Toward the Exploitation of Sustainable Green Factory: Biotechnology Use of Nannochloropsis spp.
Abstract
Simple Summary
Abstract
1. Introduction
2. Biotechnological Tools
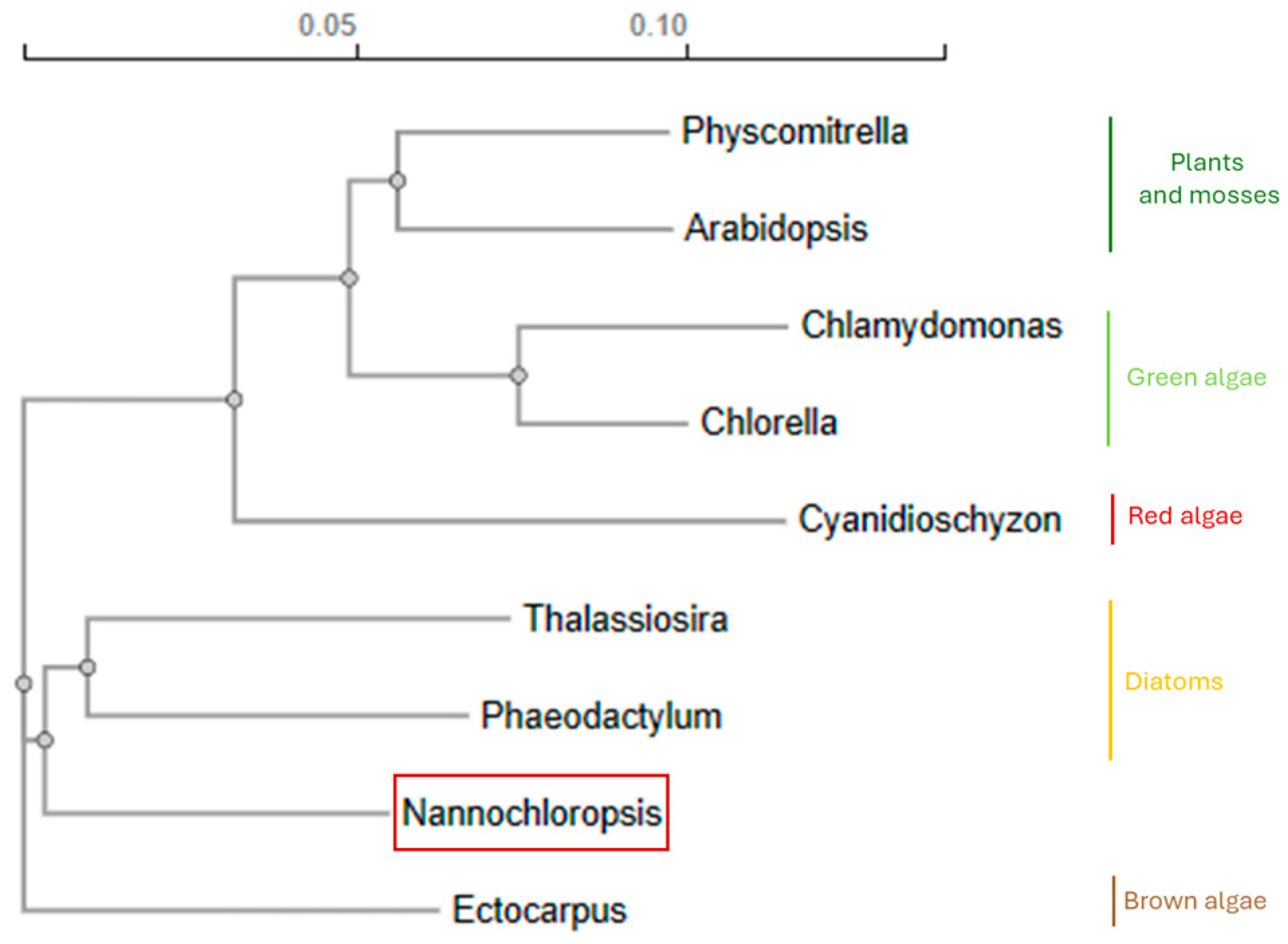
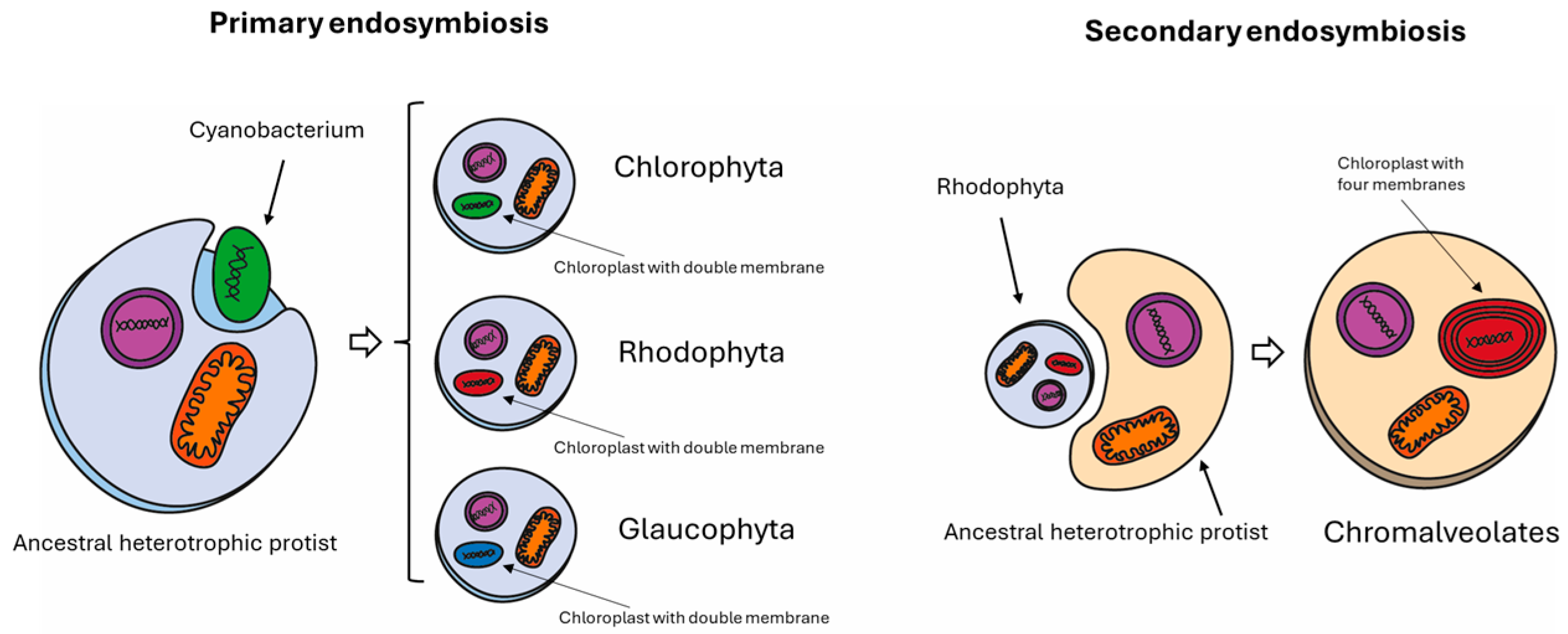
2.1. Genome Availability
2.2. Transformation Methods
2.3. Promoters
| Promoter | Nannochloropsis Strain | References |
|---|---|---|
| Lipid droplet surface protein (LDSP) | N. oceanica CCMP1779 | [39,56,72] |
| N. oceanica IMET1 | [49] | |
| β-tubulin | N. gaditana CCMP526 | [73] |
| N. oceanica IMET1 | [46,63] | |
| N. salina CCMP1776 | [64,74,75] | |
| Ubiquitin extension protein (UEP) | N. gaditana CCMP526 | [73] |
| N. salina CCMP1776 | [64,74] | |
| Violaxanthin/chlorophyll a-binding protein 1 (VCP1) | N. oceanica IMET1 | [44,46,49,58,76] |
| Violaxanthin/chlorophyll a-binding protein 2 (VCP2) | N. oceanica W2J3B | [58] |
| N. oceanica CCMP1779 | [66] | |
| N. oceanica IMET1 | [63] | |
| Elongation factor (EF) | N. oceanica CCMP1779 | [56,65] |
| Heat Shock Protein (HSP) | N. gaditana CCMP526 | [73,77] |
| N. oceanica IMET1 | [63] | |
| Ribosomal subunits (Ribi) | N. oceanica CCMP1779 | [56] |
| Extrinsic protein in photosystem II (EPPSII) | N. gaditana CCMP526 | [77] |
| ATPase | N. gaditana CCMP526 | [77] |
| Sulfoquinovosyl diacylglycerol synthase 2 (SQD2) | N. oceanica NIES-2155 | [69] |
| Nitrate reductase (NIT) | N. gaditana CCMP526 | [70] |
2.4. Selection Markers and Reporters for Gene Expression
2.5. Genetic Knockout Strategies
2.6. Multiple Gene Expression
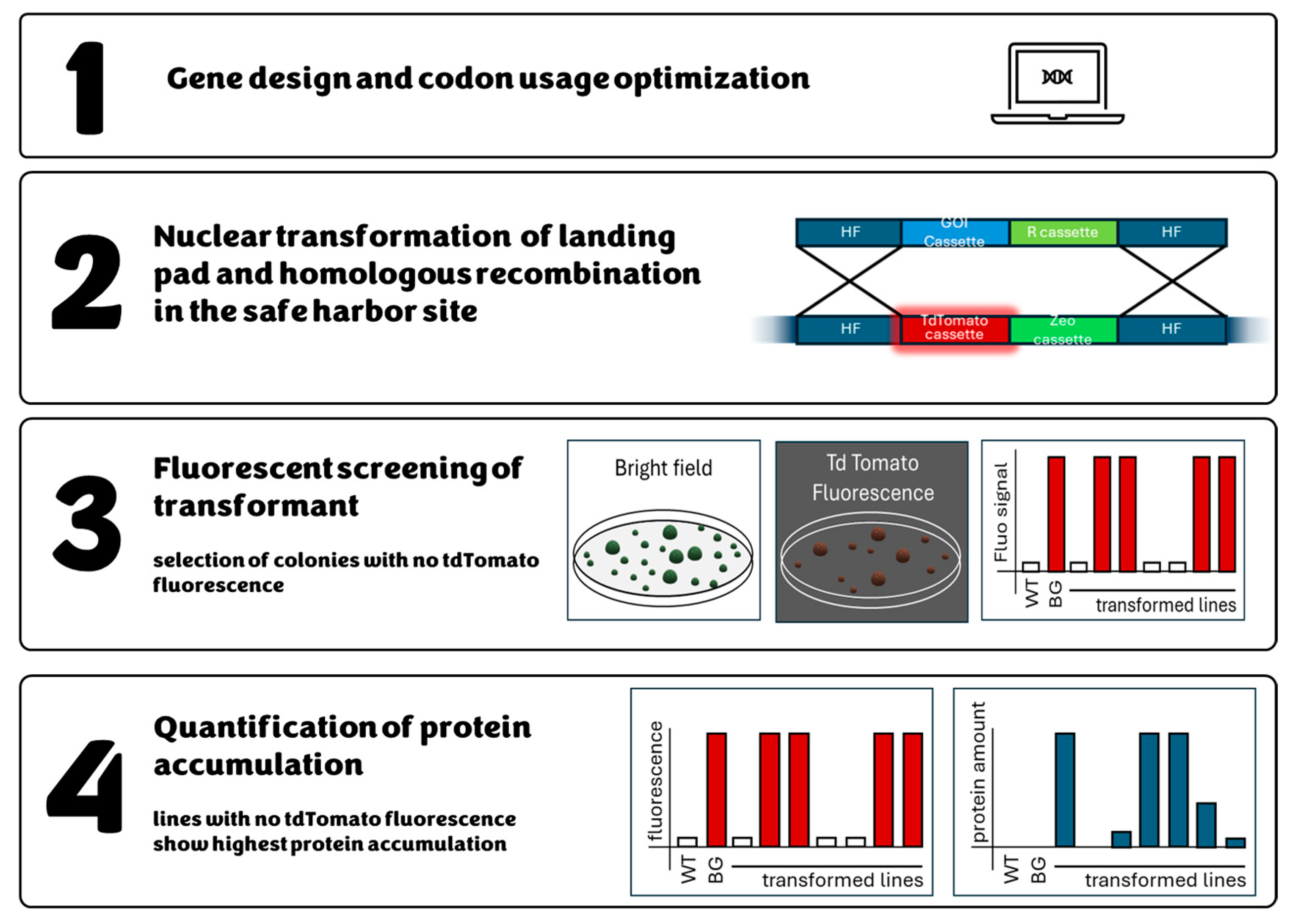
2.7. Genomic Safe Harbor as a Method of Maximizing Transgene Expression
2.8. Chloroplast Genome Manipulation
3. Biotechnological Exploitation: A Few Examples
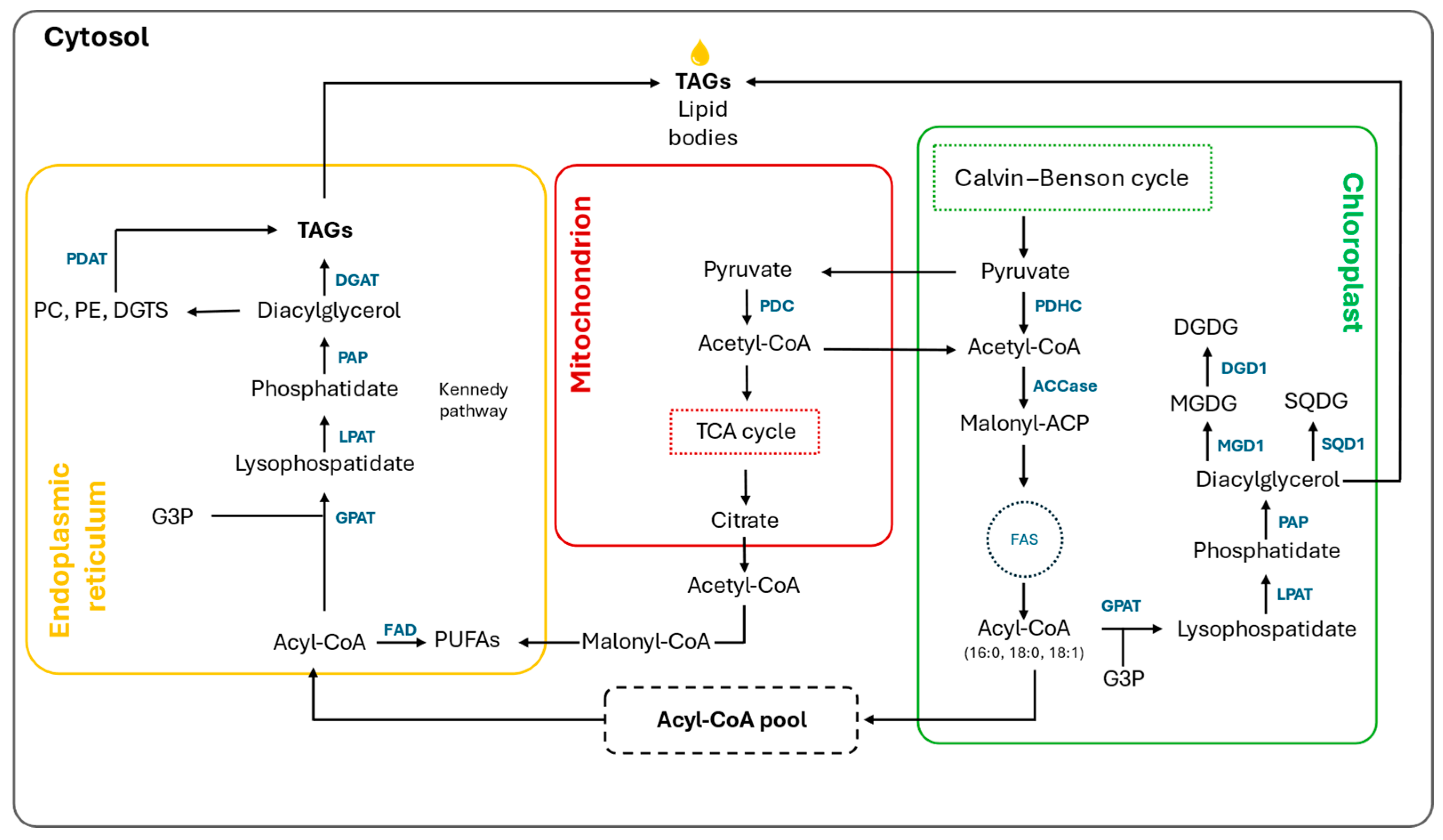
3.1. Lipids
3.2. Carotenoids and Other Pigments
3.3. Other High-Value Products and Biotechnological Manipulation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rasheed, R.; Thaher, M.; Younes, N.; Bounnit, T.; Schipper, K.; Nasrallah, G.K.; Al Jabri, H.; Gifuni, I.; Goncalves, O.; Pruvost, J. Solar cultivation of microalgae in a desert environment for the development of techno-functional feed ingredients for aquaculture in Qatar. Sci. Total Environ. 2022, 835, 155538. [Google Scholar] [CrossRef]
- Winckelmann, D.; Bleeke, F.; Thomas, B.; Elle, C.; Klöck, G. Open pond cultures of indigenous algae grown on non-arable land in an arid desert using wastewater. Int. Aquat. Res. 2015, 7, 221–233. [Google Scholar] [CrossRef]
- Revellame, E.D.; Aguda, R.; Chistoserdov, A.; Fortela, D.L.; Hernandez, R.A.; Zappi, M.E. Microalgae cultivation for space exploration: Assessing the potential for a new generation of waste to human life-support system for long duration space travel and planetary human habitation. Algal Res. 2021, 55, 102258. [Google Scholar] [CrossRef]
- Williamson, E.; Ross, I.L.; Wall, B.T.; Hankamer, B. Microalgae: Potential novel protein for sustainable human nutrition. Trends Plant Sci. 2024, 29, 370–382. [Google Scholar] [CrossRef]
- Barkia, I.; Saari, N.; Manning, S.R. Microalgae for High-Value Products Towards Human Health and Nutrition. Mar. Drugs 2019, 17, 304. [Google Scholar] [CrossRef]
- Kandilian, R.; Lee, E.; Pilon, L. Radiation and optical properties of Nannochloropsis oculata grown under different irradiances and spectra. Bioresour. Technol. 2013, 137, 63–73. [Google Scholar] [CrossRef]
- Janouskovec, J.; Horák, A.; Oborník, M.; Lukes, J.; Keeling, P.J. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl. Acad. Sci. USA 2010, 107, 10949–10954. [Google Scholar] [CrossRef]
- Guo, L.; Liang, S.; Zhang, Z.; Liu, H.; Wang, S.; Pan, K.; Xu, J.; Ren, X.; Pei, S.; Yang, G. Genome assembly of Nannochloropsis oceanica provides evidence of host nucleus overthrow by the symbiont nucleus during speciation. Commun. Biol. 2019, 2, 249. [Google Scholar] [CrossRef]
- Basso, S.; Simionato, D.; Gerotto, C.; Segalla, A.; Giacometti, G.M.; Morosinotto, T. Characterization of the photosynthetic apparatus of the Eustigmatophycean Nannochloropsis gaditana: Evidence of convergent evolution in the supramolecular organization of photosystem I. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 306–314. [Google Scholar] [CrossRef]
- Lubián, L.M. Nannochloropsis gaditana spec. nov., una nueva Eustigmatophyceae marina. Lezama 1982, 4, 287–293. [Google Scholar]
- Karlson, B.; Potter, D.; Kuylenstierna, M.; Andersen, R.A. Ultrastructure, pigment composition, and 18S rRNA gene sequence for Nannochloropsis granulata sp. nov. (Monodopsidaceae, Eustigmatophyceae), a marine ultraplankter isolated from the Skagerrak, northeast Atlantic Ocean. Phycologia 1996, 35, 253–260. [Google Scholar] [CrossRef]
- Suda, S.; Atsumi, M.; Miyashita, H. Taxonomic characterization of a marine Nannochloropsis species, N. oceanica sp. nov. (Eustigmatophyceae). Phycologia 2002, 41, 273–279. [Google Scholar] [CrossRef]
- Krienitz, L.; Hepperle, D.; Stich, H.-B.; Weiler, W. Nannochloropsis limnetica (Eustigmatophyceae), a new species of picoplankton from freshwater. Phycologia 2000, 39, 219–227. [Google Scholar] [CrossRef]
- Hibberd, D.J. Notes on the taxonomy and nomenclature of the algal classes Eustigmatophyceae and Tribophyceae (synonym Xanthophyceae). Bot. J. Linn. Soc. 2008, 82, 93–119. [Google Scholar] [CrossRef]
- Fawley, M.W.; Jameson, I.; Fawley, K.P. The phylogeny of the genus Nannochloropsis (Monodopsidaceae, Eustigmatophyceae), with descriptions of N. australis sp. nov. and Microchloropsis gen. nov. Phycologia 2015, 54, 545–552. [Google Scholar] [CrossRef]
- Starkenburg, S.R.; Kwon, K.J.; Jha, R.K.; McKay, C.; Jacobs, M.; Chertkov, O.; Twary, S.; Rocap, G.; Cattolico, R.A. A pangenomic analysis of the Nannochloropsisorganellar genomes reveals novel genetic variations in key metabolic genes. BMC Genom. 2014, 15, 212. [Google Scholar] [CrossRef]
- Jinkerson, R.E.; Radakovits, R.; Posewitz, M.C. Genomic insights from the oleaginous model alga Nannochloropsis gaditana. Bioengineered 2013, 4, 37–43. [Google Scholar] [CrossRef]
- Sukenik, A.; Carmeli, Y.; Berner, T. Regulation of Fatty Acid Composition by Irradiance Level in the Eustigmatophyte Nannochloropsis sp. J. Phycol. 1989, 25, 686–692. [Google Scholar] [CrossRef]
- Boussiba, S.; Vonshak, A.; Cohen, Z.; Avissar, Y.J.; Richmond, A.E. Lipid and biomass production by the halotolerant microalga Nannochloropsis salina. Biomass 1987, 12, 37–47. [Google Scholar] [CrossRef]
- Chini Zittelli, G.; Lavista, F.; Bastianini, A.; Rodolfi, L.; Vincenzini, M.; Tredici, M.R. Production of eicosapentaenoic acid by Nannochloropsis sp. cultures in outdoor tubular photobioreactors. J. Biotechnol. 1999, 70, 299–312. [Google Scholar] [CrossRef]
- Tonon, T.; Harvey, D.; Larson, T.R.; Graham, I.A. Long chain polyunsaturated fatty acid production and partitioning to triacylglycerols in four microalgae. Phytochemistry 2002, 61, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhang, J.; Cui, J.; Feng, Y.; Cui, Q. Metabolic profiles of Nannochloropsis oceanica IMET1 under nitrogen-deficiency stress. Bioresour. Technol. 2013, 130, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Han, D.; Gerken, H.G.; Li, Y.; Sommerfeld, M.; Hu, Q.; Xu, J. Molecular mechanisms for photosynthetic carbon partitioning into storage neutral lipids in Nannochloropsis oceanica under nitrogen-depletion conditions. Algal Res. 2015, 7, 66–77. [Google Scholar] [CrossRef]
- Li, Z.; Wakao, S.; Fischer, B.B.; Niyogi, K.K. Sensing and responding to excess light. Annu. Rev. Plant Biol. 2009, 60, 239–260. [Google Scholar] [CrossRef] [PubMed]
- Solovchenko, A.; Lukyanov, A.; Solovchenko, O.; Didi-Cohen, S.; Boussiba, S.; Khozin-Goldberg, I. Interactive effects of salinity, high light, and nitrogen starvation on fatty acid and carotenoid profiles in Nannochloropsis oceanica CCALA 804. Eur. J. Lipid Sci. Technol. 2014, 116, 635–644. [Google Scholar] [CrossRef]
- Cecchin, M.; Berteotti, S.; Paltrinieri, S.; Vigliante, I.; Iadarola, B.; Giovannone, B.; Maffei, M.E.; Delledonne, M.; Ballottari, M. Improved lipid productivity in Nannochloropsis gaditana in nitrogen-replete conditions by selection of pale green mutants. Biotechnol. Biofuels 2020, 13, 78. [Google Scholar] [CrossRef]
- Fakhry, E.M.; El Maghraby, D.M. Lipid accumulation in response to nitrogen limitation and variation of temperature in Nannochloropsis salina. Bot. Stud. 2015, 56, 6. [Google Scholar] [CrossRef]
- Barber, J.; Andersson, B. Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem. Sci. 1992, 17, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.W.; Sung, M.-G.; Nam, K.; Moon, M.; Kwon, J.-H.; Yang, J.-W. Effect of monochromatic illumination on lipid accumulation of Nannochloropsis gaditana under continuous cultivation. Bioresour. Technol. 2014, 159, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Van Wagenen, J.; Miller, T.W.; Hobbs, S.; Hook, P.; Crowe, B.; Huesemann, M. Effects of Light and Temperature on Fatty Acid Production in Nannochloropsis salina. Energies 2012, 5, 731–740. [Google Scholar] [CrossRef]
- Li, J.; Han, D.; Wang, D.; Ning, K.; Jia, J.; Wei, L.; Jing, X.; Huang, S.; Chen, J.; Li, Y.; et al. Choreography of Transcriptomes and Lipidomes of Nannochloropsis Reveals the Mechanisms of Oil Synthesis in Microalgae. Plant Cell 2014, 26, 1645–1665. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Cao, X.; Yao, C.; Xue, S.; Yang, Q. Identification of the role of polar glycerolipids in lipid metabolism and their acyl attribution for TAG accumulation in Nannochloropsis oceanica. Algal Res. 2017, 24, 122–129. [Google Scholar] [CrossRef]
- Han, D.; Jia, J.; Li, J.; Sommerfeld, M.; Xu, J.; Hu, Q. Metabolic Remodeling of Membrane Glycerolipids in the Microalga Nannochloropsis oceanica under Nitrogen Deprivation. Front. Mar. Sci. 2017, 4, 242. [Google Scholar] [CrossRef]
- Simionato, D.; Block, M.A.; La Rocca, N.; Jouhet, J.; Maréchal, E.; Finazzi, G.; Morosinotto, T. The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryot Cell 2013, 12, 665–676. [Google Scholar] [CrossRef]
- Liang, J.; Wen, F.; Liu, J. Transcriptomic and lipidomic analysis of an EPA-containing Nannochloropsis sp. PJ12 in response to nitrogen deprivation. Sci. Rep. 2019, 9, 4540. [Google Scholar] [CrossRef]
- Lubian, L.M.; Montero, O.; Moreno-Garrido, I.; Huertas, I.E.; Sobrino, C.; Gonzalez-del Valle, M.; Pares, G. Nannochloropsis (Eustigmatophyceae) as source of commercially valuable pigments. J. Appl. Phycol. 2000, 12, 249–255. [Google Scholar] [CrossRef]
- Sarker, P.K.; Kapuscinski, A.R.; McKuin, B.; Fitzgerald, D.S.; Nash, H.M.; Greenwood, C. Microalgae-blend tilapia feed eliminates fishmeal and fish oil, improves growth, and is cost viable. Sci. Rep. 2020, 10, 19328. [Google Scholar] [CrossRef]
- Kumar, P.; Jain, K.; Munilkumar, S.; Chalal, R.S. Beta glucan: Avaluable nutraceutical for promoting health in aquaculture (short review). Res. Rev. BioSci. RRBS 2014, 5, 220–227. [Google Scholar]
- Vieler, A.; Wu, G.; Tsai, C.H.; Bullard, B.; Cornish, A.J.; Harvey, C.; Reca, I.B.; Thornburg, C.; Achawanantakun, R.; Buehl, C.J.; et al. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 2012, 8, e1003064. [Google Scholar] [CrossRef]
- Liang, C.; Cao, S.; Zhang, X.; Zhu, B.; Su, Z.; Xu, D.; Guang, X.; Ye, N. De Novo Sequencing and Global Transcriptome Analysis of Nannochloropsis sp. (Eustigmatophyceae) Following Nitrogen Starvation. BioEnergy Res. 2013, 6, 494–505. [Google Scholar] [CrossRef]
- Ohan, J.A.; Hovde, B.T.; Zhang, X.L.; Davenport, K.W.; Chertkov, O.; Han, C.; Twary, S.N.; Starkenburg, S.R. Nuclear Genome Assembly of the Microalga Nannochloropsis salina CCMP1776. Microbiol. Resour. Announc. 2019, 8, e00750-19. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ning, K.; Li, J.; Hu, J.; Han, D.; Wang, H.; Zeng, X.; Jing, X.; Zhou, Q.; Su, X.; et al. Nannochloropsis Genomes Reveal Evolution of Microalgal Oleaginous Traits. PLOS Genet. 2014, 10, e1004094. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Jiang, Z.; Liu, B. A CRISPR/dCas9-based transcription activated system developed in marine microalga Nannochloropsis oceanica. Aquaculture 2022, 546, 737064. [Google Scholar] [CrossRef]
- Naduthodi, M.I.S.; Mohanraju, P.; Südfeld, C.; D’Adamo, S.; Barbosa, M.J.; van der Oost, J. CRISPR–Cas ribonucleoprotein mediated homology-directed repair for efficient targeted genome editing in microalgae Nannochloropsis oceanica IMET1. Biotechnol. Biofuels 2019, 12, 66. [Google Scholar] [CrossRef]
- Naduthodi, M.I.S.; Südfeld, C.; Avitzigiannis, E.K.; Trevisan, N.; van Lith, E.; Alcaide Sancho, J.; D’Adamo, S.; Barbosa, M.; van der Oost, J. Comprehensive Genome Engineering Toolbox for Microalgae Nannochloropsis oceanica Based on CRISPR-Cas Systems. ACS Synth. Biol. 2021, 10, 3369–3378. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lu, Y.; Xin, Y.; Wei, L.; Huang, S.; Xu, J. Genome editing of model oleaginous microalgae Nannochloropsis spp. by CRISPR/Cas9. Plant J. 2016, 88, 1071–1081. [Google Scholar] [CrossRef]
- Wang, Q.; Gong, Y.; He, Y.; Xin, Y.; Lv, N.; Du, X.; Li, Y.; Jeong, B.-R.; Xu, J. Genome engineering of Nannochloropsis with hundred-kilobase fragment deletions by Cas9 cleavages. Plant J. 2021, 106, 1148–1162. [Google Scholar] [CrossRef] [PubMed]
- Poliner, E.; Takeuchi, T.; Du, Z.-Y.; Benning, C.; Farré, E.M. Nontransgenic Marker-Free Gene Disruption by an Episomal CRISPR System in the Oleaginous Microalga, Nannochloropsis oceanica CCMP1779. ACS Synth. Biol. 2018, 7, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Südfeld, C.; Pozo-Rodríguez, A.; Manjavacas Díez, S.A.; Wijffels, R.H.; Barbosa, M.J.; D’Adamo, S. The nucleolus as a genomic safe harbor for strong gene expression in Nannochloropsis oceanica. Mol. Plant 2022, 15, 340–353. [Google Scholar] [CrossRef]
- Schwartz Ariel, S.; Brown, R.; Ajjawi, I.; McCarren, J.; Atilla, S.; Bauman, N.; Richardson Toby, H. Complete Genome Sequence of the Model Oleaginous Alga Nannochloropsis gaditana CCMP1894. Genome Announc. 2018, 6, e01448-17. [Google Scholar] [CrossRef]
- Corteggiani Carpinelli, E.; Telatin, A.; Vitulo, N.; Forcato, C.; D’Angelo, M.; Schiavon, R.; Vezzi, A.; Giacometti, G.M.; Morosinotto, T.; Valle, G. Chromosome Scale Genome Assembly and Transcriptome Profiling of Nannochloropsis gaditana in Nitrogen Depletion. Mol. Plant 2014, 7, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; El Hajjami, M.; Shen, C.; You, W.; Lu, Y.; Li, J.; Jing, X.; Hu, Q.; Zhou, W.; Poetsch, A.; et al. Transcriptomic and proteomic responses to very low CO2 suggest multiple carbon concentrating mechanisms in Nannochloropsis oceanica. Biotechnol. Biofuels 2019, 12, 168. [Google Scholar] [CrossRef] [PubMed]
- Hulatt, C.J.; Smolina, I.; Dowle, A.; Kopp, M.; Vasanth, G.K.; Hoarau, G.G.; Wijffels, R.H.; Kiron, V. Proteomic and Transcriptomic Patterns during Lipid Remodeling in Nannochloropsis gaditana. Int. J. Mol. Sci. 2020, 21, 6946. [Google Scholar] [CrossRef]
- Wei, L.; You, W.; Gong, Y.; El Hajjami, M.; Liang, W.; Xu, J.; Poetsch, A. Transcriptomic and proteomic choreography in response to light quality variation reveals key adaption mechanisms in marine Nannochloropsis oceanica. Sci. Total Environ. 2020, 720, 137667. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Wei, L.; Gong, Y.; Hajjami, M.E.; Xu, J.; Poetsch, A. Integration of proteome and transcriptome refines key molecular processes underlying oil production in Nannochloropsis oceanica. Biotechnol. Biofuels 2020, 13, 109. [Google Scholar] [CrossRef]
- Poliner, E.; Farré, E.M.; Benning, C. Advanced genetic tools enable synthetic biology in the oleaginous microalgae Nannochloropsis sp. Plant Cell Rep. 2018, 37, 1383–1399. [Google Scholar] [CrossRef]
- Chen, H.L.; Li, S.S.; Huang, R.; Tsai, H.J. Conditional production of a functional fish growth hormone in the transgenic line of Nannochloropsis oculata (Eustigmatophyceae). J. Phycol. 2008, 44, 768–776. [Google Scholar] [CrossRef]
- Kilian, O.; Benemann, C.S.; Niyogi, K.K.; Vick, B. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl. Acad. Sci. USA 2011, 108, 21265–21269. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Gao, D.; Hu, H. High-efficiency nuclear transformation of the oleaginous marine Nannochloropsis species using PCR product. Biosci. Biotechnol. Biochem. 2014, 78, 812–817. [Google Scholar] [CrossRef]
- Cha, T.-S.; Chen, C.-F.; Yee, W.; Aziz, A.; Loh, S.-H. Cinnamic acid, coumarin and vanillin: Alternative phenolic compounds for efficient Agrobacterium-mediated transformation of the unicellular green alga, Nannochloropsis sp. J. Microbiol. Methods 2011, 84, 430–434. [Google Scholar] [CrossRef]
- Kang, N.K.; Choi, G.-G.; Kim, E.K.; Shin, S.-E.; Jeon, S.; Park, M.S.; Jeong, K.J.; Jeong, B.-r.; Chang, Y.K.; Yang, J.-W.; et al. Heterologous overexpression of sfCherry fluorescent protein in Nannochloropsis salina. Biotechnol. Rep. 2015, 8, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.K.; Jeon, S.; Kwon, S.; Koh, H.G.; Shin, S.-E.; Lee, B.; Choi, G.-G.; Yang, J.-W.; Jeong, B.-r.; Chang, Y.K. Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina. Biotechnol. Biofuels 2015, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Wang, Q.T.; Xin, Y.; Lu, Y.D.; Xu, J. Enhancing photosynthetic biomass productivity of industrial oleaginous microalgae by overexpression of RuBisCO activase. Algal Res.-Biomass Biofuels Bioprod. 2017, 27, 366–375. [Google Scholar] [CrossRef]
- Kang, N.K.; Kim, E.K.; Kim, Y.U.; Lee, B.; Jeong, W.-J.; Jeong, B.-R.; Chang, Y.K. Increased lipid production by heterologous expression of AtWRI1 transcription factor in Nannochloropsis salina. Biotechnol. Biofuels 2017, 10, 231. [Google Scholar] [CrossRef] [PubMed]
- Zienkiewicz, K.; Zienkiewicz, A.; Poliner, E.; Du, Z.-Y.; Vollheyde, K.; Herrfurth, C.; Marmon, S.; Farré, E.M.; Feussner, I.; Benning, C. Nannochloropsis, a rich source of diacylglycerol acyltransferases for engineering of triacylglycerol content in different hosts. Biotechnol. Biofuels 2017, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Moog, D.; Stork, S.; Reislöhner, S.; Grosche, C.; Maier, U.G. In vivo localization studies in the stramenopile alga Nannochloropsis oceanica. Protist 2015, 166, 161–171. [Google Scholar] [CrossRef]
- Poliner, E.; Panchy, N.; Newton, L.; Wu, G.; Lapinsky, A.; Bullard, B.; Zienkiewicz, A.; Benning, C.; Shiu, S.-H.; Farré, E.M. Transcriptional coordination of physiological responses in Nannochloropsis oceanica CCMP1779 under light/dark cycles. Plant J. 2015, 83, 1097–1113. [Google Scholar] [CrossRef] [PubMed]
- Poliner, E.; Clark, E.; Cummings, C.; Benning, C.; Farre, E.M. A high-capacity gene stacking toolkit for the oleaginous microalga, Nannochloropsis oceanica CCMP1779. Algal Res. 2020, 45, 101664. [Google Scholar] [CrossRef]
- Iwai, M.; Hori, K.; Sasaki-Sekimoto, Y.; Shimojima, M.; Ohta, H. Manipulation of oil synthesis in Nannochloropsis strain NIES-2145 with a phosphorus starvation–inducible promoter from Chlamydomonas reinhardtii. Front. Microbiol. 2015, 6, 912. [Google Scholar] [CrossRef]
- Jackson, H.O.; Berepiki, A.; Baylay, A.J.; Terry, M.J.; Moore, C.M.; Bibby, T.S. An inducible expression system in the alga Nannochloropsis gaditana controlled by the nitrate reductase promoter. J. Appl. Phycol. 2019, 31, 269–279. [Google Scholar] [CrossRef]
- de Grahl, I.; Rout, S.S.; Maple-Grødem, J.; Reumann, S. Development of a constitutive and an auto-inducible high-yield expression system for recombinant protein production in the microalga Nannochloropsis oceanica. Appl. Microbiol. Biotechnol. 2020, 104, 8747–8760. [Google Scholar] [CrossRef]
- Kaye, Y.; Grundman, O.; Leu, S.; Zarka, A.; Zorin, B.; Didi-Cohen, S.; Khozin-Goldberg, I.; Boussiba, S. Metabolic engineering toward enhanced LC-PUFA biosynthesis in Nannochloropsis oceanica: Overexpression of endogenous Δ12 desaturase driven by stress-inducible promoter leads to enhanced deposition of polyunsaturated fatty acids in TAG. Algal Res. 2015, 11, 387–398. [Google Scholar] [CrossRef]
- Radakovits, R.; Jinkerson, R.E.; Fuerstenberg, S.I.; Tae, H.; Settlage, R.E.; Boore, J.L.; Posewitz, M.C. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nat. Commun. 2012, 3, 686. [Google Scholar] [CrossRef]
- Kwon, S.; Kang, N.K.; Koh, H.G.; Shin, S.E.; Lee, B.; Jeong, B.R.; Chang, Y.K. Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina. Biotechnol. Bioeng. 2018, 115, 331–340. [Google Scholar] [CrossRef]
- Ma, X.; Yao, L.; Yang, B.; Lee, Y.K.; Chen, F.; Liu, J. RNAi-mediated silencing of a pyruvate dehydrogenase kinase enhances triacylglycerol biosynthesis in the oleaginous marine alga Nannochloropsis salina. Sci. Rep. 2017, 7, 11485. [Google Scholar] [CrossRef]
- Südfeld, C.; Hubáček, M.; Figueiredo, D.; Naduthodi, M.I.S.; van der Oost, J.; Wijffels, R.H.; Barbosa, M.J.; D’Adamo, S. High-throughput insertional mutagenesis reveals novel targets for enhancing lipid accumulation in Nannochloropsis oceanica. Metab. Eng. 2021, 66, 239–258. [Google Scholar] [CrossRef]
- Ramarajan, M.; Fabris, M.; Abbriano, R.M.; Pernice, M.; Ralph, P.J. Novel endogenous promoters for genetic engineering of the marine microalga Nannochloropsis gaditana CCMP526. Algal Res. 2019, 44, 101708. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, L.; Liu, H.; Pan, X.; Liu, L.; Zong, Y.; Yang, G. Zeocin treatment significantly elevated transformation efficiency of Nannochloropsis oceanica. J. Appl. Phycol. 2022, 34, 1587–1594. [Google Scholar] [CrossRef]
- Tsien, R.Y. THE GREEN FLUORESCENT PROTEIN. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Poliner, E.; Pulman, J.A.; Zienkiewicz, K.; Childs, K.; Benning, C.; Farré, E.M. A toolkit for Nannochloropsis oceanica CCMP1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production. Plant Biotechnol. J. 2018, 16, 298–309. [Google Scholar] [CrossRef]
- Shih, C.H.; Chen, H.Y.; Lee, H.C.; Tsai, H.J. Purple chromoprotein gene serves as a new selection marker for transgenesis of the microalga Nannochloropsis oculata. PLoS ONE 2015, 10, e0120780. [Google Scholar] [CrossRef]
- Wei, H.; Shi, Y.; Ma, X.; Pan, Y.; Hu, H.; Li, Y.; Luo, M.; Gerken, H.; Liu, J. A type-I diacylglycerol acyltransferase modulates triacylglycerol biosynthesis and fatty acid composition in the oleaginous microalga, Nannochloropsis oceanica. Biotechnol. Biofuels 2017, 10, 174. [Google Scholar] [CrossRef]
- Poliner, E.; Busch, A.W.U.; Newton, L.; Kim, Y.U.; Clark, R.; Gonzalez-Martinez, S.C.; Jeong, B.-R.; Montgomery, B.L.; Farré, E.M. Aureochromes maintain polyunsaturated fatty acid content in Nannochloropsis oceanica. Plant Physiol. 2022, 189, 906–921. [Google Scholar] [CrossRef]
- Jiang, W.; Brueggeman, A.J.; Horken, K.M.; Plucinak, T.M.; Weeks, D.P. Successful Transient Expression of Cas9 and Single Guide RNA Genes in Chlamydomonas reinhardtii. Eukaryot. Cell 2014, 13, 1465–1469. [Google Scholar] [CrossRef]
- Baek, K.; Kim, D.H.; Jeong, J.; Sim, S.J.; Melis, A.; Kim, J.S.; Jin, E.; Bae, S. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 2016, 6, 30620. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.L.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.-Y.; Liu, D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Subburaj, S.; Chung, S.J.; Lee, C.; Ryu, S.-M.; Kim, D.H.; Kim, J.-S.; Bae, S.; Lee, G.-J. Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016, 35, 1535–1544. [Google Scholar] [CrossRef]
- Murakami, H.; Kakutani, N.; Kuroyanagi, Y.; Iwai, M.; Hori, K.; Shimojima, M.; Ohta, H. MYB-like transcription factor NoPSR1 is crucial for membrane lipid remodeling under phosphate starvation in the oleaginous microalga Nannochloropsis oceanica. FEBS Lett. 2020, 594, 3384–3394. [Google Scholar] [CrossRef]
- Murakami, H.; Nobusawa, T.; Hori, K.; Shimojima, M.; Ohta, H. Betaine Lipid Is Crucial for Adapting to Low Temperature and Phosphate Deficiency in Nannochloropsis. Plant Physiol. 2018, 177, 181–193. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Plucinak, T.M.; Horken, K.M.; Jiang, W.; Fostvedt, J.; Nguyen, S.T.; Weeks, D.P. Improved and versatile viral 2A platforms for dependable and inducible high-level expression of dicistronic nuclear genes in Chlamydomonas reinhardtii. Plant J. 2015, 82, 717–729. [Google Scholar] [CrossRef]
- Koh, H.G.; Kang, N.K.; Kim, E.K.; Jeon, S.; Shin, S.-E.; Lee, B.; Chang, Y.K. Advanced multigene expression system for Nannochloropsis salina using 2A self-cleaving peptides. J. Biotechnol. 2018, 278, 39–47. [Google Scholar] [CrossRef]
- Vafaee, Y.; Staniek, A.; Mancheno-Solano, M.; Warzecha, H. A Modular Cloning Toolbox for the Generation of Chloroplast Transformation Vectors. PLoS ONE 2014, 9, e110222. [Google Scholar] [CrossRef]
- Daniell, H.; Jin, S.; Zhu, X.-G.; Gitzendanner, M.A.; Soltis, D.E.; Soltis, P.S. Green giant—A tiny chloroplast genome with mighty power to produce high-value proteins: History and phylogeny. Plant Biotechnol. J. 2021, 19, 430–447. [Google Scholar] [CrossRef]
- Scranton, M.A.; Ostrand, J.T.; Fields, F.J.; Mayfield, S.P. Chlamydomonas as a model for biofuels and bio-products production. Plant J. 2015, 82, 523–531. [Google Scholar] [CrossRef]
- Larrea-Alvarez, M.; Purton, S. Multigenic engineering of the chloroplast genome in the green alga Chlamydomonas reinhardtii. Microbiology 2020, 166, 510–515. [Google Scholar] [CrossRef]
- Munjal, N.; Garzon-Sanabria, A.J.; Quinones, K.W.; Gregory, J.; Nikolov, Z.L. Light-Induced Production of an Antibody Fragment and Malaria Vaccine Antigen from Chlamydomonas reinhardtii. Processes 2014, 2, 625–638. [Google Scholar] [CrossRef]
- Gan, Q.; Jiang, J.; Han, X.; Wang, S.; Lu, Y. Engineering the Chloroplast Genome of Oleaginous Marine Microalga Nannochloropsis oceanica. Front. Plant Sci. 2018, 9, 439. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, K.; Xu, W.; Wang, Y.; Gao, Z.; Cui, H.; Meng, C.; Qin, S. Plastid Engineering of a Marine Alga, Nannochloropsis gaditana, for Co-Expression of Two Recombinant Peptides. J. Phycol. 2021, 57, 569–576. [Google Scholar] [CrossRef]
- Perozeni, F.; Baier, T. Current Nuclear Engineering Strategies in the Green Microalga Chlamydomonas reinhardtii. Life 2023, 13, 1566. [Google Scholar] [CrossRef]
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of Triacylglycerol Accumulation in Plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef]
- Li, D.-W.; Balamurugan, S.; Yang, Y.-F.; Zheng, J.-W.; Huang, D.; Zou, L.-G.; Yang, W.-D.; Liu, J.-S.; Guan, Y.; Li, H.-Y. Transcriptional regulation of microalgae for concurrent lipid overproduction and secretion. Sci. Adv. 2019, 5, eaau3795. [Google Scholar] [CrossRef]
- Chen, J.W.; Liu, W.J.; Hu, D.X.; Wang, X.; Balamurugan, S.; Alimujiang, A.; Yang, W.D.; Liu, J.S.; Li, H.Y. Identification of a malonyl CoA-acyl carrier protein transacylase and its regulatory role in fatty acid biosynthesis in oleaginous microalga Nannochloropsis oceanica. Biotechnol. Appl. Biochem. 2017, 64, 620–626. [Google Scholar] [CrossRef]
- Han, X.; Song, X.; Li, F.; Lu, Y. Improving lipid productivity by engineering a control-knob gene in the oleaginous microalga Nannochloropsis oceanica. Metab. Eng. Commun. 2020, 11, e00142. [Google Scholar] [CrossRef]
- Jeon, S.; Koh, H.G.; Cho, J.M.; Kang, N.K.; Chang, Y.K. Enhancement of lipid production in Nannochloropsis salina by overexpression of endogenous NADP-dependent malic enzyme. Algal Res. 2021, 54, 102218. [Google Scholar] [CrossRef]
- Liu, J.; Liu, M.; Pan, Y.; Shi, Y.; Hu, H. Metabolic engineering of the oleaginous alga Nannochloropsis for enriching eicosapentaenoic acid in triacylglycerol by combined pulling and pushing strategies. Metab. Eng. 2022, 69, 163–174. [Google Scholar] [CrossRef]
- Liu, M.; Ding, W.; Yu, L.; Shi, Y.; Liu, J. Functional characterization of carotenogenic genes provides implications into carotenoid biosynthesis and engineering in the marine alga Nannochloropsis oceanica. Algal Res. 2022, 67, 102853. [Google Scholar] [CrossRef]
- Perin, G.; Bellan, A.; Michelberger, T.; Lyska, D.; Wakao, S.; Niyogi, K.K.; Morosinotto, T. Modulation of xanthophyll cycle impacts biomass productivity in the marine microalga Nannochloropsis. Proc. Natl. Acad. Sci. USA 2023, 120, e2214119120. [Google Scholar] [CrossRef]
- Dautermann, O.; Lyska, D.; Andersen-Ranberg, J.; Becker, M.; Fröhlich-Nowoisky, J.; Gartmann, H.; Krämer, L.C.; Mayr, K.; Pieper, D.; Rij, L.M.; et al. An algal enzyme required for biosynthesis of the most abundant marine carotenoids. Sci. Adv. 2020, 6, eaaw9183. [Google Scholar] [CrossRef]
- Liu, M.; Ding, W.; Pan, Y.; Hu, H.; Liu, J. Zeaxanthin epoxidase is involved in the carotenoid biosynthesis and light-dependent growth of the marine alga Nannochloropsis oceanica. Biotechnol. Biofuels Bioprod. 2023, 16, 74. [Google Scholar] [CrossRef]
- Park, S.-B.; Yun, J.-H.; Ryu, A.J.; Yun, J.; Kim, J.W.; Lee, S.; Choi, S.; Cho, D.-H.; Choi, D.-Y.; Lee, Y.J.; et al. Development of a novel nannochloropsis strain with enhanced violaxanthin yield for large-scale production. Microb. Cell Factories 2021, 20, 43. [Google Scholar] [CrossRef]
- Cecchin, M.; Cazzaniga, S.; Martini, F.; Paltrinieri, S.; Bossi, S.; Maffei, M.E.; Ballottari, M. Astaxanthin and eicosapentaenoic acid production by S4, a new mutant strain of Nannochloropsis gaditana. Microb. Cell Factories 2022, 21, 117. [Google Scholar] [CrossRef]
- Koh, H.G.; Kang, N.K.; Jeon, S.; Shin, S.-E.; Jeong, B.-r.; Chang, Y.K. Heterologous synthesis of chlorophyll b in Nannochloropsis salina enhances growth and lipid production by increasing photosynthetic efficiency. Biotechnol. Biofuels 2019, 12, 122. [Google Scholar] [CrossRef]
- Li, S.S.; Tsai, H.J. Transgenic microalgae as a non-antibiotic bactericide producer to defend against bacterial pathogen infection in the fish digestive tract. Fish Shellfish. Immunol. 2009, 26, 316–325. [Google Scholar] [CrossRef]
- Rout, S.S.; de Grahl, I.; Yu, X.; Reumann, S. Production of a viral surface protein in Nannochloropsis oceanica for fish vaccination against infectious pancreatic necrosis virus. Appl. Microbiol. Biotechnol. 2022, 106, 6535–6549. [Google Scholar] [CrossRef]
- Lauersen, K.J. Eukaryotic microalgae as hosts for light-driven heterologous isoprenoid production. Planta 2019, 249, 155–180. [Google Scholar] [CrossRef]
- Wichmann, J.; Eggert, A.; Elbourne, L.D.H.; Paulsen, I.T.; Lauersen, K.J.; Kruse, O. Farnesyl pyrophosphate compartmentalization in the green microalga Chlamydomonas reinhardtii during heterologous (E)-α-bisabolene production. Microb. Cell Factories 2022, 21, 190. [Google Scholar] [CrossRef]
- Einhaus, A.; Steube, J.; Freudenberg, R.A.; Barczyk, J.; Baier, T.; Kruse, O. Engineering a powerful green cell factory for robust photoautotrophic diterpenoid production. Metab. Eng. 2022, 73, 82–90. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Overmans, S.; Wellman, G.B.; Samaras, V.G.; Oviedo, C.; Gede, M.; Szekely, G.; Lauersen, K.J. A green alternative to fragrant agarwood sesquiterpenoid production. bioRxiv 2023. [Google Scholar] [CrossRef]
- Du, Z.-Y.; Bhat, W.W.; Poliner, E.; Johnson, S.; Bertucci, C.; Farre, E.; Hamberger, B. Engineering Nannochloropsis oceanica for the production of diterpenoid compounds. mLife 2023, 2, 428–437. [Google Scholar] [CrossRef]
- Perin, G.; Bellan, A.; Segalla, A.; Meneghesso, A.; Alboresi, A.; Morosinotto, T. Generation of random mutants to improve light-use efficiency of Nannochloropsis gaditana cultures for biofuel production. Biotechnol. Biofuels 2015, 8, 161. [Google Scholar] [CrossRef]
- Arora, N.; Lo, E.; Philippidis, G.P. Dissecting Enhanced Carbohydrate and Pigment Productivity in Mutants of Nannochloropsis oculata Using Metabolomics and Lipidomics. J. Agric. Food Chem. 2022, 70, 8338–8350. [Google Scholar] [CrossRef]
- Kang, N.K.; Kim, E.K.; Sung, M.G.; Kim, Y.U.; Jeong, B.R.; Chang, Y.K. Increased biomass and lipid production by continuous cultivation of Nannochloropsis salina transformant overexpressing a bHLH transcription factor. Biotechnol. Bioeng. 2019, 116, 555–568. [Google Scholar] [CrossRef]

| Protein | Conferring Resistance to | Reference |
|---|---|---|
| Bleomycin resistance protein (Sh ble) | Zeomycin | [58] |
| Hygromycin-B phosphotransferase (AphVII) | Hygromycin | [39] |
| Blasticidin-S deaminase (BSD) | Blasticidin | [68] |
| Nourseothricin acetyltransferase (NAT) | Nourseothricin | [68] |
| Aminoglycoside 3′ phosphotransferase | G418 | [68] |
| Name | Ex λ | Em λ | Brightness | Excitation/Emission Spectra |
|---|---|---|---|---|
| Clover (GFP) | 505 | 515 | 84.36 | 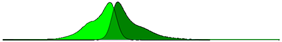 |
| mVenus (YFP) | 515 | 527 | 66.56 | 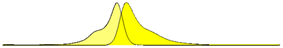 |
| mCerulean (CFP) | 433 | 475 | 34.8 | 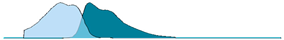 |
| sfCherry | 589 | 610 | 17.47 | 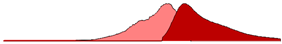 |
| tdTomato | 554 | 581 | 95.22 |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canini, D.; Ceschi, E.; Perozeni, F. Toward the Exploitation of Sustainable Green Factory: Biotechnology Use of Nannochloropsis spp. Biology 2024, 13, 292. https://doi.org/10.3390/biology13050292
Canini D, Ceschi E, Perozeni F. Toward the Exploitation of Sustainable Green Factory: Biotechnology Use of Nannochloropsis spp. Biology. 2024; 13(5):292. https://doi.org/10.3390/biology13050292
Chicago/Turabian StyleCanini, Davide, Edoardo Ceschi, and Federico Perozeni. 2024. "Toward the Exploitation of Sustainable Green Factory: Biotechnology Use of Nannochloropsis spp." Biology 13, no. 5: 292. https://doi.org/10.3390/biology13050292
APA StyleCanini, D., Ceschi, E., & Perozeni, F. (2024). Toward the Exploitation of Sustainable Green Factory: Biotechnology Use of Nannochloropsis spp. Biology, 13(5), 292. https://doi.org/10.3390/biology13050292







