Cellular Response of Adapted and Non-Adapted Tetrahymena thermophila Strains to Europium Eu(III) Compounds
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganism, Culture Conditions and Eu-Adapted Strains
2.2. Growth Kinetics and LC50 Calculation by Flow Cytometry
2.3. Oxidative Stress Detection
2.4. Transmission Electron Microscopy (TEM) and Microanalysis (TEM-XEDS)
2.5. Total RNA Isolation and cDNA Synthesis
2.6. Quantitative RT-PCR (qRT-PCR)
2.7. Statistical Analysis
3. Results
3.1. Ecotoxicological Parameters
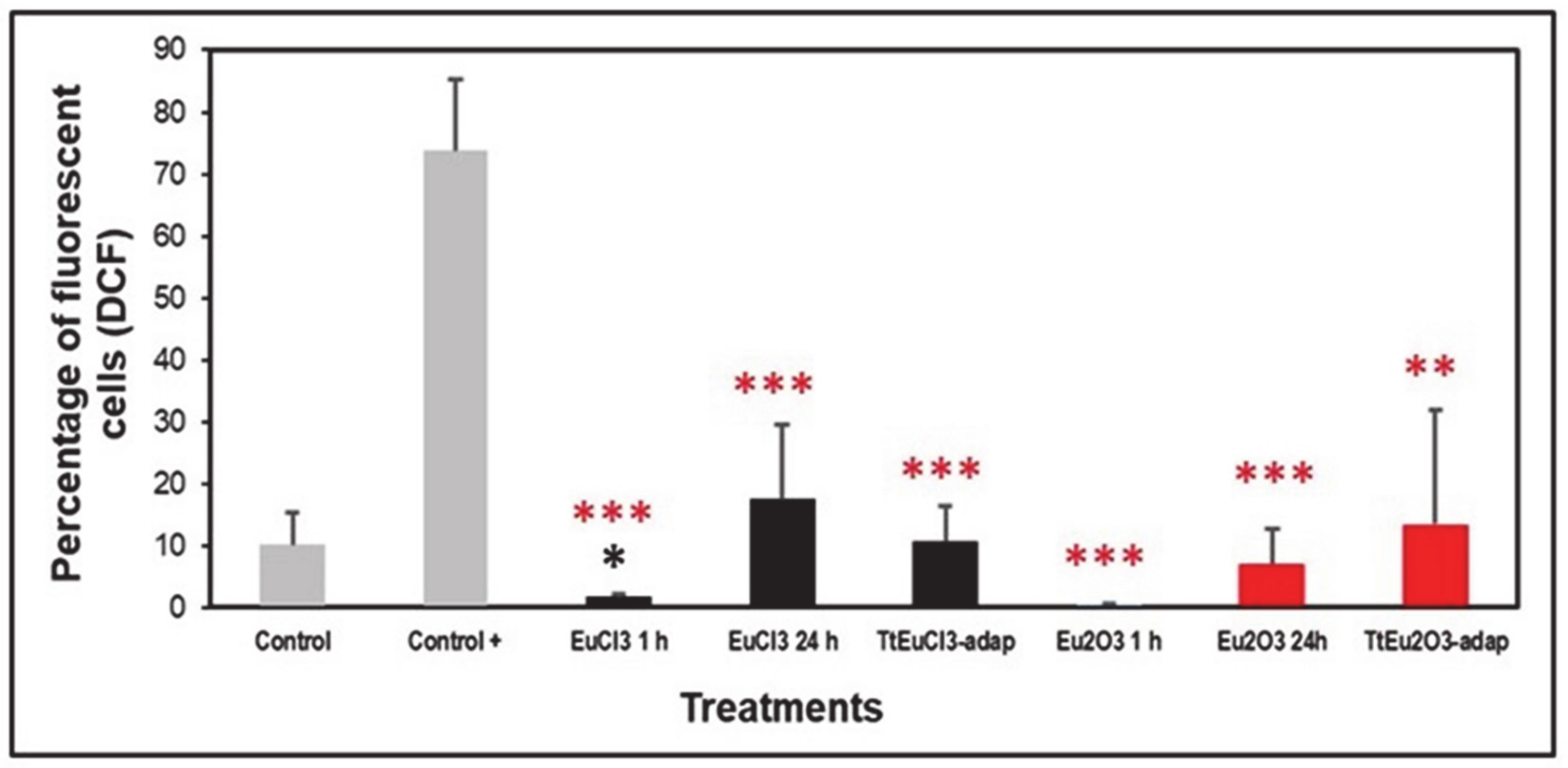
3.2. Oxidative Stress Assessment
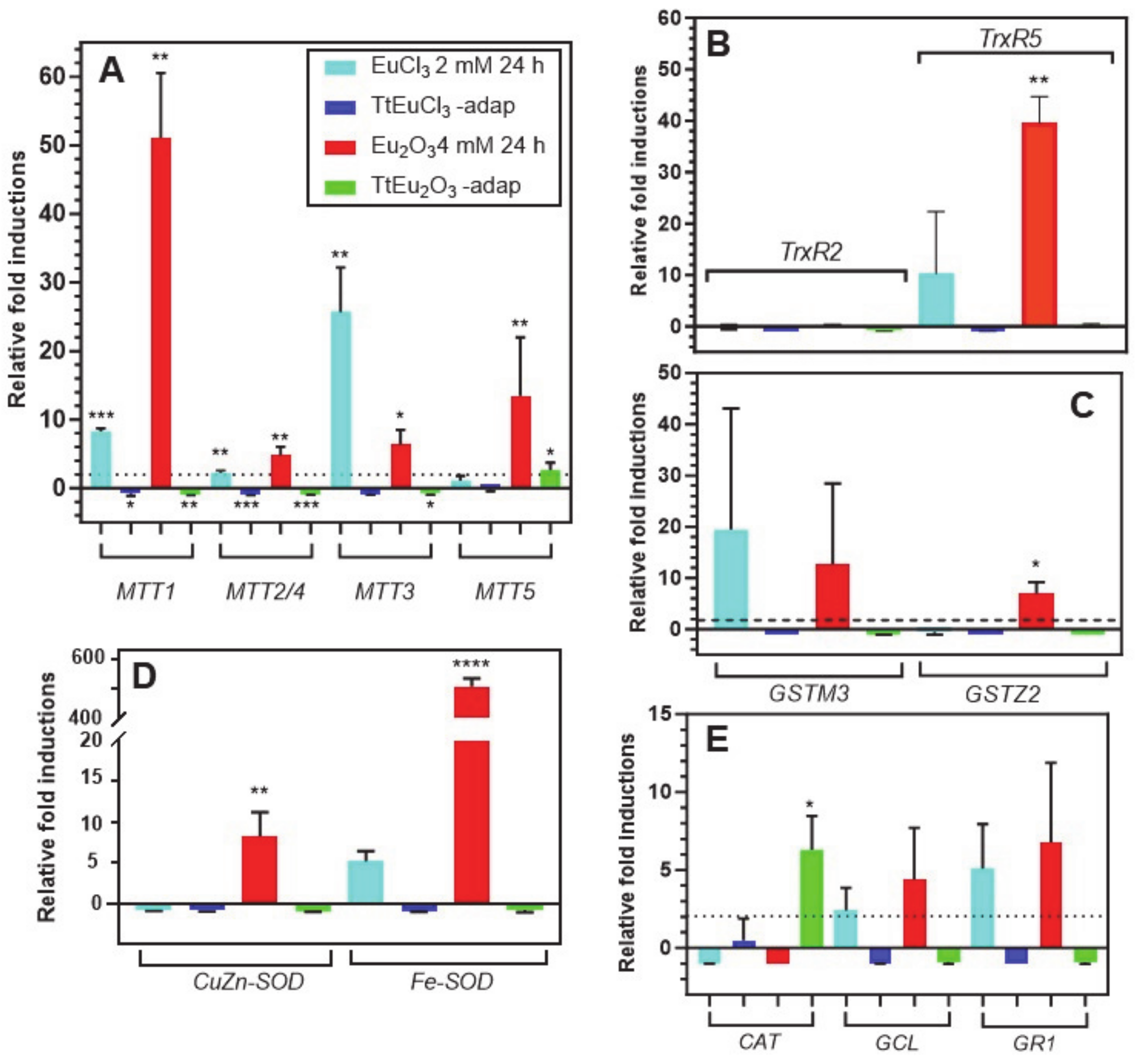
3.3. Comparative Quantitative Expression Analysis of Several Genes Involved in Oxidative and/or General Stress
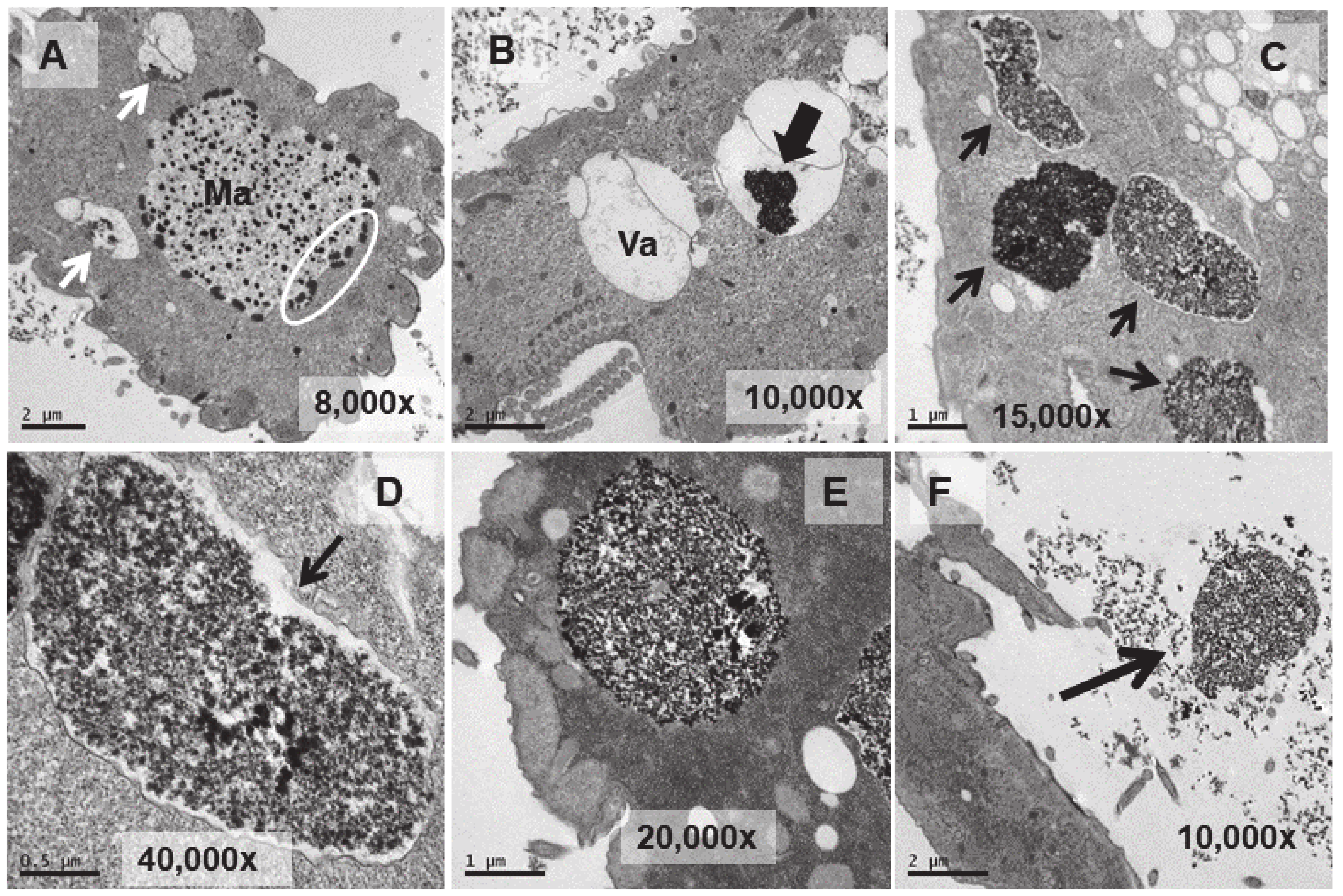
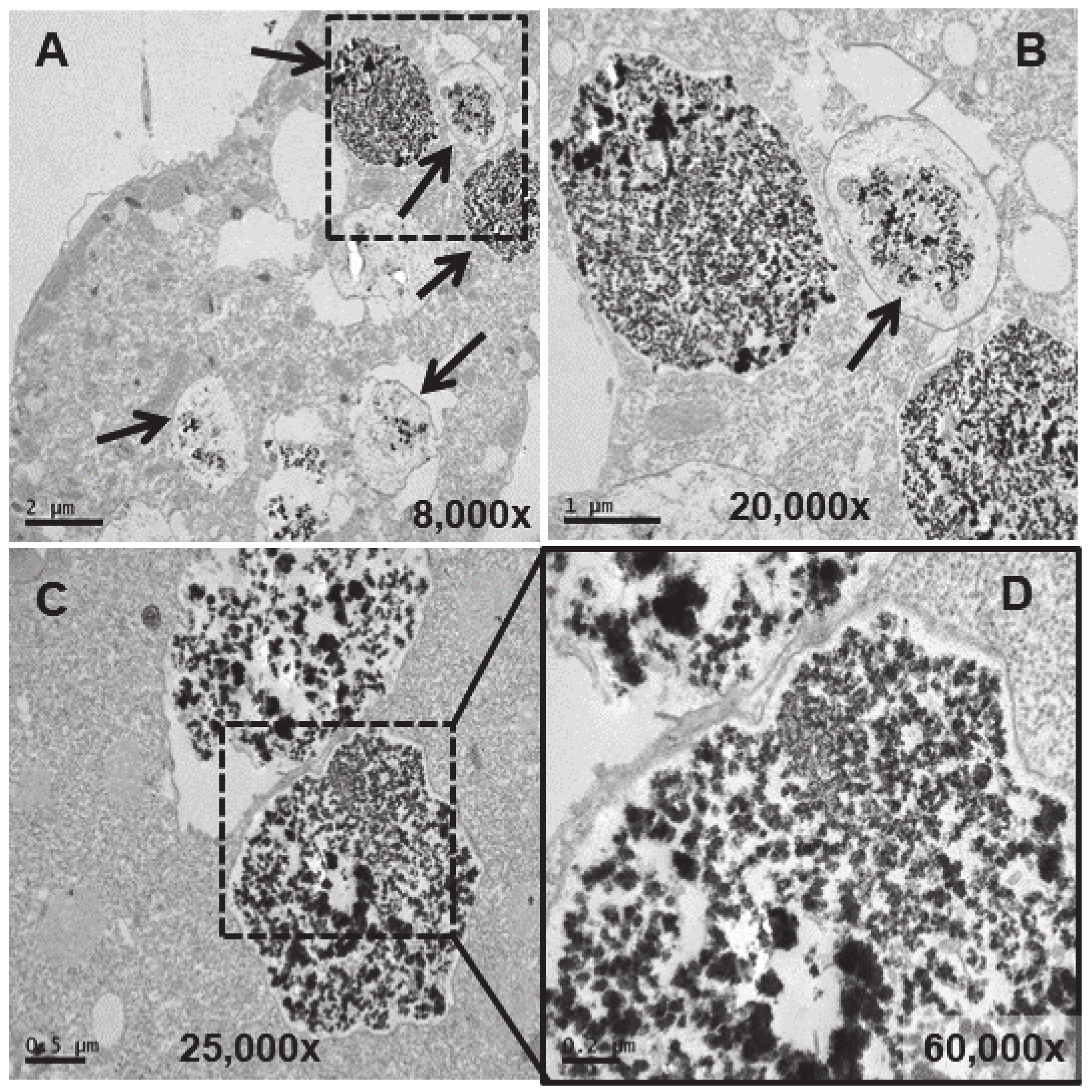
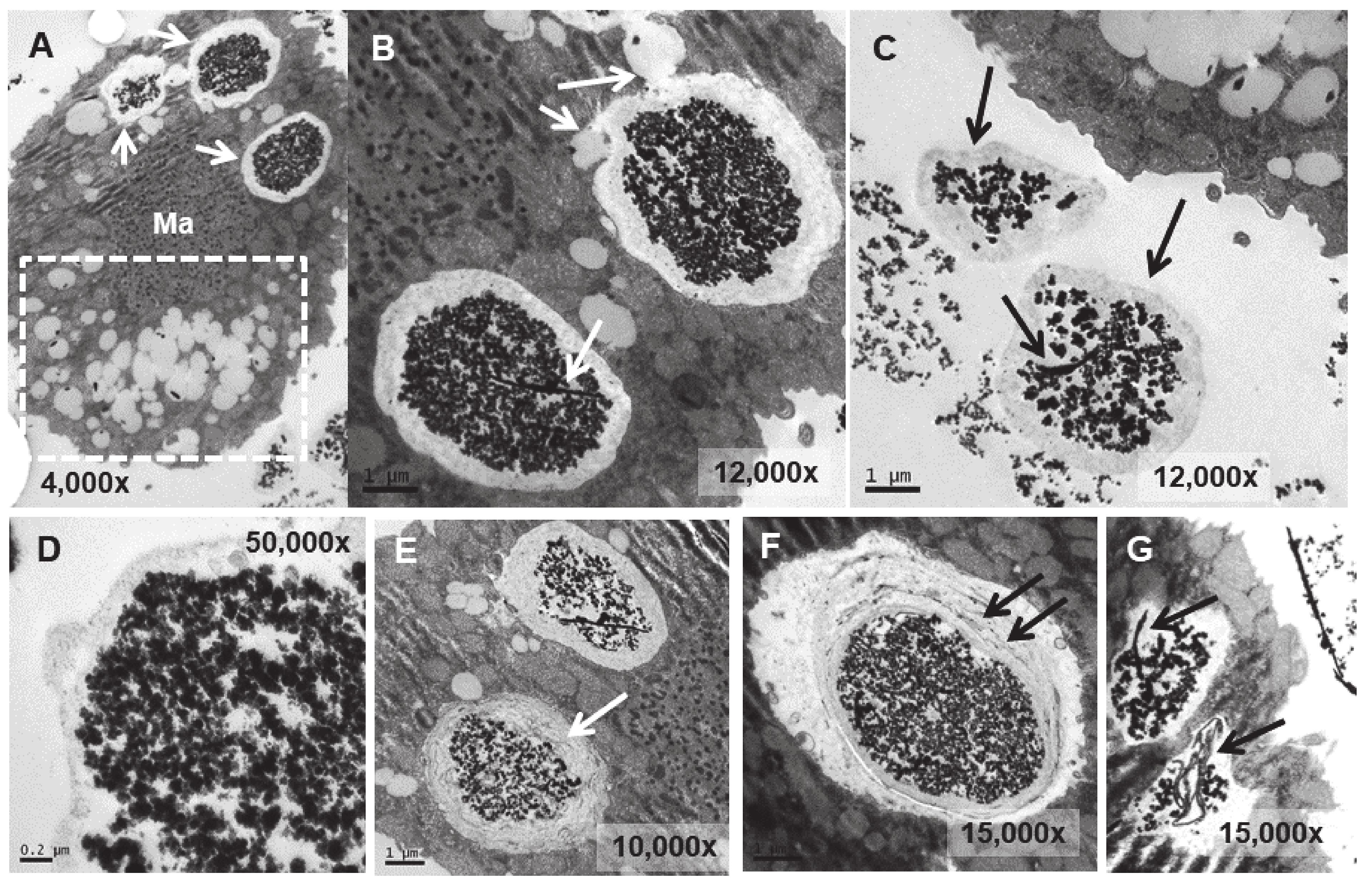
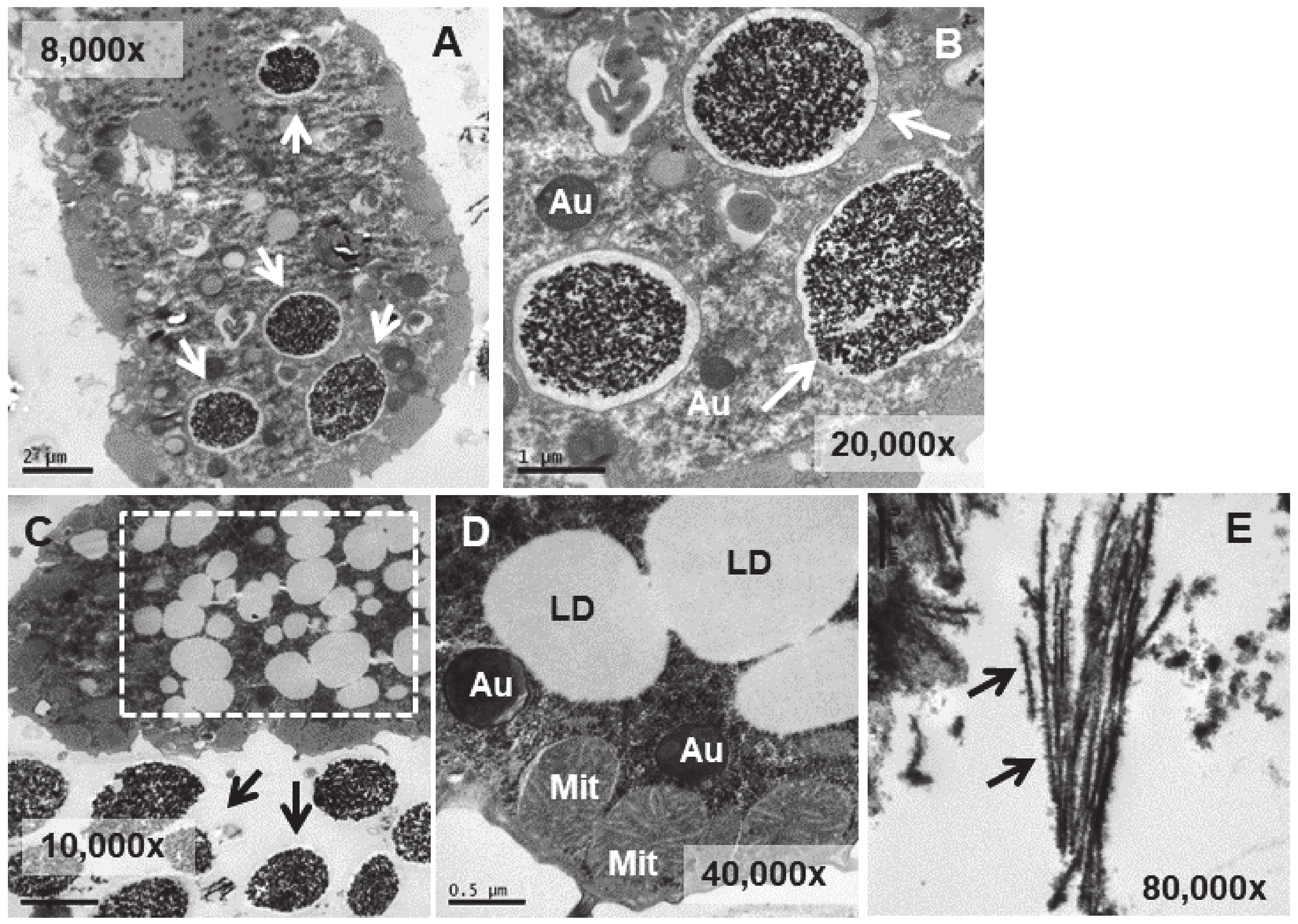
3.4. Ultrastructural Analysis
3.5. Microanalysis (TEM-XEDS)
4. Discussion
4.1. Toxicological and Growth Kinetics Parameters
4.2. Oxidative Stress Assessment
4.3. Expression Analysis of Genes Involved in General and/or Oxidative Stress Cell Response
4.4. Ultrastructural Modifications and Microanalysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blinova, I.; Muna, M.; Heinlaan, M.; Lukjanova, A.; Kahru, A. Potential hazard of lanthanides and lanthanide-based nanoparticles to aquatic ecosystems: Data gaps, challenges and future research needs derived from bibliometric analysis. Nanomaterials 2020, 10, 328. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, V.; Vignati, D.A.L.; Leyval, C.; Giamberini, L. Environmental fate and ecotoxicology of lanthanides: Are they a uniform group beyond chemistry? Environ. Int. 2014, 71, 148–157. [Google Scholar] [CrossRef]
- Gonzalez, V.; Vignati, D.A.L.; Pons, M.-N.; Montarges-Pelletier, E.; Bojic, C.; Giamberini, L. Lanthanide ecotoxicity: First attempt to measure environmental risk for aquatic organisms. Environ. Pollut. 2015, 199, 139–147. [Google Scholar] [CrossRef]
- Martino, C.; Chianese, T.; Chiarelli, R.; Roccheri, M.C.; Scudiero, R. Toxicological impact of rare earth elements (REEs) on the reproduction and development of aquatic organisms using sea urchins as biological models. Int. J. Mol. Sci. 2022, 23, 2876. [Google Scholar] [CrossRef] [PubMed]
- Pagano, G.; Thomas, P.J.; Nunzio, A.D.; Trifuoggi, M. Human exposures to rare earth elements: Present knowledge and research prospects. Environ. Res. 2019, 171, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Balaram, V. Rare earth elements: A review of applications, occurrence, exploration, analysis, recycling, and environmental impact. Geosci. Front. 2019, 10, 1285–1303. [Google Scholar] [CrossRef]
- Cotruvo, J.A., Jr. The chemistry of lanthanides in biology: Recent discoveries, emerging principles, and technological applications. ACS Cent. Sci. 2019, 5, 1496–1506. [Google Scholar] [CrossRef]
- Bünzli, J.-C.G. Lanthanide light for biology and medical diagnosis. J. Lumin. 2016, 170, 866–878. [Google Scholar] [CrossRef]
- Qin, X.; Wang, J.; Yuan, Q. Synthesis and biomedical applications of lanthanides-doped persistent luminescence phosphors with NIR emissions. Front. Chem. 2020, 8, 608578. [Google Scholar] [CrossRef]
- Syamchand, S.S.; Sony, G. Europium enabled luminescent nanoparticles for biomedical applications. J. Lumin. 2015, 165, 190–215. [Google Scholar] [CrossRef]
- Liang, T.; Zhang, S.; Wang, L.; Kung, H.-T.; Wang, Y.; Hu, A.; Ding, S. Environmental biogeochemical behaviors of rare earth elements in soil–plant systems. Environ. Geochem. Health 2005, 27, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Adeel, M.; Lee, J.Y.; Zain, M.; Rizwan, M.; Nawab, A.; Ahmad, M.A.; Shafiq, M.; Yi, H.; Jilani, G.; Javed, R.; et al. Cryptic footprints of rare earth elements on natural resources and living organisms. Environ. Int. 2019, 127, 785–800. [Google Scholar] [CrossRef] [PubMed]
- Turra, C. Sustainability of rare earth elements chain: From production to food-a review. Int. J. Environ. Health Res. 2017, 28, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Gwenzi, W.; Mangori, L.; Danha, C.; Chaukura, N.; Dunjana, N.; Sanganyadoe, E. Sources, behaviour, and environmental and human health risks of high technology rare earth elements as emerging contaminants. Sci. Total Environ. 2018, 636, 289–313. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, F.; Thomas, P.J.; Pagano, G.; Perono, G.A.; Oral, R.; Lyons, D.M.; Toscanesi, M.; Trifuoggi, M. Review of rare earth elements as fertilizers and feed additives: A knowledge gap analysis. Arch. Environ. Contam. Toxicol. 2021, 81, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Rim, K.T.; Koo, K.H.; Park, J.S. Toxicological evaluations of rare earths and their health impacts to workers: A literature review. Saf. Health Work 2013, 4, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, N.; Hsu, H.-S.; Liang, S.-T.; Roldan, M.J.M.; Lee, J.-S.; Ger, T.-R.; Hsiao, C.-D. An update review of toxicity effect of the rare earth elements (REEs) on aquatic organisms. Animals 2020, 10, 1663. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Prakash, R.; Singh, V.K. Synthesis, characterization, and applications of europium oxide: A review. Rev. Adv. Sci. Eng. 2015, 4, 247–257. [Google Scholar] [CrossRef]
- Silva, R.; Chojnacki, J.; Falcão, E.H.; Alves, S. New coordination polymers based on a V-shaped ligand and lanthanides: Structural description and symmetry-luminescence correlation using europium as a probe. J. Lumin. 2017, 182, 29–38. [Google Scholar] [CrossRef]
- Avila, J.N.; Ireland, T.R.; Lugaro, M.; Gyngard, F.; Zinner, E.; Cristallo, S.; Holden, P.; Rauscher, T. Europium s-process signature at close-to-solar metallicity in stardust sic grains from asymptotic giant branch stars. Astrophysic. J. Lett. 2013, 768, L18. [Google Scholar] [CrossRef]
- Trifuoggi, M.; Pagano, G.; Guida, M.; Palumbo, A.; Siciliano, A.; Gravina, M.; Lyons, D.M.; Burié, P.; Levak, M.; Thomas, P.J.; et al. Comparative toxicity of seven rare earth elements in sea urchin early life stages. Environ. Sci. Poll. Res. 2017, 24, 20803–20810. [Google Scholar] [CrossRef]
- Bollu, V.S.; Nethi, S.K.; Dasari, R.K.; Rao, S.S.N.; Misra, S.; Patra, C.R. Evaluation of in vivo cytogenetic toxicity of europium hydroxide nanorods (EHNs) in male and female Swiss albino mice. Nanotoxicology 2016, 10, 413–425. [Google Scholar] [CrossRef]
- Zhuang, W.Q.; Fitts, J.P.; Ajo-Franklin, C.M.; Maes, S.; Alvarez-Cohen, L.; Hennebel, T. Recovery of critical metals using biometallurgy. Curr. Opin. Biotechnol. 2015, 33, 327–335. [Google Scholar] [CrossRef]
- Maleke, M.; Valverde, A.; Vermeulen, J.G.; Cason, E.; Gomez-Arias, A.; Moloantoa, K.; Coetsee-Hugo, L.; Swart, H.; Heerden, E.; Castillo, J. Biomineralization and bioaccumulation of europium by a thermophilic metal resistant bacterium. Front. Microbiol. 2019, 10, 81. [Google Scholar] [CrossRef]
- Das, N.; Das, D. Recovery of rare earth metals through biosorption: An overview. J. Rare Earths 2013, 31, 933–943. [Google Scholar] [CrossRef]
- Furuhashi, Y.; Honda, R.; Noguchi, M.; Hara-Yamamura, H.; Kobayashi, S.; Higashimine, K.; Hasegawa, H. Optimum conditions of pH, temperatura and preculture for biosorption of europiu by microalgae Acutodesmus acuminatus. Biochem. Eng. J. 2019, 143, 58–64. [Google Scholar] [CrossRef]
- Jena, A.; Pradhan, S.; Mishra, S.; Sahoo, N.K. Evaluation of europium biosorption using Deinococcus radiodurans. Environ. Process. 2021, 8, 251–265. [Google Scholar] [CrossRef]
- Philip, L.; Iyengar, L.; Venkobachar, C. Biosorption of U, La, Pr, Nd, Eu, and Dy by Pseudomonas aeruginosa. J. Ind. Microbiol. Biotechmol. 2000, 25, 1–7. [Google Scholar] [CrossRef]
- Liang, J.; Li, L.; Song, W. Improved Eu(III) immobilization by Cladosporium sphaerospermum induced by low-temperature plasma. J. Radioanal. Nucl. Chem. 2018, 316, 963–970. [Google Scholar] [CrossRef]
- Arunraj, B.; Sathvika, T.; Rajesh, V.; Rajesh, N. Cellulose and Saccharomyces cerevisiae embark to recover europium from phosphor powder. ACS Omega 2019, 4, 940–952. [Google Scholar] [CrossRef]
- Kozai, N.; Skamoto, F.; Tanaka, K.; Ohnuki, T.; Satoh, T.; Kamiya, T.; Grambow, B. Complexation of Eu(III), Pb(II) and U(VI) with a Paramecium glycoprotein: Microbial transformation of heavy elements in the aquatic environment. Chemosphere 2018, 196, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, J.-L.; Dusser, M.; Bohatier, J.; Laffosse, J. Cytotoxicity assessment of three therapeutic agents, cyclosporine-A, cisplatin and doxorubicin, with the ciliated protozoan Tetrahymena pyriformis. Res. Microbiol. 2003, 154, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Dunlop, S.; Chapman, G. Detoxification of zinc and cadmium by the freshwater protozoan Tetrahymena pyriformis: II. Growth experiments and ultrastructural studies on sequestration of heavy metals. Environ. Res. 1981, 24, 264–274. [Google Scholar] [CrossRef]
- Diaz, S.; Martin-Gonzalez, A.; Cubas, L.; Ortega, R.; Amaro, F.; Rodriguez-Martin, D.; Gutierrez, J.C. High resistance of Tetrahymena thermophila to paraquat: Mitocondrial alterations, oxidative stress and antioxidant genes expression. Chemosphere 2016, 144, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Miao, W.; Yu, T.; Orias, E.; Wan, M.L.; Fu, C.J. Identification of differentially expressed genes in Tetrahymena thermophila in response to Dichlorodiphenyltrichloroethane (DDT) by suppression subtractive hybridization. Environ. Microbiol. 2006, 8, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, L.R. 1989. Dose- and pH-dependent effects of chloroquine on Tetrahymena. Eur. J. Protistol. 1989, 24, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Romero, I.; De Francisco, P.; Gutierrez, J.C.; Martin-Gonzalez, A. Selenium cytotoxicity in Tetrahymena thermophila: New clues about its biological effects and cellular resistance mechanisms. Sci. Total Environ. 2019, 671, 850–865. [Google Scholar] [CrossRef]
- Sauvant, N.P.; Pepin, D.; Piccinni, E. Tetrahymena pyriformis: A tool for toxicological studies. A review. Chemosphere 1999, 38, 1631–1669. [Google Scholar] [CrossRef] [PubMed]
- Eisen, J.A.; Coyne, R.S.; Wu, M.; Wu, D.; Thiagarajan, M.; Wortman, J.R.; Badger, J.H.; Ren, Q.; Amedeo, P.; Jones, K.M.; et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006, 4, e286. [Google Scholar] [CrossRef]
- De Francisco, P.; Martin-Gonzalez, A.; Turkewitz, A.P.; Gutierrez, J.C. Genome plasticity in response to stress in Tetrahymena thermophila: Selective and reversible chromosome amplification and paralogous expansion of metallothionein genes. Environ. Microbiol. 2018, 20, 2410–2421. [Google Scholar] [CrossRef]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Martin, D.; Murciano, A.; Herraiz, M.; De Francisco, P.; Amaro, F.; Gutierrez, J.C.; Martin-Gonzalez, A.; Diaz, S. Arsenate and arsenite differential toxicity in Tetrahymena thermophila. J. Hazard Mater. 2022, 431, 128532. [Google Scholar] [CrossRef] [PubMed]
- Garner, D.L.; Johnson, L.A.; Yue, S.T.; Roth, B.L.; Haugland, R.P. Dual DNA staining assessment of bovine sperm viability using SYBR-14 and propidium iodide. J. Androl. 1994, 15, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Eruslanov, E.; Kusmartsev, S. Identification of ROS using oxidized DCFDA and flowcytometry. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2010; pp. 57–72. [Google Scholar] [CrossRef]
- Cubas-Gaona, L.L.; De Francisco, P.; Martin-Gonzalez, A.; Gutierrez, J.C. Tetrahymena glutathione peroxidase family: A comparative analysis of these antioxidant enzymes and differential gene expression to metals and oxidizing agents. Microorganisms 2020, 8, 1008. [Google Scholar] [CrossRef] [PubMed]
- Dentler, W. Fixation of Tetrahymena cells for electron microscopy. Methods Cell Biol. 2000, 62, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.; Tsai, S.M.; Gomes-Caldas, D.G. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 2012, 31, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Larionov, A.; Krause, A.; Miller, W. A standard curve-based method for relative real time PCR data processing. BMC Bioinform. 2005, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, J.C.; Amaro, F.; Diaz, S.; De Francisco, P.; Cubas, L.L.; Martin-Gonzalez, A. Ciliate metallothioneins: Unique microbial eukaryotic heavy-metal-binder molecules. J. Biol. Inorg. Chem. 2011, 16, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Amaro, F.; Rico, D.; Campos, V.; Benítez, L.; Martín-González, A.; Hamilton, E.P.; Orias, E.; Gutierrez, J.C. Tetrahymena metallothioneins fall into two discrete subfamilies. PLoS ONE 2007, 2, e291. [Google Scholar] [CrossRef]
- Brzo’ska, M.M.; Moniuszko-Jakoniuk, J. Interactions between cadmium and zinc in the organism. Food Chem. Toxicol. 2001, 39, 967–980. [Google Scholar] [CrossRef]
- Pallares, R.M.; An, D.D.; Hebert, S.; Faulkner, D.; Loguinov, A.; Proctor, M.; Villalobos, J.A.; Bjornstad, K.A.; Rosen, C.J.R.; Vulpe, C.; et al. Multidimensional genome-wide screening in yeast provides mechanistic insights into europium toxicity. Metallomics 2021, 13, mfab061. [Google Scholar] [CrossRef] [PubMed]
- Tai, P.; Zhao, Q.; Su, D.; Li, P.; Stagnitti, F. Biological toxicity of lanthanide elements on algae. Chemosphere 2010, 80, 1031–1033. [Google Scholar] [CrossRef] [PubMed]
- Kurvet, I.; Juganson, K.; Vija, H.; Sihtmae, M.; Blinova, I.; Syvertsen-Wiig, G.; Kahru, A. Toxicity of nine (doped) rare earth metal oxides and respective individual metals to aquatic microorganisms Vibrio fischeri and Tetrahymena thermophila. Materials 2017, 10, 754. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, M.; Wang, X. Population growth responses of Tetrahymena shanghaiensis in exposure to rare earth elements. Biol. Trace Elem. Res. 2000, 75, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Maleke, M.; Valverde, A.; Gomez-Arias, A.; Cason, E.D.; Vermeulen, J.-G.; Coetsee-Hugo, L.; Swart, H.; van Heerden, E.; Castillo, J. Anaerobic reduction of europium by a Clostridium strain as a strategy for rare earth biorecovery. Sci. Rep. 2019, 9, 14339. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Osborne, O.J.; Lin, S.; Ji, Z.; Damoiseux, R.; Wang, Y.; Nel, A.E.; Lin, S. Lanthanide hydroxide nanoparticles induce angiogenesis via ROS-sensitive signaling. Small 2016, 12, 4404–4411. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.B.; Forsythe, A.B. Robust tests for the equality of variances. J. Amer. Statis. Assoc. 1974, 69, 364–367. [Google Scholar] [CrossRef]
- Dede, A.F.U.; Arslanyolu, M. Genome-wide analysis of the Tetrahymena thermophila glutathione S-transferase gene superfamily. Genomics 2019, 111, 534–548. [Google Scholar] [CrossRef]
- Sheng, Y.; Abreu, I.A.; Cabelli, D.E.; Maroney, M.J.; Miller, A.-F.; Teixeira, M.; Valentine, J.S. Superoxide dismutases and superoxide reductases. Chem. Rev. 2014, 114, 3854–3918. [Google Scholar] [CrossRef]
- Wei, W.; Wang, H.; Jiang, C. Spectrofluorimetric determination of superoxide dismutase using a europium-tetracycline probe. Spectrochim. Acta Part A 2008, 70, 389–393. [Google Scholar] [CrossRef]
- Dondero, F.; Cavaletto, M.; Chezzi, A.R.; La Terza, A.; Banni, M.; Viarengo, A. Biochemical characterization and quantitative gene expression analysis of the multi-stress inducible metallothionein from Tetrahymena thermophila. Protist 2004, 155, 157–168. [Google Scholar] [CrossRef] [PubMed]
- De Francisco, P.; Martin-Gonzalez, A.; Turkewitz, A.P.; Gutierrez, J.C. Extreme metal adapted, knockout and knockdown strains reveal a coordinated gene expression among different Tetrahymena thermophila metallothionein isoforms. PLoS ONE 2017, 12, e0189076. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xu, J.; Zhu, Y.; Chai, B.; Liang, A.; Wang, W. Lanthanum (III) impacts on metallothionein MTT1 and MTT2 from Tetrahymena thermophila. Biol. Trace Elem. Res. 2011, 143, 1808–1818. [Google Scholar] [CrossRef] [PubMed]
- Geng, M.-J.; Liang, S.-X.; Liu, W.; Jin, Y. Quantification of metallothioneins in the earthworm by lomefloxacin-europium(III) fluorescent probe. Environ. Sci. Process. Impacts 2014, 16, 1923–1929. [Google Scholar] [CrossRef] [PubMed]
- Silber, H.B.; Paquette, S.J. Complexes of lanthanide ions with amino acids, nucleotides, and other ligands of biological interest in solution. In Metal ions in biological systems. In The Lanthanides and Their Interrelations with Biosystems; Sigel, A., Sigel, H., Eds.; CRC Press: Boca Raton, FL, USA, 2003; Volume 40, p. 799. ISBN 978-0-8247-4245-4. [Google Scholar]
- Lafita-Navarro, M.C.; Conacci-Sorrell, M. Nucleolar stress: From development to cancer. Semin. Cell Develop. Biol. 2023, 136, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Bahadori, M. New insights into connection of nucleolar functions and cancer. Tanaffos 2019, 18, 173–179. [Google Scholar] [PubMed]
- Weeks, S.; Metge, B.J.; Samant, R.S. The nucleolus: A central response hub for the stressors that drive cancer progression. Cell. Mol. Life Sci. 2019, 76, 4511–4524. [Google Scholar] [CrossRef] [PubMed]
- Neuburger, M.; Herget, G.W.; Plaumann, L.; Falk, A.; Schwab, H.; Adler, C.-P. Change in size, number, and morphology of the nucleoli in human hearts as a result of hyperfunction. Pathol. Res. Pract. 1998, 194, 385–390. [Google Scholar] [CrossRef] [PubMed]
- MacMillan, G.A.; Clayden, M.G.; Chetelat, J.; Richardson, M.C.; Ponton, D.E.; Perron, T.; Amyot, M. Environmental drivers of rare earth element bioaccumulation in freshwater zooplankton. Environ. Sci. Technol. 2019, 53, 1650–1660. [Google Scholar] [CrossRef]
- Henne, W.M.; Reese, M.L.; Goodman, J.M. 2018. The assembly of lipid droplets and their roles in challenged cells. EMBO J. 2018, 37, e98947. [Google Scholar] [CrossRef]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef]
- Cui, L.; Liu, P. Two types of contact between lipid droplets and mitochondria. Front. Cell. Dev. Biol. 2020, 8, 618322. [Google Scholar] [CrossRef]
- Amen, T.; Kaganovich, D. Dynamic droplets: The role of cytoplasmic inclusions in stress, function, and disease. Cell. Mol. Life Sci. 2015, 72, 401–415. [Google Scholar] [CrossRef]
- Rajakumar, S.; Nachiappan, V. Lipid droplets alleviate cadmium-induced cytotoxicity in Saccharomyces cerevisiae. Toxicol. Res. 2017, 6, 30–41. [Google Scholar] [CrossRef]
- Kennedy, D.C.; Lyn, R.K.; Pezacki, J.P. Cellular lipid metabolism is influenced by the coordination environment of copper. J. Am. Chem. Soc. 2009, 131, 2444–2445. [Google Scholar] [CrossRef] [PubMed]
- Moron, A.; Martin-Gonzalez, A.; Diaz, S.; Gutierrez, J.C.; Amaro, F. Autophagy and lipid droplets are a defense mechanism against toxic copper oxide nanotubes in the eukaryotic microbial model Tetrahymena thermophila. Sci. Total Environ. 2022, 847, 157580. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Ohnuki, T.; Utsunomiya, S. Biomineralization of middle rare earth element samarium in yeast and bacteria systems. Geomicrobiol. J. 2018, 35, 375–384. [Google Scholar] [CrossRef]
- Nadella, S.; Sahoo, J.; Subramanian, P.S.; Sahu, A.; Mishra, S.; Albrecht, M. Sensing of phosphates by using luminescent Eu(III) and Tb(III) complexes: Application to the microalgal cell Chlorella vulgaris. Chem. Eur. J. 2014, 20, 6047–6053. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Ohnuki, T.; Kozai, N.; Tanaka, K.; Suzuki, Y.; Sakamoto, F.; Kamiishi, E.; Utsunomiya, S. Biological nano-mineralization of Ce phosphate by Saccharomyces cerevisiae. Chem. Geol. 2010, 277, 61–69. [Google Scholar] [CrossRef]
- Jiang, M.; Ohnuki, T.; Kozai, N.; Tanaka, K.; Kozai, N.; Kamiishi, E.; Utsunomiya, S. Post-adsorption process of Yb phosphate nano-particle formation by Saccharomyces cerevisiae. Geochim. Cosmochim. Acta 2012, 93, 30–46. [Google Scholar] [CrossRef]
- De Francisco, P.; Amaro, F.; Martin-Gonzalez, A.; Serrano, A.; Gutierrez, J.C. Quantitative proteomic analyses of a Pb-adapted Tetrahymena thermophila strain reveal the cellular strategy to Pb(II) stress including lead biomineralization to chloropyromorphite. Sci. Total Environ. 2023, 891, 164252. [Google Scholar] [CrossRef] [PubMed]



| Medium | EuCl3 (μM) | Eu2O3 (μM) |
|---|---|---|
| Tris-HCl | 127.93 | 173.32 |
| PP210 | 4830.68 | 7916.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alonso, P.; Blas, J.; Amaro, F.; de Francisco, P.; Martín-González, A.; Gutiérrez, J.C. Cellular Response of Adapted and Non-Adapted Tetrahymena thermophila Strains to Europium Eu(III) Compounds. Biology 2024, 13, 285. https://doi.org/10.3390/biology13050285
Alonso P, Blas J, Amaro F, de Francisco P, Martín-González A, Gutiérrez JC. Cellular Response of Adapted and Non-Adapted Tetrahymena thermophila Strains to Europium Eu(III) Compounds. Biology. 2024; 13(5):285. https://doi.org/10.3390/biology13050285
Chicago/Turabian StyleAlonso, Patricia, Javier Blas, Francisco Amaro, Patricia de Francisco, Ana Martín-González, and Juan Carlos Gutiérrez. 2024. "Cellular Response of Adapted and Non-Adapted Tetrahymena thermophila Strains to Europium Eu(III) Compounds" Biology 13, no. 5: 285. https://doi.org/10.3390/biology13050285
APA StyleAlonso, P., Blas, J., Amaro, F., de Francisco, P., Martín-González, A., & Gutiérrez, J. C. (2024). Cellular Response of Adapted and Non-Adapted Tetrahymena thermophila Strains to Europium Eu(III) Compounds. Biology, 13(5), 285. https://doi.org/10.3390/biology13050285