Genome-Wide Analysis of the HSF Gene Family Reveals Its Role in Astragalus mongholicus under Different Light Conditions
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Soilless Cultivation of A. mongholicus Culture and Light Treatment
2.2. Identification and Physicochemical Properties Analysis of the HSF Gene Family in A. mongholicus
2.3. Phylogenetic Analysis of AmHSF Proteins
2.4. Conserved Motifs and Gene Structure Analysis of the AmHSF Gene Family
2.5. Analysis of Cis-Elements of the AmHSF Gene Family
2.6. Chromosomal Mapping and Collinearity Analysis of AmHSF Gene
2.7. Analysis of AmHSF Gene Family Expression Pattern in Different Tissue Sites and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Validation
3. Results
3.1. Identification and Physicochemical Properties Analysis of the HSF Gene Family in A. mongholicus
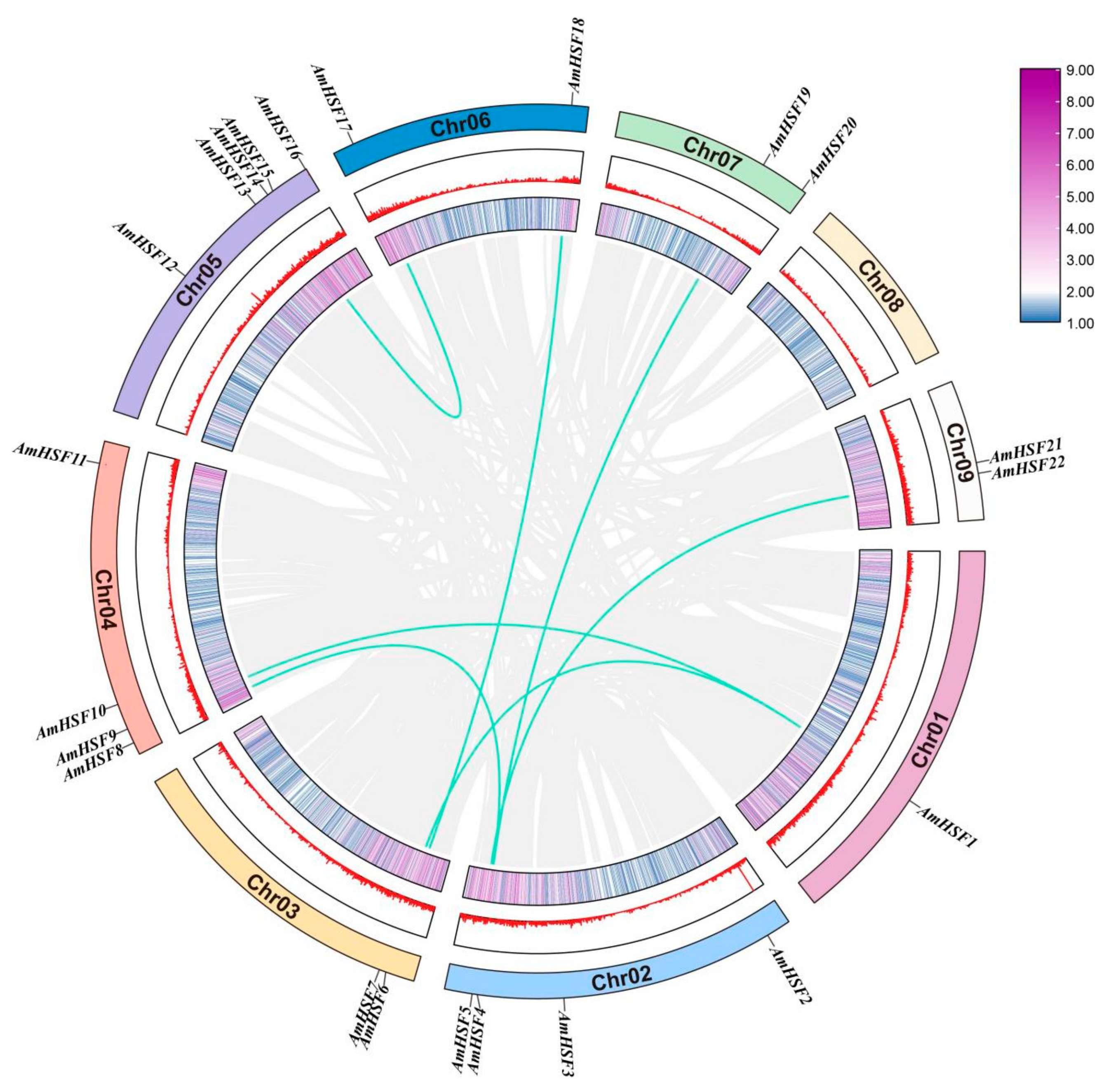
3.2. Chromosomal Distribution and Gene Duplication Event Analysis of AmHSF Genes
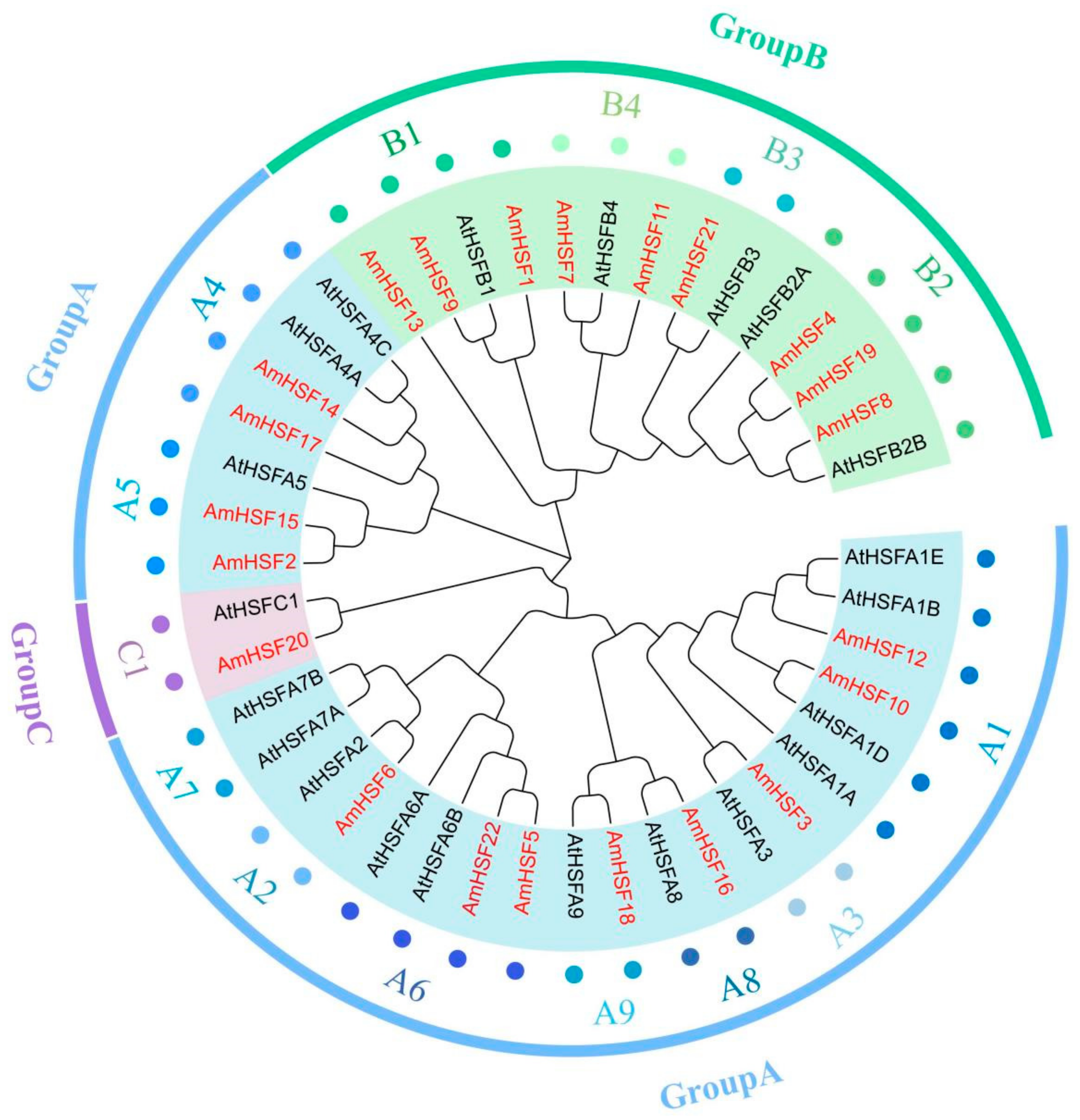
3.3. Phylogenetic Analysis of AmHSF Gene Family
3.4. Analysis of Conserved Motifs and Gene Structures in AmHSF
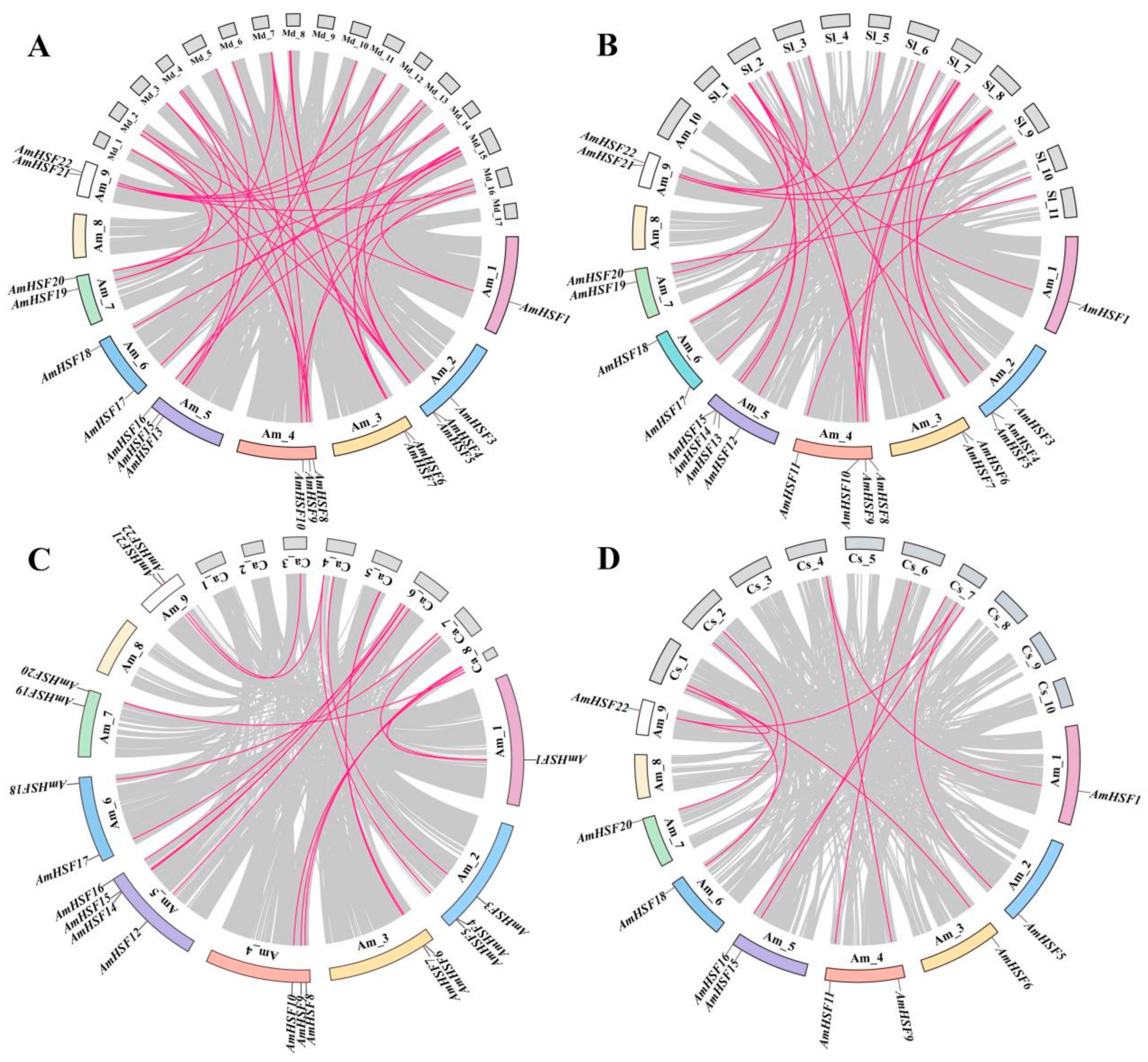
3.5. Syntenic Analysis of AmHSF Genes in Different Species
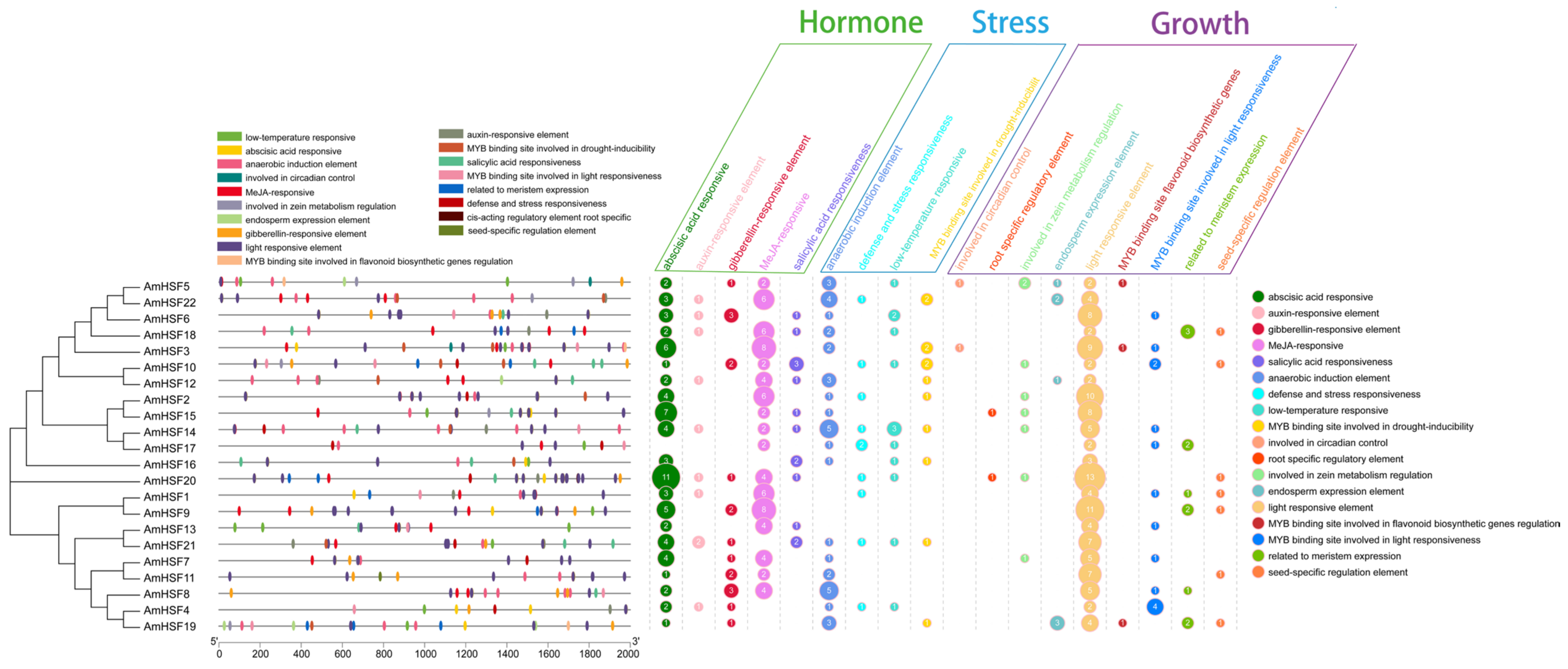
3.6. Cis-Element Analysis of AmHsf Genes
3.7. Expression Profile and qRT-PCR Verification of AmHsf Genes in Different Tissues of A. mongholicus
3.8. Analysis of AmHSF Expression Patterns after Different Light Treatments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- von Koskull-Döring, P.; Scharf, K.D.; Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Baniwal, S.K.; Bharti, K.; Chan, K.Y.; Fauth, M.; Ganguli, A.; Kotak, S.; Mishra, S.K.; Nover, L.; Port, M.; Scharf, K.D.; et al. Heat stress response in plants: A complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 2004, 29, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; von Koskull-Döring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wu, J.; Ji, Q.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Döring, P.; Treuter, E.; Kistner, C.; Lyck, R.; Chen, A.; Nover, L. The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 2000, 12, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, J.; Kim, H.; Son, N.; Kim, E.J.; Kim, J.; Byun, D.; Lee, Y.; Park, Y.M.; Nageswaran, D.C.; et al. Arabidopsis heat shock factor binding protein is required to limit meiotic crossovers and HEI10 transcription. EMBO J. 2022, 41, e109958. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Khurana, N.; Agarwal, P.; Khurana, J.P.; Khurana, P. A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment. PLoS ONE 2013, 8, e79577. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, Z.; Zhang, Z.; Cao, H.; Kong, L.; Ma, W.; Ren, W. A review of the botany, phytochemistry, traditional uses, pharmacology, toxicology, and quality control of the Astragalus memeranaceus. Front. Pharmacol. 2023, 14, 1242318. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zheng, S.; Brinckmann, J.A.; Fu, J.; Zeng, R.; Huang, L.; Chen, S. Chemical and genetic diversity of Astragalus mongholicus grown in different eco-climatic regions. PLoS ONE 2017, 12, e184791. [Google Scholar] [CrossRef] [PubMed]
- Tseng, A.; Yang, C.H.; Chen, C.H.; Chen, C.H.; Hsu, S.L.; Lee, M.H.; Lee, H.C.; Su, L.J. An in vivo molecular response analysis of colorectal cancer treated with Astragalus membranaceus extract. Oncol. Rep. 2016, 35, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, L.; Wang, J.; Ren, B.; Zhang, L.; Li, W. Formononetin, an isoflavone from Astragalus membranaceus inhibits proliferation and metastasis of ovarian cancer cells. J. Ethnopharmacol. 2018, 221, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Wang, Y.; Zhang, Y.; Zhou, T.; Fang, C.; Nan, P.; Wang, X.; Li, X.; Wei, Y.; Chen, J. Phenylalanine ammonia lyase functions as a switch directly controlling the accumulation of calycosin and calycosin-7-O-beta-D-glucoside in Astragalus membranaceus var. mongholicus plants. J. Exp. Bot. 2008, 59, 3027–3037. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhang, H.; Li, P.; Jin, J.; Li, Z. The bacterial consortia promote plant growth and secondary metabolite accumulation in Astragalus mongholicus under drought stress. BMC Plant Biol. 2022, 22, 475. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Wang, Y.; Abozeid, A.; Tian, D.M.; Zhang, X.N.; Tang, Z.H. The Different Resistance of Two Astragalus Plants to UV-B Stress is Tightly Associated with the Organ-specific Isoflavone Metabolism. Photochem. Photobiol. 2018, 94, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fang, T.; Su, H.; Duan, S.; Ma, R.; Wang, P.; Wu, L.; Sun, W.; Hu, Q.; Zhao, M.; et al. A reference-grade genome assembly for Astragalus mongholicus and insights into the biosynthesis and high accumulation of triterpenoids and flavonoids in its roots. Plant Commun. 2023, 4, 100469. [Google Scholar] [CrossRef]
- Gomez-Pastor, R.; Burchfiel, E.T.; Thiele, D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, A.; Yabuta, Y.; Yoshida, E.; Maruta, T.; Yoshimura, K.; Shigeoka, S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006, 48, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Raza, A.; Iqbal, J.; Shaukat, M.; Mahmood, T. Analyzing the regulatory role of heat shock transcription factors in plant heat stress tolerance: A brief appraisal. Mol. Biol. Rep. 2022, 49, 5771–5785. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Huang, Q.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide investigation of the heat shock transcription factor (Hsf) gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 871. [Google Scholar] [CrossRef] [PubMed]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Dong, Q.; Jiang, H.; Zhu, S.; Chen, B.; Xiang, Y. Genome-wide analysis of the heat shock transcription factors in Populus trichocarpa and Medicago truncatula. Mol. Biol. Rep. 2012, 39, 1877–1886. [Google Scholar] [CrossRef] [PubMed]
- Talaee, L.; Fathipour, Y.; Talebi, A.A.; Khajehali, J. Screening of Potential Sources of Resistance to Spodoptera exigua (Lepidoptera: Noctuidae) in 24 Sugar Beet Genotypes. J. Econ. Entomol. 2017, 110, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Luehrsen, K.R.; Taha, S.; Walbot, V. Nuclear pre-mRNA processing in higher plants. Prog. Nucleic Acid Res. Mol. Biol. 1994, 47, 149–193. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, N.; Deng, T.; Zhang, L.; Zuo, K. Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC Genom. 2014, 15, 961. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, B.; Li, J.; Zhang, L.; Wang, Y.; Zheng, H.; Lu, M.; Chen, J. Hsf and Hsp gene families in Populus: Genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genom. 2015, 16, 181. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Shu, J.; Xu, L.; Ya, H.; Yang, L.; Li, S.; Wang, D. Genome-wide identification of R2R3-MYB transcription factor family in Docynia delavayi (Franch.) Schneid and its expression analysis during the fruit development. Food Biosci. 2023, 54, 102878. [Google Scholar] [CrossRef]
- Wunderlich, M.; Gross-Hardt, R.; Schöffl, F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 2014, 85, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Begum, T.; Reuter, R.; Schöffl, F. Overexpression of AtHsfB4 induces specific effects on root development of Arabidopsis. Mech. Dev. 2013, 130, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Vierling, E.; Bäumlein, H.; von Koskull-Döring, P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 2007, 19, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.M.; Kim, D.W.; Woo, M.S.; Jeong, H.S.; Son, Y.S.; Akhter, S.; Choi, G.J.; Bahk, J.D. Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant Cell Environ. 2014, 37, 1202–1222. [Google Scholar] [CrossRef] [PubMed]
- Li, P.S.; Yu, T.F.; He, G.H.; Chen, M.; Zhou, Y.B.; Chai, S.C.; Xu, Z.S.; Ma, Y.Z. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1009. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, R.; Jin, H.; Ding, Y.; Cai, C. Molecular Characterization and Expression Profile Analysis of Heat Shock Transcription Factors in Mungbean. Front. Genet. 2018, 9, 736. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Meng, X.; Wang, S.; Mi, Y.; Qin, Z.; Liu, T.; Zhang, Y.; Wan, H.; Chen, W.; Sun, W.; et al. Genome-wide identification of HSF gene family and their expression analysis in vegetative tissue of young seedlings of hemp under different light treatments. Ind. Crops Prod. 2023, 204, 117375. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa-Yokoi, A.; Nosaka, R.; Hayashi, H.; Tainaka, H.; Maruta, T.; Tamoi, M.; Ikeda, M.; Ohme-Takagi, M.; Yoshimura, K.; Yabuta, Y.; et al. HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 2011, 52, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Xu, X. The influence of LEDs light on Growth and Biosynthesis of Formononetin and Calycosin in the Astragalus membranaceus (Fisch.) Bunge Hairy Root Cultures. Master’s Thesis, Northeast Forestry University, Harbin, China, 2022. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Wang, P.; He, J.; Kong, L.; Zhang, W.; Liu, W.; Liu, X.; Ma, W. Genome-Wide Analysis of the HSF Gene Family Reveals Its Role in Astragalus mongholicus under Different Light Conditions. Biology 2024, 13, 280. https://doi.org/10.3390/biology13040280
Wang Z, Wang P, He J, Kong L, Zhang W, Liu W, Liu X, Ma W. Genome-Wide Analysis of the HSF Gene Family Reveals Its Role in Astragalus mongholicus under Different Light Conditions. Biology. 2024; 13(4):280. https://doi.org/10.3390/biology13040280
Chicago/Turabian StyleWang, Zhen, Panpan Wang, Jiajun He, Lingyang Kong, Wenwei Zhang, Weili Liu, Xiubo Liu, and Wei Ma. 2024. "Genome-Wide Analysis of the HSF Gene Family Reveals Its Role in Astragalus mongholicus under Different Light Conditions" Biology 13, no. 4: 280. https://doi.org/10.3390/biology13040280
APA StyleWang, Z., Wang, P., He, J., Kong, L., Zhang, W., Liu, W., Liu, X., & Ma, W. (2024). Genome-Wide Analysis of the HSF Gene Family Reveals Its Role in Astragalus mongholicus under Different Light Conditions. Biology, 13(4), 280. https://doi.org/10.3390/biology13040280





