, T.A.A.d.M.; de Azevedo, J.C.V.; Cobucci, R.N.O.; de Carvalho, M.G.F.; Andrade, V.S.; de Araújo, J.M.G. Link between Chronic Inflammation and Human Papillomavirus-Induced Carcinogenesis (Review). Oncol. Lett. 2015, 9, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Magata, F.; Tsukamura, H.; Matsuda, F. The Impact of Inflammatory Stress on Hypothalamic Kisspeptin Neurons: Mechanisms Underlying Inflammation-Associated Infertility in Humans and Domestic Animals. Peptides 2023, 162, 170958. [Google Scholar] [CrossRef] [PubMed]
- Manuel, M.T.A.; Tayo, L.L. Navigating the Gene Co-Expression Network and Drug Repurposing Opportunities for Brain Disorders Associated with Neurocognitive Impairment. Brain Sci. 2023, 13, 1564. [Google Scholar] [CrossRef] [PubMed]
- Mailem, R.C.; Tayo, L.L. Drug Repurposing Using Gene Co-Expression and Module Preservation Analysis in Acute Respiratory Distress Syndrome (ARDS), Systemic Inflammatory Response Syndrome (SIRS), Sepsis, and COVID-19. Biology 2022, 11, 1827. [Google Scholar] [CrossRef] [PubMed]
- Wickramarachchi, D.C.; Theofilopoulos, A.N.; Kono, D.H. Immune Pathology Associated with Altered Actin Cytoskeleton Regulation. Autoimmunity 2010, 43, 64–75. [Google Scholar] [CrossRef]
- Yang, H.; Beutler, B.; Zhang, D. Emerging Roles of Spliceosome in Cancer and Immunity. Protein Cell 2022, 13, 559–579. [Google Scholar] [CrossRef]
- Al-Badawi, I.A.; Abu-Zaid, A.; Alomar, O.; Alsabban, M.; Alsehaimi, S.O.; Alqarni, S.M.; Alabdrabalamir, S.N.; Baradwan, S.; Al Baalharith, M.; AlOdaini, A.A.; et al. Association between Endometriosis and the Risk of Ovarian, Endometrial, Cervical, and Breast Cancer: A Population-Based Study from the U.S. National Inpatient Sample 2016–2019. Curr. Oncol. 2024, 31, 472–481. [Google Scholar] [CrossRef]
- Liu, H.; Bebu, I.; Li, X. Microarray Probes and Probe Sets. Front. Biosci.-Elite 2010, 2, 325–338. [Google Scholar] [CrossRef]
- Junet, V.; Farrés, J.; Mas, J.M.; Daura, X. CuBlock: A Cross-Platform Normalization Method for Gene-Expression Microarrays. Bioinformatics 2021, 37, 2365–2373. [Google Scholar] [CrossRef] [PubMed]
- goodSamplesGenes Function—Rdocumentation. Available online: https://www.rdocumentation.org/packages/WGCNA/versions/1.72-5/topics/goodSamplesGenes (accessed on 20 May 2024).
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yang, X.; Zu, Y. Integrated Analysis of WGCNA and Machine Learning Identified Diagnostic Biomarkers in Dilated Cardiomyopathy with Heart Failure. Front. Cell Dev. Biol. 2022, 10, 1089915. [Google Scholar] [CrossRef] [PubMed]
- Fuller, T.; Langfelder, P.; Presson, A.; Horvath, S. Review of Weighted Gene Coexpression Network Analysis. In Handbook of Statistical Bioinformatics; Lu, H.H.-S., Schölkopf, B., Zhao, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 369–388. ISBN 978-3-642-16345-6. [Google Scholar]
- iterativeWGCNA: Iterative Refinement to Improve Module Detection from WGCNA Co-Expression Networks. bioRxiv. 2017. Available online: https://www.biorxiv.org/content/10.1101/234062v1.full (accessed on 20 May 2024).
- Sánchez-Baizán, N.; Ribas, L.; Piferrer, F. Improved Biomarker Discovery through a Plot Twist in Transcriptomic Data Analysis. BMC Biol. 2022, 20, 208. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiarizadeh, M.R.; Hosseinpour, B.; Shahhoseini, M.; Korte, A.; Gifani, P. Weighted Gene Co-Expression Network Analysis of Endometriosis and Identification of Functional Modules Associated With Its Main Hallmarks. Front. Genet. 2018, 9, 453. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Karev, G.; Koonin, E.V. Scale-Free Networks in Biology: New Insights into the Fundamentals of Evolution? BioEssays 2002, 24, 105–109. [Google Scholar] [CrossRef]
- Lv, Y.; Xie, B.; Bai, B.; Shan, L.; Zheng, W.; Huang, X.; Zhu, H. Weighted Gene Coexpression Analysis Indicates That PLAGL2 and POFUT1 Are Related to the Differential Features of Proximal and Distal Colorectal Cancer. Oncol. Rep. 2019, 42, 2473–2485. [Google Scholar] [CrossRef]
- Langfelder, P.; Luo, R.; Oldham, M.C.; Horvath, S. Is My Network Module Preserved and Reproducible? PLoS Comput. Biol. 2011, 7, e1001057. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Cong, Y.; Shintani, M.; Imanari, F.; Osada, N.; Endo, T. A New Approach to Drug Repurposing with Two-Stage Prediction, Machine Learning, and Unsupervised Clustering of Gene Expression. OMICS J. Integr. Biol. 2022, 26, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Siu, M.K.Y.; Ngan, H.Y.S.; Chan, K.K.L. Molecular Biomarkers for the Early Detection of Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 12041. [Google Scholar] [CrossRef] [PubMed]
- Holcakova, J.; Bartosik, M.; Anton, M.; Minar, L.; Hausnerova, J.; Bednarikova, M.; Weinberger, V.; Hrstka, R. New Trends in the Detection of Gynecological Precancerous Lesions and Early-Stage Cancers. Cancers 2021, 13, 6339. [Google Scholar] [CrossRef] [PubMed]
- Giannella, L.; Ciavattini, A. Screening and Early Diagnosis in Gynecological Cancers. Cancers 2023, 15, 5152. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-Y.; Ma, K.S.-K.; Wang, L.-T.; Chiang, C.-H.; Yang, S.-F.; Wang, C.-H.; Wang, P.-H. The Risk of Endometrial Cancer and Uterine Sarcoma Following Endometriosis or Pelvic Inflammatory Disease. Cancers 2023, 15, 833. [Google Scholar] [CrossRef] [PubMed]
- Orda, M.A.; Fowler, P.M.P.T.; Tayo, L.L. Modular Hub Genes in DNA Microarray Suggest Potential Signaling Pathway Interconnectivity in Various Glioma Grades. Biology 2024, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Ganeshan, K.; Chawla, A. Metabolic Regulation of Immune Responses. Annu. Rev. Immunol. 2014, 32, 609–634. [Google Scholar] [CrossRef] [PubMed]
- Ronzier, E.; Laurenson, A.J.; Manickam, R.; Liu, S.; Saintilma, I.M.; Schrock, D.C.; Hammer, J.A.; Rotty, J.D. The Actin Cytoskeleton Responds to Inflammatory Cues and Alters Macrophage Activation. Cells 2022, 11, 1806. [Google Scholar] [CrossRef] [PubMed]
- Monin, L.; Gaffen, S.L. Interleukin 17 Family Cytokines: Signaling Mechanisms, Biological Activities, and Therapeutic Implications. Cold Spring Harb. Perspect. Biol. 2018, 10, a028522. [Google Scholar] [CrossRef]
- Reedquist, K.A.; Tak, P.P. Signal Transduction Pathways in Chronic Inflammatory Autoimmune Disease: Small GTPases. Open Rheumatol. J. 2012, 6, 259–272. [Google Scholar] [CrossRef]
- Ng, H.H.; Shen, M.; Samuel, C.S.; Schlossmann, J.; Bennett, R.G. Relaxin and Extracellular Matrix Remodeling: Mechanisms and Signaling Pathways. Mol. Cell. Endocrinol. 2019, 487, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; Walter, M.R. Cytokine-Receptor Interactions as Drug Targets. Curr. Opin. Chem. Biol. 2010, 14, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Liu, T.; Yao, Y. Implications of Viral Infections and Oncogenesis in Uterine Cervical Carcinoma Etiology and Pathogenesis. Front. Microbiol. 2023, 14, 1194431. [Google Scholar] [CrossRef] [PubMed]
- Herpes Simplex Virus and the Chemokines That Mediate the Inflammation. SpringerLink. Available online: https://link.springer.com/chapter/10.1007/978-3-540-33397-5_3 (accessed on 21 April 2024).
- Gudkov, A.V.; Gurova, K.V.; Komarova, E.A. Inflammation and P53: A Tale of Two Stresses. Genes Cancer 2011, 2, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, L.; Li, P.; Zheng, Y.; Yang, Y.; Ji, S. Apelin/APJ System in Inflammation. Int. Immunopharmacol. 2022, 109, 108822. [Google Scholar] [CrossRef] [PubMed]
- Pollyea, D.A.; Harris, C.; Rabe, J.L.; Hedin, B.R.; Arras, L.D.; Katz, S.; Wheeler, E.; Bejar, R.; Walter, M.J.; Jordan, C.T.; et al. Myelodysplastic Syndrome-Associated Spliceosome Gene Mutations Enhance Innate Immune Signaling. Haematologica 2019, 104, e388–e392. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T. Cytokines and Cytokine Receptors as Targets of Immune-Mediated Inflammatory Diseases—RA as a Role Model. Inflamm. Regen. 2022, 42, 35. [Google Scholar] [CrossRef] [PubMed]
- Jurewicz, M.M.; Stern, L.J. Class II MHC Antigen Processing in Immune Tolerance and Inflammation. Immunogenetics 2019, 71, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Moens, U.; Kostenko, S.; Sveinbjørnsson, B. The Role of Mitogen-Activated Protein Kinase-Activated Protein Kinases (MAPKAPKs) in Inflammation. Genes 2013, 4, 101–133. [Google Scholar] [CrossRef]
- Chen, Y.; Williams, V.; Filippova, M.; Filippov, V.; Duerksen-Hughes, P. Viral Carcinogenesis: Factors Inducing DNA Damage and Virus Integration. Cancers 2014, 6, 2155–2186. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB Signaling in Inflammation. Signal Transduction and Targeted Therapy. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Zhou, C.; Li, C.; Zheng, Y.; Liu, X. Identification of Pyroptosis-Related Signature for Cervical Cancer Predicting Prognosis. Aging 2021, 13, 24795–24814. [Google Scholar] [CrossRef] [PubMed]
- Gudkov, A.V.; Komarova, E.A. P53 and the Carcinogenicity of Chronic Inflammation. Cold Spring Harb. Perspect. Med. 2016, 6, a026161. [Google Scholar] [CrossRef]
- Seif, F.; Khoshmirsafa, M.; Aazami, H.; Mohsenzadegan, M.; Sedighi, G.; Bahar, M. The Role of JAK-STAT Signaling Pathway and Its Regulators in the Fate of T Helper Cells. Cell Commun. Signal. CCS 2017, 15, 23. [Google Scholar] [CrossRef]
- Chew, T.; Taylor, K.E.; Mossman, K.L. Innate and Adaptive Immune Responses to Herpes Simplex Virus. Viruses 2009, 1, 979–1002. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR Signaling in Growth Control and Disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lu, Y.; Piao, W.; Jin, H. The Translational Regulation in mTOR Pathway. Biomolecules 2022, 12, 802. [Google Scholar] [CrossRef] [PubMed]
- Whitmarsh, A.J. Regulation of Gene Transcription by Mitogen-Activated Protein Kinase Signaling Pathways. Biochim. Biophys. Acta 2007, 1773, 1285–1298. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT Signaling Pathway: From Bench to Clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Giuliani, C.; Bucci, I.; Napolitano, G. The Role of the Transcription Factor Nuclear Factor-Kappa B in Thyroid Autoimmunity and Cancer. Front. Endocrinol. 2018, 9, 471. [Google Scholar] [CrossRef]
- Zheng, D.; Worthington, J.; Timms, J.F.; Woo, P. HNRNPA1 Interacts with a 5′-Flanking Distal Element of Interleukin-6 and Upregulates Its Basal Transcription. Genes Immun. 2013, 14, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-F.; Lang, A.; Rothhaar, P.; Grünvogel, O.; Colasanti, O.; Ugarte, S.M.O.; Traut, J.; Piras, A.; Acosta-Rivero, N.; Magalhães, V.G.; et al. RBM39 Shapes Innate Immunity through Transcriptional and Splicing Control of IRF3 and Other Key Factors. bioRxiv 2023. [Google Scholar] [CrossRef]
- Longworth, M.S.; Laimins, L.A. Pathogenesis of Human Papillomaviruses in Differentiating Epithelia. Microbiol. Mol. Biol. Rev. MMBR 2004, 68, 362–372. [Google Scholar] [CrossRef]
- Zhang, H.; Cai, S.; Xia, Y.; Lin, Y.; Zhou, G.; Yu, Y.; Feng, M. Association between Human Herpesvirus Infection and Cervical Carcinoma: A Systematic Review and Meta-Analysis. Virol. J. 2023, 20, 288. [Google Scholar] [CrossRef] [PubMed]
- Senba, M.; Buziba, N.; Mori, N.; Fujita, S.; Morimoto, K.; Wada, A.; Toriyama, K. Human Papillomavirus Infection Induces NF-κB Activation in Cervical Cancer: A Comparison with Penile Cancer. Oncol. Lett. 2011, 2, 65–68. [Google Scholar] [CrossRef]
- Branca, M.; Ciotti, M.; Santini, D.; Di Bonito, L.; Benedetto, A.; Giorgi, C.; Paba, P.; Favalli, C.; Costa, S.; Agarossi, A.; et al. Activation of the ERK/MAP Kinase Pathway in Cervical Intraepithelial Neoplasia Is Related to Grade of the Lesion but Not to High-Risk Human Papillomavirus, Virus Clearance, or Prognosis in Cervical Cancer. Am. J. Clin. Pathol. 2004, 122, 902–911. [Google Scholar] [CrossRef]
- Morgan, E.L.; Macdonald, A. Manipulation of JAK/STAT Signalling by High-Risk HPVs: Potential Therapeutic Targets for HPV-Associated Malignancies. Viruses 2020, 12, 977. [Google Scholar] [CrossRef] [PubMed]
- Chuluunbaatar, U.; Roller, R.; Mohr, I. Suppression of Extracellular Signal-Regulated Kinase Activity in Herpes Simplex Virus 1-Infected Cells by the Us3 Protein Kinase. J. Virol. 2012, 86, 7771–7776. [Google Scholar] [CrossRef]
- Ezeonwumelu, I.J.; Garcia-Vidal, E.; Ballana, E. JAK-STAT Pathway: A Novel Target to Tackle Viral Infections. Viruses 2021, 13, 2379. [Google Scholar] [CrossRef]
- Maclean, J.A.; Hayashi, K. Progesterone Actions and Resistance in Gynecological Disorders. Cells 2022, 11, 647. [Google Scholar] [CrossRef]
- Ho, S.-M. Estrogen, Progesterone and Epithelial Ovarian Cancer. Reprod. Biol. Endocrinol. 2003, 1, 73. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Dai, X.; Gao, Y.; Shen, F.; Ding, J.; Chen, Q. The Positivity of Estrogen Receptor and Progesterone Receptor May Not Be Associated with Metastasis and Recurrence in Epithelial Ovarian Cancer. Sci. Rep. 2017, 7, 16922. [Google Scholar] [CrossRef] [PubMed]
- Motohara, T.; Masuda, K.; Morotti, M.; Zheng, Y.; El-Sahhar, S.; Chong, K.Y.; Wietek, N.; Alsaadi, A.; Karaminejadranjbar, M.; Hu, Z.; et al. An Evolving Story of the Metastatic Voyage of Ovarian Cancer Cells: Cellular and Molecular Orchestration of the Adipose-Rich Metastatic Microenvironment. Oncogene 2019, 38, 2885–2898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, G. Progesterone Resistance in Endometriosis: Current Evidence and Putative Mechanisms. Int. J. Mol. Sci. 2023, 24, 6992. [Google Scholar] [CrossRef]
- Chantalat, E.; Valera, M.C.; Vaysse, C.; Noirrit, E.; Rusidze, M.; Weyl, A.; Vergriete, K.; Buscail, E.; Lluel, P.; Fontaine, C.; et al. Estrogen Receptors and Endometriosis. Int. J. Mol. Sci. 2020, 21, 2815. [Google Scholar] [CrossRef]
- Song, R.X.-D.; McPherson, R.A.; Adam, L.; Bao, Y.; Shupnik, M.; Kumar, R.; Santen, R.J. Linkage of Rapid Estrogen Action to MAPK Activation by ERα-Shc Association and Shc Pathway Activation. Mol. Endocrinol. 2002, 16, 116–127. [Google Scholar] [CrossRef][Green Version]
- Clusan, L.; Ferrière, F.; Flouriot, G.; Pakdel, F. A Basic Review on Estrogen Receptor Signaling Pathways in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 6834. [Google Scholar] [CrossRef]
- Coughlan, N.; Thillainadesan, G.; Andrews, J.; Isovic, M.; Torchia, J. β-Estradiol-Dependent Activation of the JAK/STAT Pathway Requires p/CIP and CARM1. Biochim. Biophys. Acta BBA—Mol. Cell Res. 2013, 1833, 1463–1475. [Google Scholar] [CrossRef]
- Ng, C.W.; Tsang, Y.T.; Gershenson, D.M.; Wong, K.K. The Prognostic Value of MEK Pathway–Associated Estrogen Receptor Signaling Activity for Female Cancers. Br. J. Cancer 2024, 130, 1875–1884. [Google Scholar] [CrossRef]
- Schüler-Toprak, S.; Weber, F.; Skrzypczak, M.; Ortmann, O.; Treeck, O. Expression of Estrogen-Related Receptors in Ovarian Cancer and Impact on Survival. J. Cancer Res. Clin. Oncol. 2021, 147, 2555–2567. [Google Scholar] [CrossRef]
- Marla, S.; Mortlock, S.; Houshdaran, S.; Fung, J.; McKinnon, B.; Holdsworth-Carson, S.J.; Girling, J.E.; Rogers, P.A.W.; Giudice, L.C.; Montgomery, G.W. Genetic Risk Factors for Endometriosis near Estrogen Receptor 1 and Coexpression of Genes in This Region in Endometrium. Mol. Hum. Reprod. 2021, 27, gaaa082. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-M.; Wu, Z.-M.; Huang, H.; Chu, X.-Y.; Lou, J.; Xu, L.-X.; Chen, Y.-T.; Wang, L.-Q.; Huang, O.-P. Estrogen Receptor 1 Mutations in 260 Cervical Cancer Samples from Chinese Patients. Oncol. Lett. 2019, 18, 2771–2776. [Google Scholar] [CrossRef] [PubMed]
- Dessources, K.; Miller, K.M.; Kertowidjojo, E.; Paula, A.D.C.; Zou, Y.; Selenica, P.; da Silva, E.M.; Benayed, R.; Ashley, C.W.; Abu-Rustum, N.R.; et al. ESR1 Hotspot Mutations in Endometrial Stromal Sarcoma with High-Grade Transformation and Endocrine Treatment. Mod. Pathol. 2022, 35, 972–978. [Google Scholar] [CrossRef]
- Kato, S.; McFall, T.; Takahashi, K.; Bamel, K.; Ikeda, S.; Eskander, R.N.; Plaxe, S.; Parker, B.; Stites, E.; Kurzrock, R. KRAS-Mutated, Estrogen Receptor-Positive Low-Grade Serous Ovarian Cancer: Unraveling an Exceptional Response Mystery. Oncologist 2021, 26, e530–e536. [Google Scholar] [CrossRef]
- Chibbar, R.; Foerstner, S.; Suresh, J.; Chibbar, R.; Piche, A.; Kundapur, D.; Kanthan, R.; Kundapur, V.; Lee, C.H.; Agrawal, A.; et al. Estrogen/Progesterone Receptor Loss, CTNNB1 and KRAS Mutations Are Associated with Local Recurrence or Distant Metastasis in Low-Grade Endometrial Endometrioid Carcinoma. Appl. Immunohistochem. Mol. Morphol. 2023, 31, 181. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Ye, X.; Dai, J.; Sun, F. SOS1 Promotes Epithelial-Mesenchymal Transition of Epithelial Ovarian Cancer(EOC) Cells through AKT Independent NF-κB Signaling Pathway. Transl. Oncol. 2021, 14, 101160. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, R.A.; Zyju, D.P.; Subramaniam, A.G.; Nautiyal, J.; Laloraya, M. Son of Sevenless 1 (SOS1), the RasGEF, Interacts with ERα and STAT3 during Embryo Implantation. J. Mol. Endocrinol. 2023, 70, e220089. [Google Scholar] [CrossRef] [PubMed]
- Binai, N.A.; Carra, G.; Löwer, J.; Löwer, R.; Wessler, S. Differential Gene Expression in ERα-Positive and ERα-Negative Breast Cancer Cells upon Leptin Stimulation. Endocrine 2013, 44, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, H.; Yao, J. ERα, A Key Target for Cancer Therapy: A Review. OncoTargets Ther. 2020, 13, 2183–2191. [Google Scholar] [CrossRef]
- Kurnit, K.C.; Kim, G.N.; Fellman, B.M.; Urbauer, D.L.; Mills, G.B.; Zhang, W.; Broaddus, R.R. CTNNB1 (Beta-Catenin) Mutation Identifies Low Grade, Early Stage Endometrial Cancer Patients at Increased Risk of Recurrence. Mod. Pathol. 2017, 30, 1032–1041. [Google Scholar] [CrossRef]
- Ali, M.; Shahin, S.M.; Sabri, N.A.; Al-Hendy, A.; Yang, Q. Activation of β-Catenin Signaling and Its Crosstalk With Estrogen and Histone Deacetylases in Human Uterine Fibroids. J. Clin. Endocrinol. Metab. 2020, 105, e1517–e1535. [Google Scholar] [CrossRef] [PubMed]
- Montemorano, L.; Shultz, Z.B.; Farooque, A.; Hyun, M.; Chappell, R.J.; Hartenbach, E.M.; Lang, J.D. TP53 Mutations and the Association with Platinum Resistance in High Grade Serous Ovarian Carcinoma. Gynecol. Oncol. 2024, 186, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Hurson, A.N.; Abubakar, M.; Hamilton, A.M.; Conway, K.; Hoadley, K.A.; Love, M.I.; Olshan, A.F.; Perou, C.M.; Garcia-Closas, M.; Troester, M.A. TP53 Pathway Function, Estrogen Receptor Status, and Breast Cancer Risk Factors in the Carolina Breast Cancer Study. Cancer Epidemiol. Biomarkers Prev. 2022, 31, 124–131. [Google Scholar] [CrossRef]
- Berger, C.; Qian, Y.; Chen, X. The P53-Estrogen Receptor Loop in Cancer. Curr. Mol. Med. 2013, 13, 1229–1240. [Google Scholar] [CrossRef]
- Berger, C.E.; Qian, Y.; Liu, G.; Chen, H.; Chen, X. P53, a Target of Estrogen Receptor (ER) α, Modulates DNA Damage-Induced Growth Suppression in ER-Positive Breast Cancer Cells. J. Biol. Chem. 2012, 287, 30117–30127. [Google Scholar] [CrossRef]
- Chou, J.-L.; Su, H.-Y.; Chen, L.-Y.; Liao, Y.-P.; Hartman-Frey, C.; Lai, Y.-H.; Yang, H.-W.; Deatherage, D.E.; Kuo, C.-T.; Huang, Y.-W.; et al. Promoter Hypermethylation of FBXO32, a Novel TGF-Beta/SMAD4 Target Gene and Tumor Suppressor, Is Associated with Poor Prognosis in Human Ovarian Cancer. Lab. Investig. 2010, 90, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.W.; Huang, Y.W.; Hartman-Frey, C.; Kuo, C.T.; Deatherage, D.; Qin, H.; Cheng, A.S.; Yan, P.S.; Davuluri, R.V.; Huang, T.H.; et al. Aberrant Transforming Growth Factor Β1 Signaling and SMAD4 Nuclear Translocation Confer Epigenetic Repression of ADAM19 in Ovarian Cancer. Neoplasia 2008, 10, 908–919. [Google Scholar] [CrossRef]
- Sanjabi, S.; Zenewicz, L.A.; Kamanaka, M.; Flavell, R.A. Anti-Inflammatory and pro-Inflammatory Roles of TGF-β, IL-10, and IL-22 in Immunity and Autoimmunity. Curr. Opin. Pharmacol. 2009, 9, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Jridi, I.; Canté-Barrett, K.; Pike-Overzet, K.; Staal, F.J.T. Inflammation and Wnt Signaling: Target for Immunomodulatory Therapy? Front. Cell Dev. Biol. 2021, 8, 615131. [Google Scholar] [CrossRef]
- Lee, Y.-R.; Park, J.; Yu, H.-N.; Kim, J.-S.; Youn, H.J.; Jung, S.H. Up-Regulation of PI3K/Akt Signaling by 17β-Estradiol through Activation of Estrogen Receptor-α, but Not Estrogen Receptor-β, and Stimulates Cell Growth in Breast Cancer Cells. Biochem. Biophys. Res. Commun. 2005, 336, 1221–1226. [Google Scholar] [CrossRef]
- Elhasnaoui, J.; Ferrero, G.; Miano, V.; Cutrupi, S.; De Bortoli, M. The Estrogen Receptor α Signaling Pathway Controls Alternative Splicing in the Absence of Ligands in Breast Cancer Cells. Cancers 2021, 13, 6261. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, C.B.; Kjaer, S.K.; Albieri, V.; Bandera, E.V.; Doherty, J.A.; Høgdall, E.; Webb, P.M.; Jordan, S.J.; Rossing, M.A.; Wicklund, K.G.; et al. Pelvic Inflammatory Disease and the Risk of Ovarian Cancer and Borderline Ovarian Tumors: A Pooled Analysis of 13 Case-Control Studies. Am. J. Epidemiol. 2017, 185, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, C.B.; Jensen, A.; Albieri, V.; Andersen, K.K.; Kjaer, S.K. Is Pelvic Inflammatory Disease a Risk Factor for Ovarian Cancer? Cancer Epidemiol. Biomark. Prev. 2017, 26, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Lin, K.Y.; Huang, C.C.; Lin, W.C. Association of Pelvic Inflammatory Disease (PID) with Ovarian Cancer: A Nationwide Population-Based Retrospective Cohort Study from Taiwan. BMC Women’s Health 2021, 21, 274. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Sheng, Z.; Cheng, C.; Zheng, H.; Lanuti, M.; Liu, R.; Wang, P.; Shen, Y.; Xie, Z. Anesthetic Propofol Promotes Tumor Metastasis in Lungs via GABAAR-Dependent TRIM21 Modulation of Src Expression. Adv. Sci. 2021, 8, 2102079. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.-C.; Liao, K.-S.; Yeh, L.-R.; Wang, Y.-K. Drug Repurposing: The Mechanisms and Signaling Pathways of Anti-Cancer Effects of Anesthetics. Biomedicines 2022, 10, 1589. [Google Scholar] [CrossRef] [PubMed]
- Tsurumi, K.; Hayashi, M.; Abe, A.; Go, K.; Fujimura, H. Anti-inflammatory and analgesic effects of benzodiazepine derivatives. Nihon Yakurigaku Zasshi 1973, 69, 343–358. (In Japanese) [Google Scholar] [CrossRef]
- Nair, A.S.; Paliwal, A. 12—Systems Pharmacology and Molecular Docking Strategies Prioritize Natural Molecules as Antiinflammatory Agents. In Inflammation and Natural Products; Gopi, S., Amalraj, A., Kunnumakkara, A., Thomas, S., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 239–275. ISBN 978-0-12-819218-4. [Google Scholar]
- Hong, W.; Wang, Y.; Chang, Z.; Yang, Y.; Pu, J.; Sun, T.; Kaur, S.; Sacchettini, J.C.; Jung, H.; Lin Wong, W.; et al. The Identification of Novel Mycobacterium Tuberculosis DHFR Inhibitors and the Investigation of Their Binding Preferences by Using Molecular Modelling. Sci. Rep. 2015, 5, 15328. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, V.A.; Wang, H.; Czura, C.J.; Friedman, S.G.; Tracey, K.J. The Cholinergic Anti-Inflammatory Pathway: A Missing Link in Neuroimmunomodulation. Mol. Med. 2003, 9, 125–134. [Google Scholar] [CrossRef]
- Moosa, M.-Y.S.; Sobel, J.D.; Elhalis, H.; Du, W.; Akins, R.A. Fungicidal Activity of Fluconazole against Candida Albicans in a Synthetic Vagina-Simulative Medium. Antimicrob. Agents Chemother. 2004, 48, 161–167. [Google Scholar] [CrossRef]
- Khalilzadeh, M.; Shayan, M.; Jourian, S.; Rahimi, M.; Sheibani, M.; Dehpour, A.R. A Comprehensive Insight into the Anti-Inflammatory Properties of Dapsone. Naunyn. Schmiedebergs Arch. Pharmacol. 2022, 395, 1509–1523. [Google Scholar] [CrossRef] [PubMed]
- Escoter-Torres, L.; Greulich, F.; Quagliarini, F.; Wierer, M.; Uhlenhaut, N.H. Anti-Inflammatory Functions of the Glucocorticoid Receptor Require DNA Binding. Nucleic Acids Res. 2020, 48, 8393–8407. [Google Scholar] [CrossRef] [PubMed]
- Kanova, M.; Kohout, P. Serotonin—Its Synthesis and Roles in the Healthy and the Critically Ill. Int. J. Mol. Sci. 2021, 22, 4837. [Google Scholar] [CrossRef] [PubMed]
- Livshits, A.; Seidman, D.S. Role of Non-Steroidal Anti-Inflammatory Drugs in Gynecology. Pharmaceuticals 2010, 3, 2082–2089. [Google Scholar] [CrossRef]
- Radman, H.M. Postoperative Treatment of the Gynecologic Patient: Use of Phenylbutazone. Am. J. Surg. 1961, 101, 431–434. [Google Scholar] [CrossRef]
| Accession No. | GSE51981 | GSE63885 | GSE63514 | GSE17025 |
|---|---|---|---|---|
| Condition | Endometriosis | Ovarian Cancer | Cervical Cancer | Endometrial Cancer |
| Year Published | 2014 | 2014 | 2015 | 2011 |
| Type | Expression Profiling by Array | |||
| Platform | GPL570-HG-U133 Plus 2 Affymetrix Human Genome U133 Plus 2.0 Array | |||
| Source | Endometrial Tissue | Primary Cancer Tissue | ||
| No. of Samples | 77 | 101 | 104 | 91 |
| Expression | Genes | Drug | Mechanism | Tau | FDR |
|---|---|---|---|---|---|
| Upregulated | KRAS, HNRNPA1, SOS1, YWHAZ, AKAP6, LARP7, PTK2, UBE2D3, SMAD4, CASP3, SQSTM1, ESR1, NRXN1, ENPEP, GPX7, CDK1, CCNB1, TOP2A, CCNA2, BUB1B, EXO1, AURKB, CDC20, BUB1, CDC45, EGFR, MRPS15, GMPS, MARS1, ELOC, POLR1B, CTNNB1, RBM39, HNRNPA2B1, RBM25, DDX17, SRSF11, RACK1, and STAT3 | Thiamphenicol | bacterial 50S ribosomal subunit inhibitor | −99.8 | 5.24 × 10−3 |
| Trimethoprim | dihydrofolate reductase inhibitor | −99.7 | 9.69 × 10−3 | ||
| Medrysone | glucocorticoid receptor agonist | −99.1 | 5.79 × 10−3 | ||
| Pentolinium | cholinergic receptor agonist | −98.9 | 4.72 × 10−4 | ||
| Paroxetine | selective serotonin reuptake inhibitor | −98.8 | 1.50× 10−3 | ||
| Downregulated | CALML6, ITIH4, CDK6, DISC1, CD9, TP53, PES1, VAMP2, FUS, and SNRNP70 | Propofol | benzodiazepine receptor agonist | −99.5 | 2.11 × 10−3 |
| Fluconazole | sterol demethylase inhibitor | −99.4 | 1.74 × 10−3 | ||
| Dapsone | bacterial antifolate | −99.1 | 8.96 × 10−3 | ||
| Hydrocortisone | glucocorticoid receptor agonist | −98.8 | 8.22 × 10−3 | ||
| MDL73005EF | serotonin receptor antagonist | −98.7 | 4.68 × 10−3 |
| Module/s | KEGG Pathway | p-Value | Reference |
|---|---|---|---|
| Cyan | Ras signaling pathway | 1.0 × 10−1 | [36] |
| Midnightblue, Green | Herpes simplex virus infection | 3.6 × 10−7 | [40] |
| Pink | p53 signaling pathway | 9.1 × 10−5 | [41] |
| Purple | Apelin signaling pathway | 3.0 × 10−2 | [42] |
| Tan | Spliceosome | 1.2 × 10−4 | [43] |
| Brown | Cytokine-cytokine receptor interaction | 5.3 × 10−8 | [44] |
| Darkgrey | Relaxin signaling pathway | 1.01 × 10−1 | [37] |
| Lightcyan | Antigen processing and presentation | 9.5 × 10−23 | [45] |
| Magenta, Skyblue | MAPK signaling pathway | 2.1 × 10−3 | [46] |
| Orange | IL-17 signaling pathway | 4.0 × 10−15 | [35] |
| Steelblue | Viral carcinogenesis | 1.1 × 10−18 | [47] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasamba, E.C.; Orda, M.A.; Villanueva, B.H.A.; Tsai, P.-W.; Tayo, L.L. Transcriptomic Analysis of Hub Genes Reveals Associated Inflammatory Pathways in Estrogen-Dependent Gynecological Diseases. Biology 2024, 13, 397. https://doi.org/10.3390/biology13060397
Pasamba EC, Orda MA, Villanueva BHA, Tsai P-W, Tayo LL. Transcriptomic Analysis of Hub Genes Reveals Associated Inflammatory Pathways in Estrogen-Dependent Gynecological Diseases. Biology. 2024; 13(6):397. https://doi.org/10.3390/biology13060397
Chicago/Turabian StylePasamba, Elaine C., Marco A. Orda, Brian Harvey Avanceña Villanueva, Po-Wei Tsai, and Lemmuel L. Tayo. 2024. "Transcriptomic Analysis of Hub Genes Reveals Associated Inflammatory Pathways in Estrogen-Dependent Gynecological Diseases" Biology 13, no. 6: 397. https://doi.org/10.3390/biology13060397
APA StylePasamba, E. C., Orda, M. A., Villanueva, B. H. A., Tsai, P.-W., & Tayo, L. L. (2024). Transcriptomic Analysis of Hub Genes Reveals Associated Inflammatory Pathways in Estrogen-Dependent Gynecological Diseases. Biology, 13(6), 397. https://doi.org/10.3390/biology13060397









