Distinct Expression Profiles of Neuroblastoma-Associated mRNAs in Peripheral Blood and Bone Marrow of Non-High-Risk and High-Risk Neuroblastoma Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. RNA Isolation and cDNA Synthesis
2.3. 7NB-mRNAs ddPCR Assay
3. Results
3.1. Characteristics of Non-HR and HR Cases
3.2. Levels of Seven NB-mRNAs in PB and BM Samples of Non-HR and HR Cases
3.3. Levels of Tumor Markers in Non-HR and HR Cases
3.4. Association between Tumor Markers and 7NB-mRNAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maris, J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef] [PubMed]
- Tolbert, V.P.; Matthay, K.K. Neuroblastoma: Clinical and biological approach to risk stratification and treatment. Cell Tissue Res. 2018, 372, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Tomolonis, J.A.; Agarwal, S.; Shohet, J.M. Neuroblastoma pathogenesis: Deregulation of embryonic neural crest development. Cell Tissue Res. 2018, 372, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Delloye-Bourgeois, C.; Castellani, V. Hijacking of Embryonic Programs by Neural Crest-Derived Neuroblastoma: From Physiological Migration to Metastatic Dissemination. Front. Mol. Neurosci. 2019, 12, 52. [Google Scholar] [CrossRef]
- Irwin, M.S.; Naranjo, A.; Zhang, F.F.; Cohn, S.L.; London, W.B.; Gastier-Foster, J.M.; Ramirez, N.C.; Pfau, R.; Reshmi, S.; Wagner, E.; et al. Revised Neuroblastoma Risk Classification System: A Report From the Children’s Oncology Group. J. Clin. Oncol. 2021, 39, 3229–3241. [Google Scholar] [CrossRef] [PubMed]
- Cohn, S.L.; Pearson, A.D.J.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Monclair, T.; Brodeur, G.M.; Ambros, P.F.; Brisse, H.J.; Cecchetto, G.; Holmes, K.; Kaneko, M.; London, W.B.; Matthay, K.K.; Nuchtern, J.G.; et al. The International Neuroblastoma Risk Group (INRG) Staging System: An INRG Task Force Report. J. Clin. Oncol. 2009, 27, 298–303. [Google Scholar] [CrossRef]
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H.; Ambros, I.M.; Dehner, L.P.; Hata, J.; Joshi, V.V.; Roald, B. Terminology and morphologic criteria of neuroblastic tumors: Recommendations by the International Neuroblastoma Pathology Committee. Cancer 1999, 86, 349–363. [Google Scholar] [CrossRef]
- Shimada, H.; Ambros, I.M.; Dehner, L.P.; Hata, J.; Joshi, V.V.; Roald, B.; Stram, D.O.; Gerbing, R.B.; Lukens, J.N.; Matthay, K.K.; et al. The International Neuroblastoma Pathology Classification (the Shimada system). Cancer 1999, 86, 364–372. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Pritchard, J.; Berthold, F.; Carlsen, N.L.; Castel, V.; Castelberry, R.P.; De Bernardi, B.; Evans, A.E.; Favrot, M.; Hedborg, F.; et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J. Clin. Oncol. 1993, 11, 1466–1477. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M.; Seeger, R.C.; Barrett, A.; Berthold, F.; Castleberry, R.P.; D’Angio, G.; De Bernardi, B.; Evans, A.E.; Favrot, M.; Freeman, A.I. International criteria for diagnosis, staging, and response to treatment in patients with neuroblastoma. J. Clin. Oncol. 1988, 6, 1874–1881. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Bagatell, R.; London, W.B.; Maris, J.M.; Cohn, S.L.; Mattay, K.K.; Hogarty, M.; Committee COGN. Children’s Oncology Group’s 2013 blueprint for research: Neuroblastoma. Pediatr. Blood Cancer 2013, 60, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Pinto, N.R.; Applebaum, M.A.; Volchenboum, S.L.; Matthay, K.K.; London, W.B.; Ambros, P.F.; Nakagawara, A.; Berthold, F.; Schleiermacher, G.; Park, J.R.; et al. Advances in risk classification and treatment strategies for neuroblastoma. J. Clin. Oncol. 2015, 33, 3008–3017. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.H.; Federico, S.M.; London, W.B.; Naranjo, A.; Irwin, M.S.; Volchenboum, S.L.; Cohn, S.L. Tailoring Therapy for Children With Neuroblastoma on the Basis of Risk Group Classification: Past, Present, and Future. JCO Clin. Cancer Inform. 2020, 4, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Boterberg, T.; Lucas, J.; Panoff, J.; Valteau-Couanet, D.; Hero, B.; Bagatell, R.; Hill-Kayser, C.E. Neuroblastoma. Pediatr. Blood Cancer 2021, 68 (Suppl. S2), e28473. [Google Scholar] [CrossRef] [PubMed]
- Pinto, N.; Naranjo, A.; Hibbitts, E.; Kreissman, S.G.; Granger, M.M.; Irwin, M.S.; Bagatell, R.; London, W.B.; Greengard, E.G.; Park, J.R.; et al. Predictors of differential response to induction therapy in high-risk neuroblastoma: A report from the Children’s Oncology Group (COG). Eur. J. Cancer 2019, 112, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Stutterheim, J.; Gerritsen, A.; Zappeij-Kannegieter, L.; Yalcin, B.; Dee, R.; van Noesel, M.M.; Berthold, F.; Versteeg, R.; Caron, H.N.; van der Schoot, C.E.; et al. Detecting minimal residual disease in neuroblastoma: The superiority of a panel of real-time quantitative PCR markers. Clin. Chem. 2009, 55, 1316–1326. [Google Scholar] [CrossRef]
- Uemura, S.; Ishida, T.; Thwin, K.K.M.; Yamamoto, N.; Tamura, A.; Kishimoto, K.; Hasegawa, D.; Kosaka, Y.; Nino, N.; Lin, K.S.; et al. Dynamics of Minimal Residual Disease in Neuroblastoma Patients. Front. Oncol. 2019, 9, 455. [Google Scholar] [CrossRef]
- Yanez, Y.; Grau, E.; Oltra, S.; Canete, A.; Martinez, F.; Orellana, C.; Noguera, R.; Palanca, S.; Castel, V. Minimal disease detection in peripheral blood and bone marrow from patients with non-metastatic neuroblastoma. J. Cancer Res. Clin. Oncol. 2011, 137, 1263–1272. [Google Scholar] [CrossRef]
- Stutterheim, J.; Zappeij-Kannegieter, L.; Versteeg, R.; Caron, H.N.; van der Schoot, C.E.; Tytgat, G.A. The prognostic value of fast molecular response of marrow disease in patients aged over 1 year with stage 4 neuroblastoma. Eur. J. Cancer 2011, 47, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Viprey, V.F.; Gregory, W.M.; Corrias, M.V.; Tchirkov, A.; Swerts, K.; Vicha, A.; Dallorso, S.; Brock, P.; Luksch, R.; Valteau-Couanet, D.; et al. Neuroblastoma mRNAs predict outcome in children with stage 4 neuroblastoma: A European HR-NBL1/SIOPEN study. J. Clin. Oncol. 2014, 32, 1074–1083. [Google Scholar] [CrossRef]
- Cheung, N.K.; Ostrovnaya, I.; Kuk, D.; Cheung, I.Y. Bone marrow minimal residual disease was an early response marker and a consistent independent predictor of survival after anti-GD2 immunotherapy. J. Clin. Oncol. 2015, 33, 755–763. [Google Scholar] [CrossRef]
- van Wezel, E.M.; Decarolis, B.; Stutterheim, J.; Zappeij-Kannegieter, L.; Berthold, F.; Schumacher-Kuckelkorn, R.; Simon, T.; Fiocco, M.; van Noesel, M.M.; Caron, H.N.; et al. Neuroblastoma messenger RNA is frequently detected in bone marrow at diagnosis of localised neuroblastoma patients. Eur. J. Cancer 2016, 54, 149–158. [Google Scholar] [CrossRef]
- Marachelian, A.; Villablanca, J.G.; Liu, C.W.; Liu, B.; Goodarzian, F.; Lai, H.A.; Shimada, H.; Tran, H.C.; Parra, J.A.; Gallego, R.; et al. Expression of Five Neuroblastoma Genes in Bone Marrow or Blood of Patients with Relapsed/Refractory Neuroblastoma Provides a New Biomarker for Disease and Prognosis. Clin. Cancer Res. 2017, 23, 5374–5383. [Google Scholar] [CrossRef] [PubMed]
- Corrias, M.V.; Parodi, S.; Tchirkov, A.; Lammens, T.; Vicha, A.; Pasqualini, C.; Trager, C.; Yanez, Y.; Dallorso, S.; Varesio, L.; et al. Event-free survival of infants and toddlers enrolled in the HR-NBL-1/SIOPEN trial is associated with the level of neuroblastoma mRNAs at diagnosis. Pediatr. Blood Cancer 2018, 65, e27052. [Google Scholar] [CrossRef]
- Fan, H.; Xing, T.; Hong, H.; Duan, C.; Zhao, W.; Zhao, Q.; Wang, X.; Huang, C.; Zhu, S.; Jin, M.; et al. The expression of PHOX2B in bone marrow and peripheral blood predicts adverse clinical outcome in non-high-risk neuroblastoma. Pediatr. Hematol. Oncol. 2022, 39, 343–356. [Google Scholar] [CrossRef]
- Thwin, K.K.M.; Ishida, T.; Uemura, S.; Yamamoto, N.; Lin, K.S.; Tamura, A.; Kozaki, A.; Saito, A.; Kishimoto, K.; Mori, T.; et al. Level of Seven Neuroblastoma-Associated mRNAs Detected by Droplet Digital PCR Is Associated with Tumor Relapse/Regrowth of High-Risk Neuroblastoma Patients. J. Mol. Diagn. 2020, 22, 236–246. [Google Scholar] [CrossRef]
- Boeva, V.; Louis-Brennetot, C.; Peltier, A.; Durand, S.; Pierre-Eugene, C.; Raynal, V.; Etchevers, H.C.; Thomas, S.; Lermine, A.; Daudigeos-Dubus, E.; et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 2017, 49, 1408–1413. [Google Scholar] [CrossRef] [PubMed]
- van Groningen, T.; Koster, J.; Valentijn, L.J.; Zwijnenburg, D.A.; Akogul, N.; Hasselt, N.E.; Broekmans, M.; Haneveld, F.; Nowakowska, N.E.; Bras, J.; et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 2017, 49, 1261–1266. [Google Scholar] [CrossRef]
- Gautier, M.; Thirant, C.; Delattre, O.; Janoueix-Lerosey, I. Plasticity in Neuroblastoma Cell Identity Defines a Noradrenergic-to-Mesenchymal Transition (NMT). Cancers 2021, 13, 2904. [Google Scholar] [CrossRef]
- Nishimura, N.; Ishida, T.; Yokota, I.; Matsumoto, K.; Shichino, H.; Fujisaki, H.; Sarashina, T.; Kamijo, T.; Takimoto, T.; Iehara, T.; et al. Minimal Residual Disease Detected by the 7NB-mRNAs ddPCR Assay Is Associated with Disease Progression in High-Risk Neuroblastoma Patients: A Prospective Multicenter Observational Study in Japan. Biology 2023, 12, 1350. [Google Scholar] [CrossRef] [PubMed]
- Hartomo, T.B.; Kozaki, A.; Hasegawa, D.; Van Huyen Pham, T.; Yamamoto, N.; Saitoh, A.; Ishida, T.; Kawasaki, K.; Kosaka, Y.; Ohashi, H.; et al. Minimal residual disease monitoring in neuroblastoma patients based on the expression of a set of real-time RT-PCR markers in tumor-initiating cells. Oncol. Rep. 2013, 29, 1629–1636. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The digital MIQE guidelines: Minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef]
- Simon, T.; Hero, B.; Hunneman, D.H.; Berthold, F. Tumour markers are poor predictors for relapse or progression in neuroblastoma. Eur. J. Cancer 2003, 39, 1899–1903. [Google Scholar] [CrossRef]
- London, W.B.; Castel, V.; Monclair, T.; Ambros, P.F.; Pearson, A.D.; Cohn, S.L.; Berthold, F.; Nakagawara, A.; Ladenstein, R.L.; Iehara, T.; et al. Clinical and biologic features predictive of survival after relapse of neuroblastoma: A report from the International Neuroblastoma Risk Group project. J. Clin. Oncol. 2011, 29, 3286–3292. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.D. A Systematic Review of Molecular and Biological Tumor Markers in Neuroblastoma. Clin. Cancer Res. 2004, 10, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Uemura, S.; Lin, K.S.; Mon Thwin, K.K.; Nakatani, N.; Ishida, T.; Yamamoto, N.; Tamura, A.; Saito, A.; Mori, T.; Hasegawa, D.; et al. Limited correlation between tumor markers and minimal residual disease detected by seven neuroblastoma-associated mRNAs in high-risk neuroblastoma patients. Mol. Clin. Oncol. 2021, 15, 137. [Google Scholar] [CrossRef]
- Burchill, S.A.; Selby, P.J. Molecular detection of low-level disease in patients with cancer. J. Pathol. 2000, 190, 6–14. [Google Scholar] [CrossRef]
- Yamamoto, N.; Kozaki, A.; Hartomo, T.B.; Yanai, T.; Hasegawa, D.; Kawasaki, K.; Kosaka, Y.; Matsuo, M.; Hirase, S.; Mori, T.; et al. Differential expression of minimal residual disease markers in peripheral blood and bone marrow samples from high-risk neuroblastoma patients. Oncol. Lett. 2015, 10, 3228–3232. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.S.; Uemura, S.; Thwin, K.K.M.; Nakatani, N.; Ishida, T.; Yamamoto, N.; Tamura, A.; Saito, A.; Mori, T.; Hasegawa, D.; et al. Minimal residual disease in high-risk neuroblastoma shows a dynamic and disease burden-dependent correlation between bone marrow and peripheral blood. Transl. Oncol. 2021, 14, 101019. [Google Scholar] [CrossRef] [PubMed]
- van Wezel, E.M.; van Zogchel, L.M.J.; van Wijk, J.; Timmerman, I.; Vo, N.K.; Zappeij-Kannegieter, L.; deCarolis, B.; Simon, T.; van Noesel, M.M.; Molenaar, J.J.; et al. Mesenchymal Neuroblastoma Cells Are Undetected by Current mRNA Marker Panels: The Development of a Specific Neuroblastoma Mesenchymal Minimal Residual Disease Panel. JCO Precis. Oncol. 2019, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
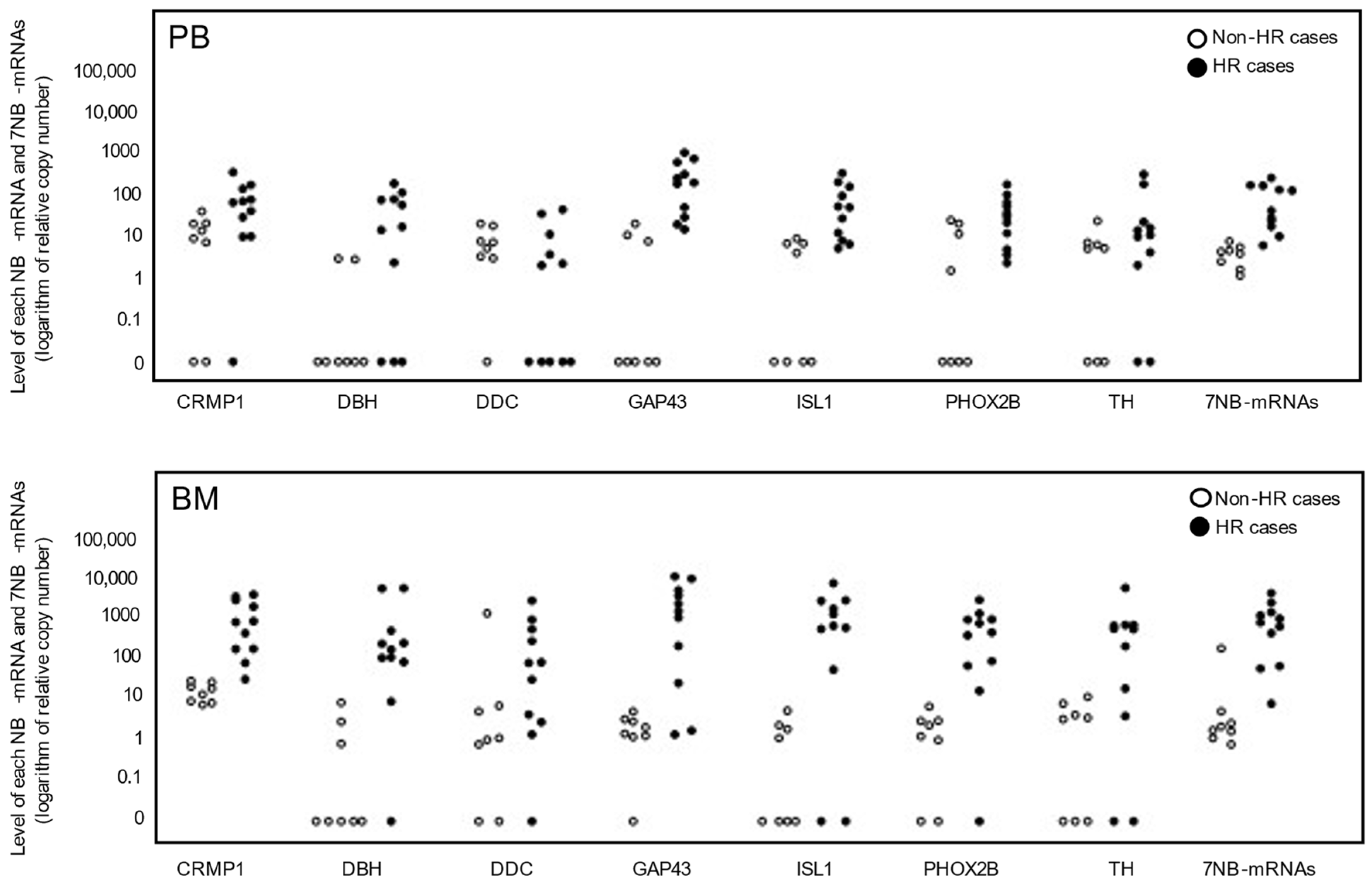
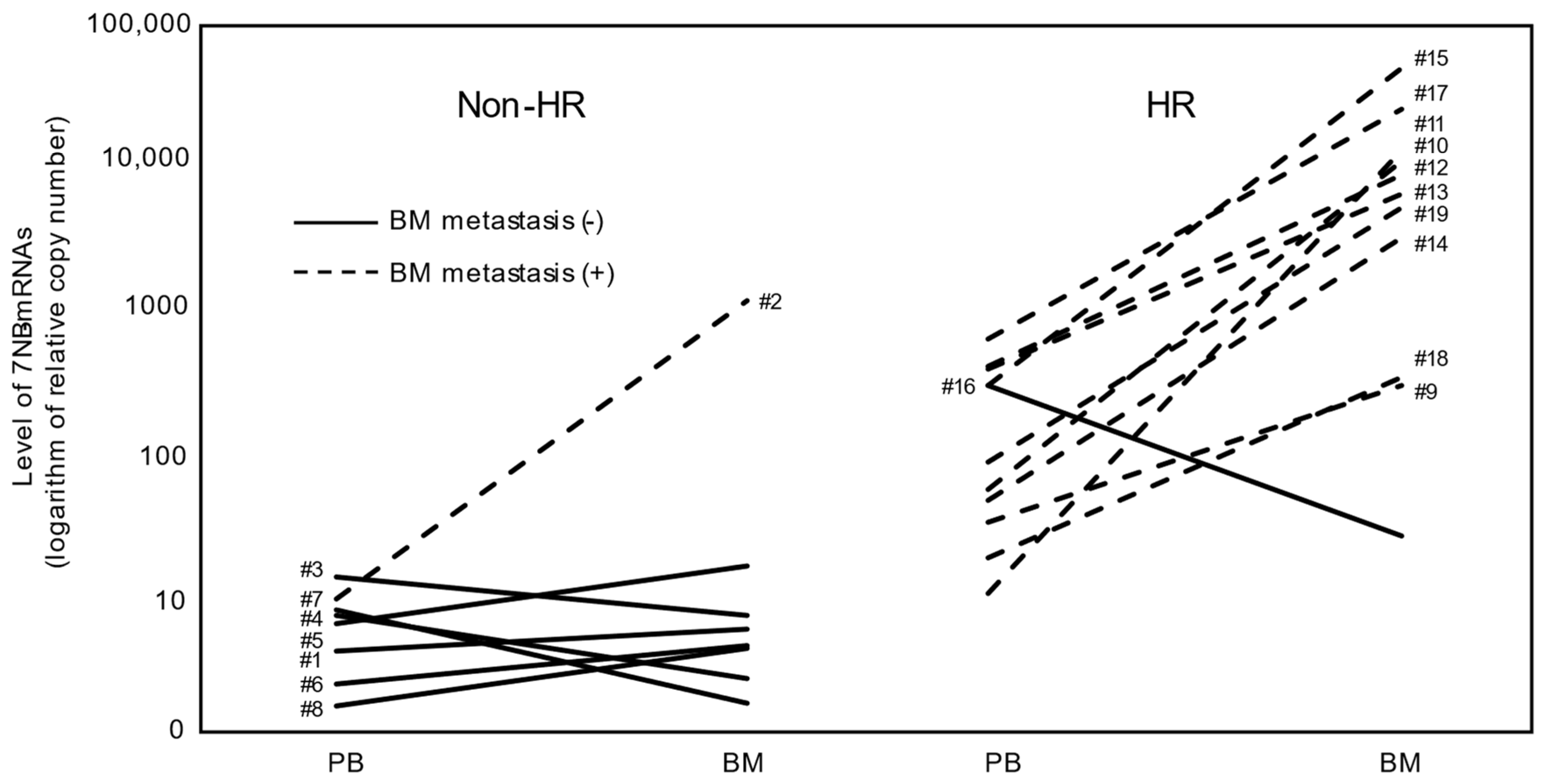
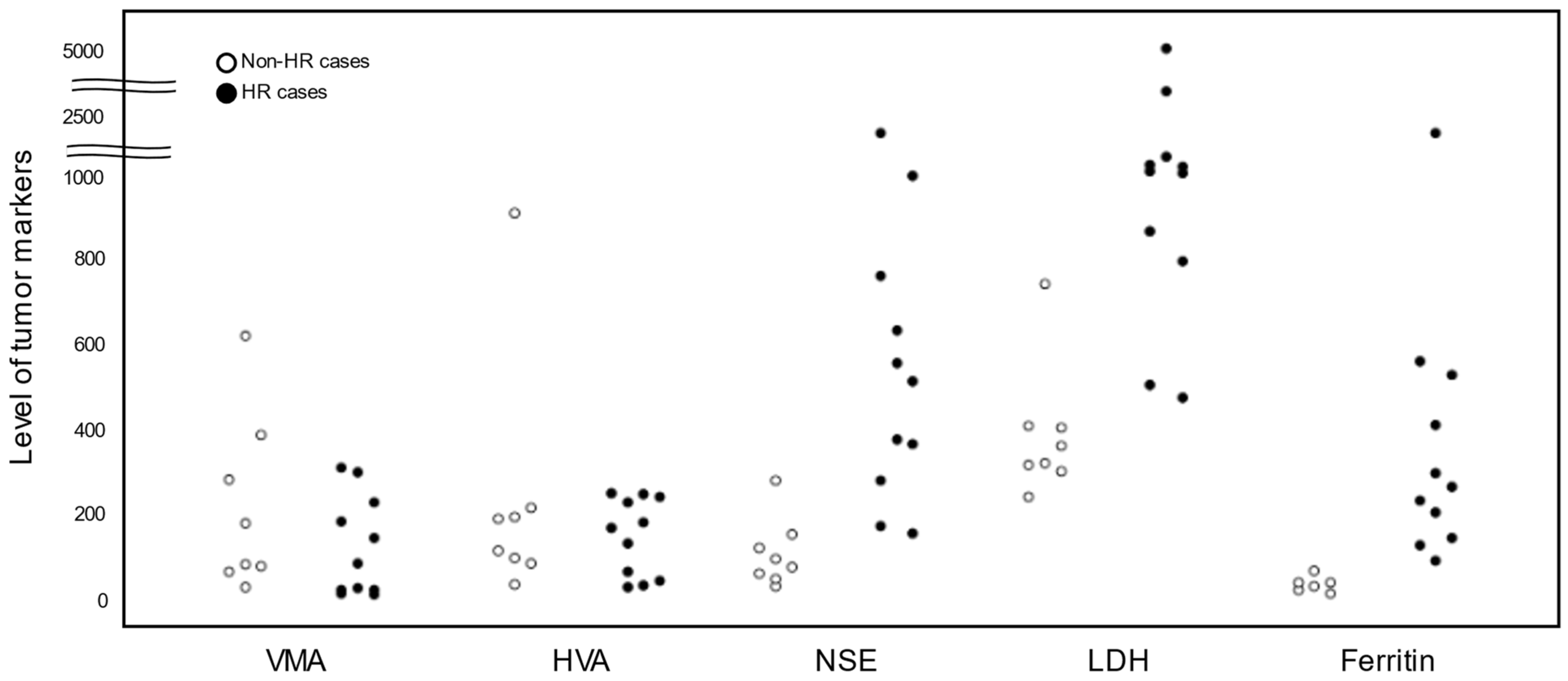
| Case | Risk | Age | Sex | Primary Tumor Site | Pathology | BM Metastasis | MYCN Status | DNA Ploidy | Relapse | Present Status |
|---|---|---|---|---|---|---|---|---|---|---|
| #1 | IR | 11 m | M | Adrenal grand | NB | (−) | non-Amp | Diploid and hyperdiploid | (−) | Alive |
| #2 | LR | 11 m | M | Adrenal grand | NB | (+) | non-Amp | Diploid and hyperdiploid | (−) | Alive |
| #3 | LR | 4 m | M | Adrenal grand | NB | (−) | non-Amp | Diploid and hyperdiploid | (−) | Alive |
| #4 | VLR | 68 m | M | Adrenal grand | GNB intermixed | (−) | non-Amp | Diploid | (−) | Alive |
| #5 | IR | 12 m | M | Adrenal grand | NB | (−) | non-Amp | Diploid | (−) | Alive |
| #6 | IR | 17 m | F | Mediastinum | NB | (−) | non-Amp | Diploid | (−) | Alive |
| #7 | IR | 9 m | M | Neck | NB | (−) | non-Amp | Diploid and hyperdiploid | (−) | Alive |
| #8 | VLR | 20 m | F | Adrenal grand | NB | (−) | non-Amp | Diploid and hyperdiploid | (−) | Alive |
| #9 | HR | 46 m | F | Adrenal grand | NB | (+) | Amp | Diploid | (+) | Dead |
| #10 | HR | 11 m | M | Adrenal grand | NB | (+) | Amp | Diploid | (+) | Dead |
| #11 | HR | 14 m | M | Adrenal grand | NB | (+) | Amp | Diploid | (+) | Dead |
| #12 | HR | 17 m | F | Adrenal grand | NB | (+) | Amp | Diploid | (+) | Dead |
| #13 | HR | 25 m | M | Adrenal grand | GNB nodular | (+) | non-Amp | Diploid | (+) | Dead |
| #14 | HR | 30 m | M | Adrenal grand | NB | (+) | non-Amp | Diploid | (+) | Dead |
| #15 | HR | 36 m | M | Adrenal grand | GNB nodular | (+) | non-Amp | Diploid | (−) | Alive |
| #16 | HR | 39 m | F | Neck | NB | (−) | non-Amp | Diploid | (+) | Dead |
| #17 | HR | 40 m | M | Adrenal grand | GNB nodular | (+) | Amp | Diploid | (−) | Alive |
| #18 | HR | 56 m | F | Mediastinum | NB | (+) | non-Amp | Diploid | (+) | Dead |
| #19 | HR | 19 m | M | Adrenal grand | NB | (+) | Amp | Diploid and near-tetraploid | (−) | Alive |
| NB-mRNAs | Non-HR Cases | HR Cases | ||||
|---|---|---|---|---|---|---|
| PB | BM | PB/BM Ratio | PB | BM | PB/BM Ratio | |
| Median | Median | Median | Median | Median | Median | |
| (IQR) | (IQR) | (IQR) | (IQR) | (IQR) | (IQR) | |
| CRMP1 | 16.6 | 48.7 | 0.23 | 105.3 | 4349.4 | 0.02 |
| (7.7–30.7) | (24.4–71.0) | (0.08–0.60) | (29.2–180.0) | (759.7–15348.1) | (0.01–0.04) | |
| DBH | 0.0 | 0.0 | 0.61 | 25.3 | 734.2 | 0.01 |
| (0.0–1.0) | (0.0–2.8) | (0.31–1.53) | (1.60–123.9) | (372.4–1798.5) | (0.00–0.10) | |
| DDC | 8.7 | 2.1 | 1.45 | 2.7 | 307.7 | 0.01 |
| (4.4–15.0) | (1.1–14.4) | (0.14–2.64) | (0.0–10.8) | (8.50–1989.1) | (0.00–0.13) | |
| GAP43 | 0.0 | 3.8 | 0.00 | 333.3 | 8892.1 | 0.06 |
| (0.0–12.2) | (2.6–6.8) | (0.00–1.20) | (61.1–812.3) | (506.2–29655.8) | (0.02–2.67) | |
| ISL1 | 2.8 | 1.1 | 0.48 | 78.8 | 3436.8 | 0.03 |
| (0.0–9.6) | (0.0–4.3) | (0.00–1.32) | (14.6–209.3) | (1512.0–13919.8) | (0.00–0.05) | |
| PHOX2B | 1.0 | 3.9 | 0.39 | 46.1 | 2213.8 | 0.02 |
| (0.0–20.1) | (1.5–7.0) | (0.00–0.90) | (12.3–93.5) | (299.1–5100.5) | (0.01–0.07) | |
| TH | 7.2 | 8.1 | 0.47 | 15.8 | 2712.3 | 0.02 |
| (0.0–9.3) | (0.0–13.0) | (0.27–0.85) | (4.25–29.1) | (33.3–3508.3) | (0.00–0.08) | |
| 7NB-mRNAs | 5.7 | 4.3 | 0.64 | 65.0 | 4194.7 | 0.03 |
| (3.1–6.8) | (3.2–7.7) | (0.42–2.03) | (31.1–247.8) | (1170.2–7560.0) | (0.01–0.06) | |
| Tumor Markers | Non-HR Cases | HR Cases |
|---|---|---|
| Median | Median | |
| (IQR) | (IQR) | |
| Urinary VMA | 128.1 | 81.4 |
| (μg/mg/cr) | (73.3–306.7) | (19.1–203.4) |
| Urinary HVA | 149.8 | 165.9 |
| (μg/mg/cr) | (92.5–197.9) | (52.5–232.7) |
| Serum NSE | 83.8 | 513.0 |
| (ng/mL) | (54.6–126.0) | (321.0–698.0) |
| Serum LDH | 339.0 | 1225.0 |
| (U/L) | (310.3–404.0) | (831.5–1732.5) |
| Serum Ferritin | 32.3 | 263.0 |
| (ng/mL) | (22.2–37.0) | (172.3–467.5) |
| Non-HR Cases | HR Cases | ||||
|---|---|---|---|---|---|
| PB | BM | PB | BM | ||
| High 7NB-mRNAs § | High 7NB-mRNAs # | High 7NB-mRNAs † | High 7NB-mRNAs ‡ | ||
| Urinary VMA (μg/mg/cr) | ≥100 | 1/4 (25%) | 3/4 (75%) | 3/5 (60%) | 3/5 (60%) |
| <100 | 3/4 (75%) | 1/4 (25%) | 3/6 (50%) | 3/6 (50%) | |
| Urinary HVA (μg/mg/cr) | ≥100 | 2/5 (40%) | 3/5 (60%) | 3/7 (43%) | 5/7 (71%) |
| <100 | 2/3 (67%) | 1/3 (33%) | 3/4 (75%) | 1/4 (25%) | |
| Serum NSE (ng/mL) | ≥100 | 1/3 (33%) | 2/3 (67%) | 6/11 (55%) | 6/11 (55%) |
| <100 | 3/5 (60%) | 2/5 (40%) | 0/0 (0%) | 0/0 (0%) | |
| Serum LDH (U/L) | ≥500 | 1/1 (100%) | 1/1 (100%) | 6/10 (60%) | 6/10 (60%) |
| <500 | 3/7 (43%) | 3/7 (43%) | 0/1 (0%) | 0/1 (0%) | |
| Serum Ferritin (ng/mL) | ≥300 | 0/0 (0%) | 0/0 (0%) | 2/4 (50%) | 2/4 (50%) |
| <300 | 3/6 (50%) | 4/6 (67%) | 4/7 (57%) | 4/7 (57%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakatani, N.; Win, K.H.N.; Mon, C.Y.; Fujikawa, T.; Uemura, S.; Saito, A.; Ishida, T.; Mori, T.; Hasegawa, D.; Kosaka, Y.; et al. Distinct Expression Profiles of Neuroblastoma-Associated mRNAs in Peripheral Blood and Bone Marrow of Non-High-Risk and High-Risk Neuroblastoma Patients. Biology 2024, 13, 345. https://doi.org/10.3390/biology13050345
Nakatani N, Win KHN, Mon CY, Fujikawa T, Uemura S, Saito A, Ishida T, Mori T, Hasegawa D, Kosaka Y, et al. Distinct Expression Profiles of Neuroblastoma-Associated mRNAs in Peripheral Blood and Bone Marrow of Non-High-Risk and High-Risk Neuroblastoma Patients. Biology. 2024; 13(5):345. https://doi.org/10.3390/biology13050345
Chicago/Turabian StyleNakatani, Naoko, Kaung Htet Nay Win, Cho Yee Mon, Tomoko Fujikawa, Suguru Uemura, Atsuro Saito, Toshiaki Ishida, Takeshi Mori, Daiichiro Hasegawa, Yoshiyuki Kosaka, and et al. 2024. "Distinct Expression Profiles of Neuroblastoma-Associated mRNAs in Peripheral Blood and Bone Marrow of Non-High-Risk and High-Risk Neuroblastoma Patients" Biology 13, no. 5: 345. https://doi.org/10.3390/biology13050345
APA StyleNakatani, N., Win, K. H. N., Mon, C. Y., Fujikawa, T., Uemura, S., Saito, A., Ishida, T., Mori, T., Hasegawa, D., Kosaka, Y., Inoue, S., Nishimura, A., Nino, N., Tamura, A., Yamamoto, N., Nozu, K., & Nishimura, N. (2024). Distinct Expression Profiles of Neuroblastoma-Associated mRNAs in Peripheral Blood and Bone Marrow of Non-High-Risk and High-Risk Neuroblastoma Patients. Biology, 13(5), 345. https://doi.org/10.3390/biology13050345





