The Female-Biased General Odorant Binding Protein 2 of Semiothisa cinerearia Displays Binding Affinity for Biologically Active Host Plant Volatiles
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Culture, Tissue Collection, RNA Extraction, and cDNA Synthesis
2.2. Gene Cloning and Sequence Analysis
2.3. RT–PCR and qRT–PCR
2.4. Recombinant ScinGOBP2 Expression
2.5. Fluorescence-Based Competitive Binding Assays
2.6. Three-Dimensional Modeling of ScinGOBP2 and Ligand Docking
2.7. Electrophysiological Recordings
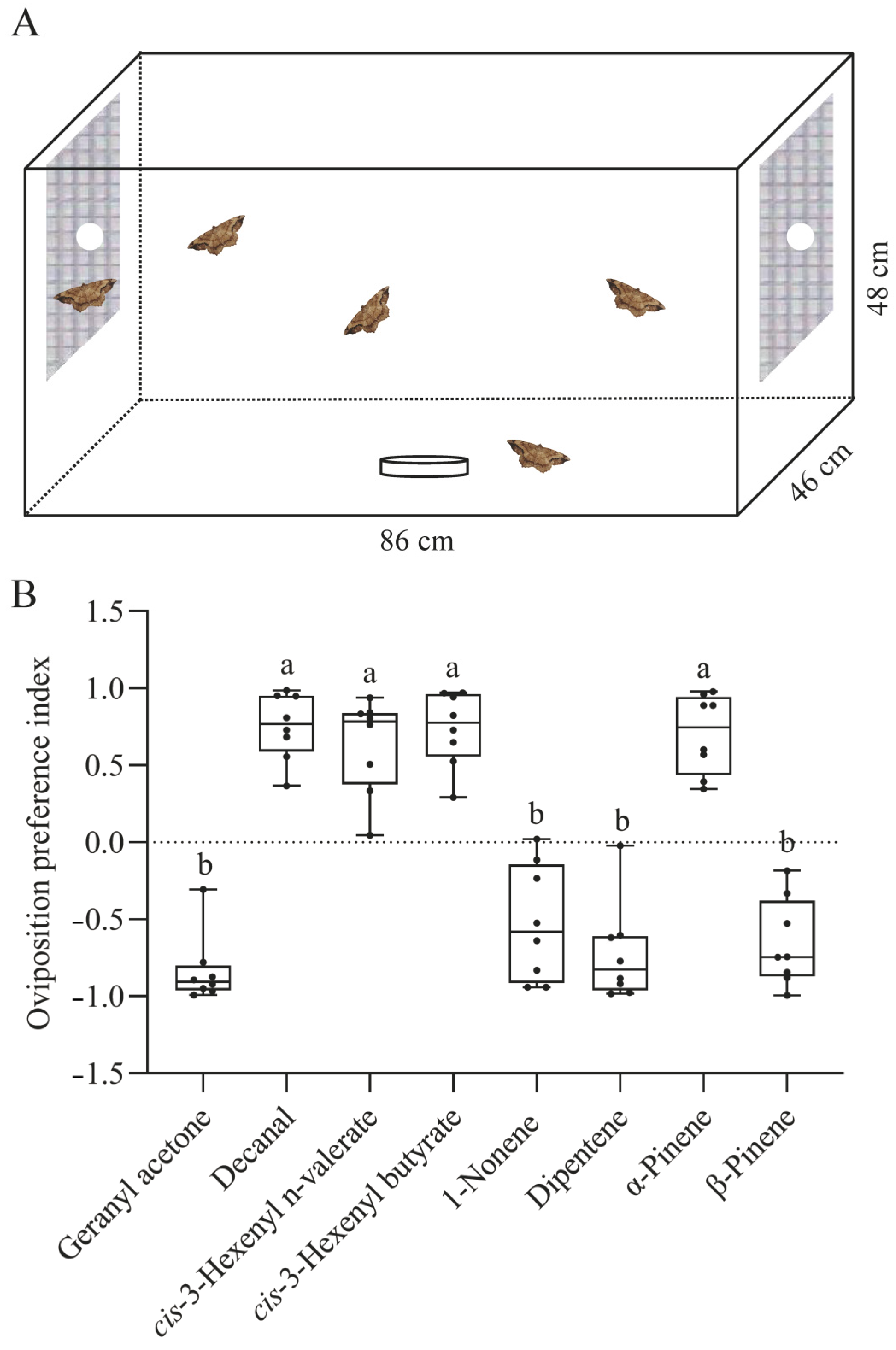
2.8. Two-Choice Oviposition Preference Assay
2.9. Statistical Analysis
3. Results
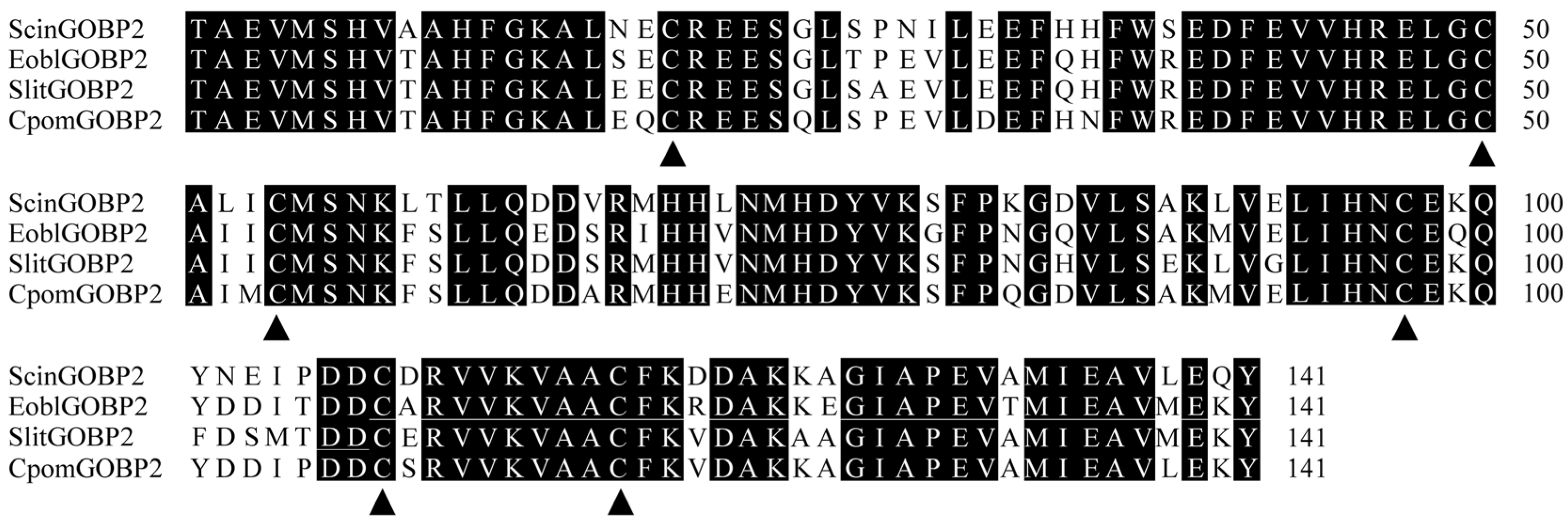
3.1. Gene Cloning and Sequence Analysis of ScinGOBP2
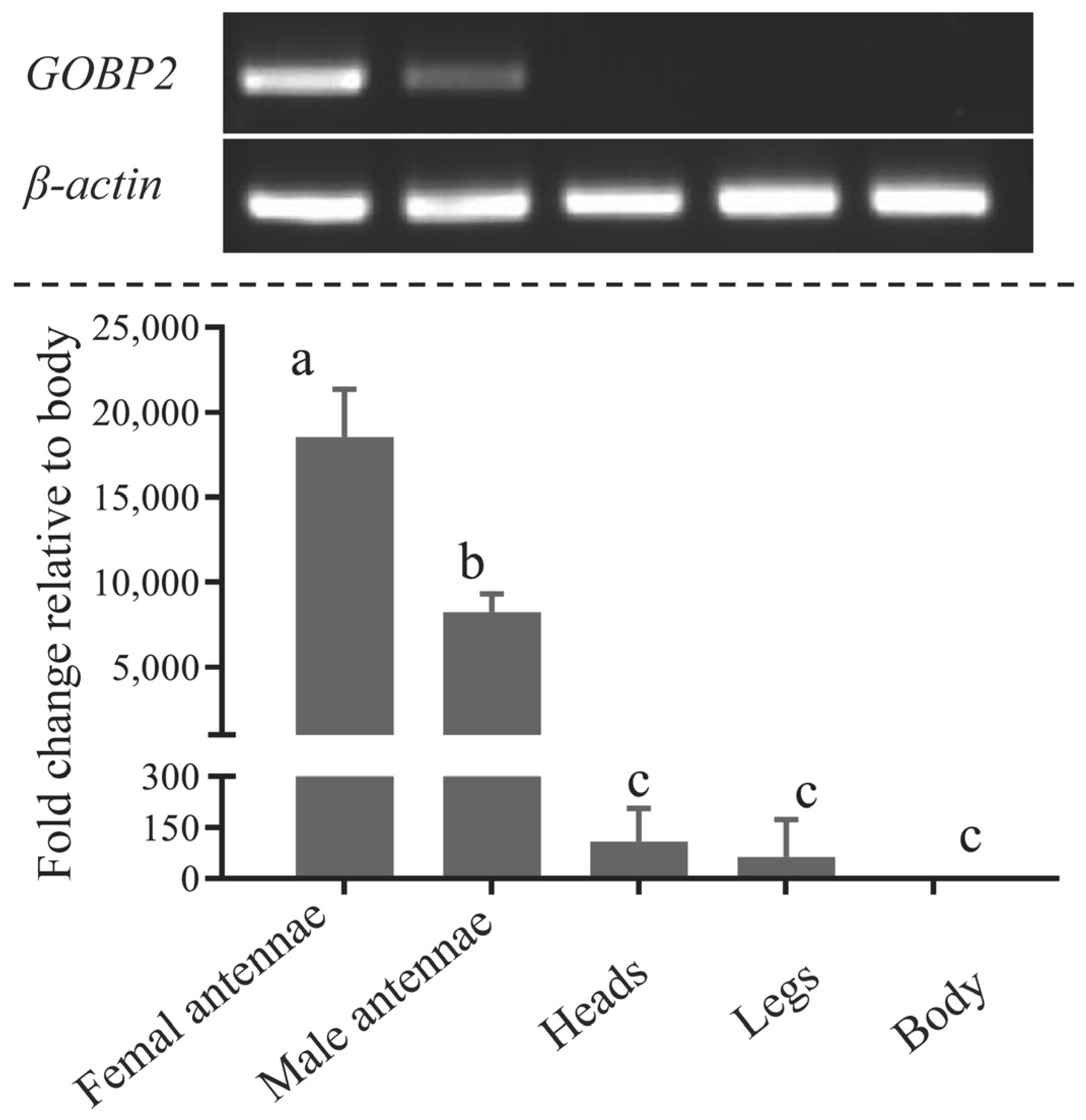
3.2. Tissue Expression Patterns of ScinGOBP2
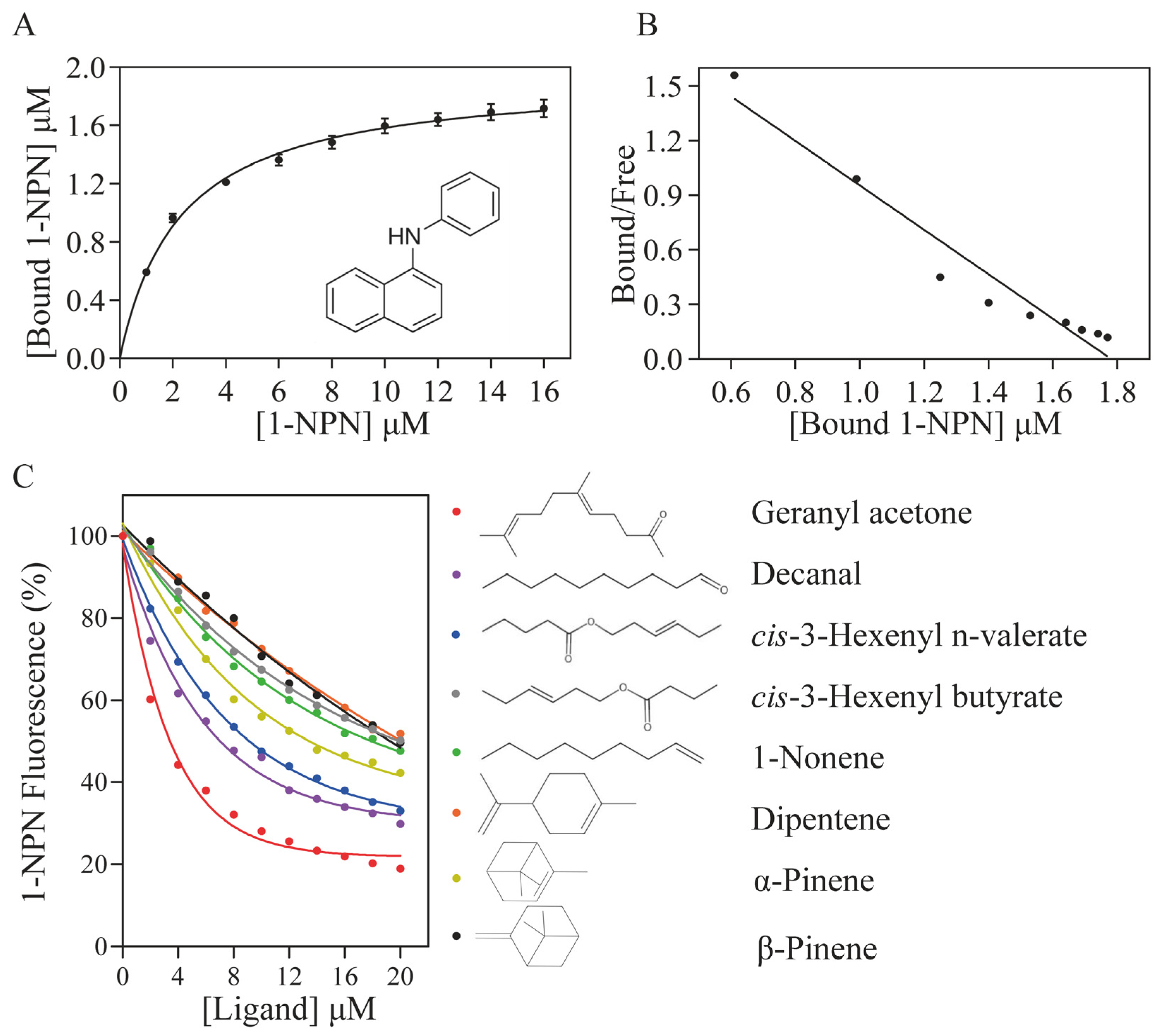
3.3. Binding Characteristics of Recombinant ScinGOBP2
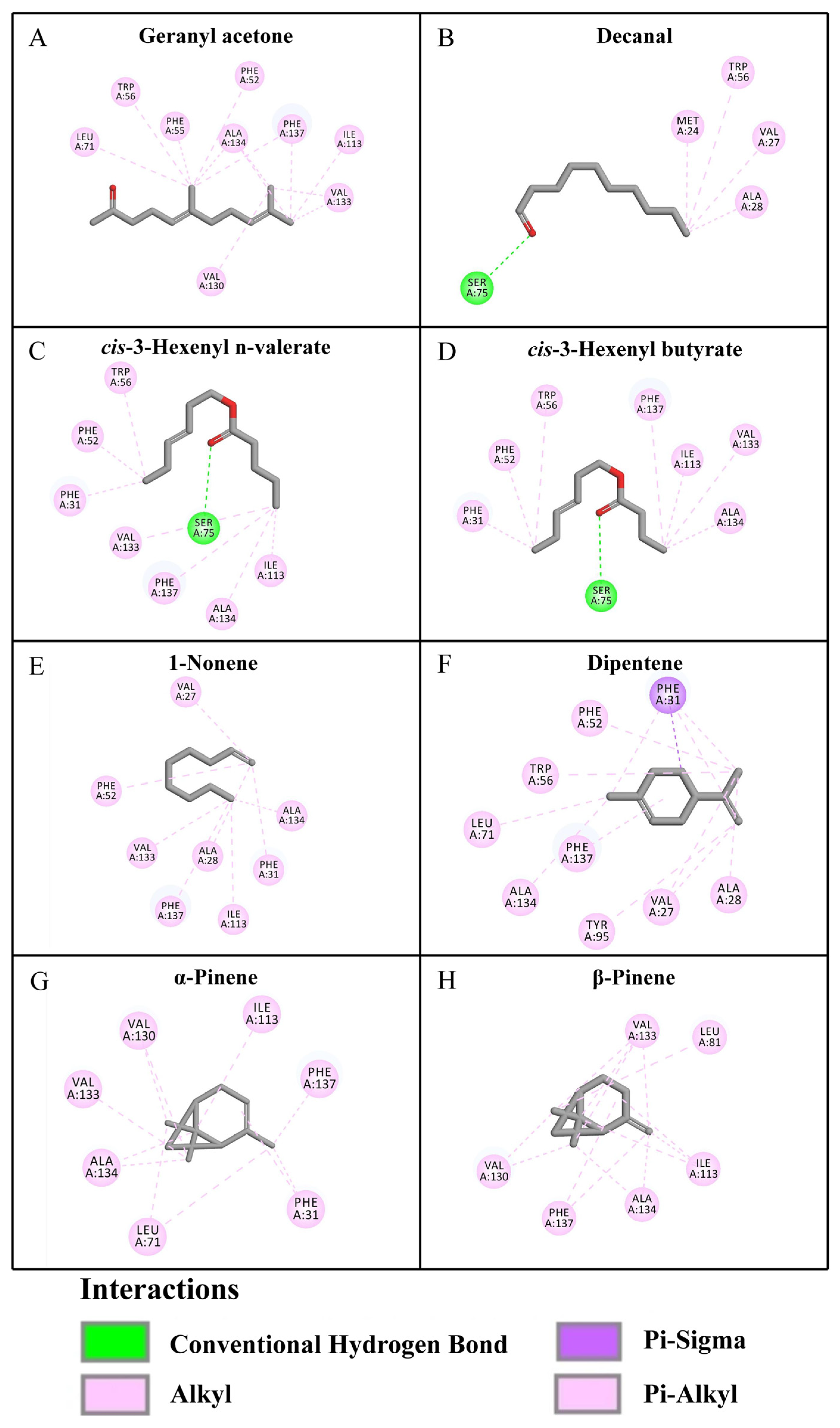
3.4. Protein Structure Prediction and Molecular Docking
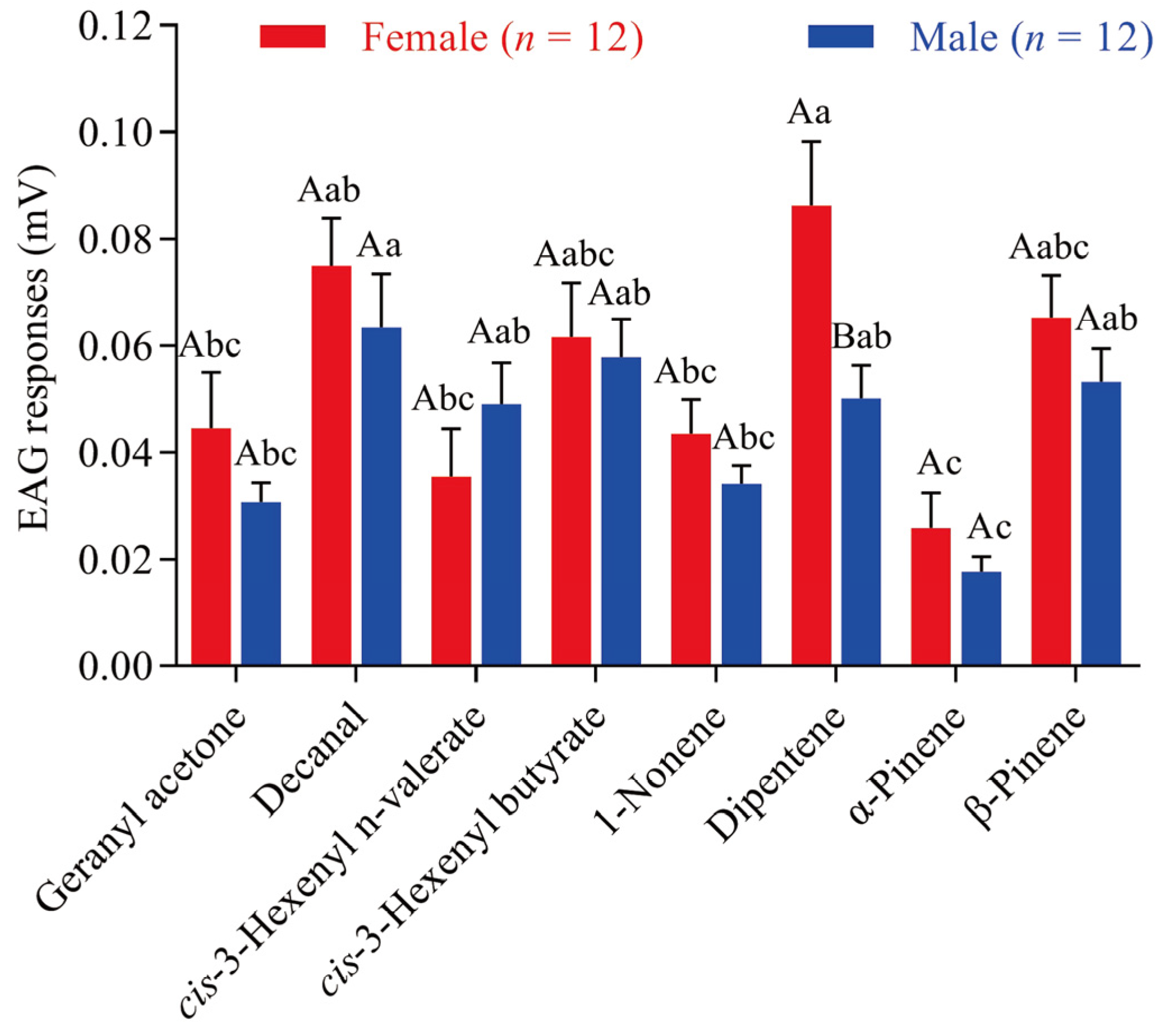
3.5. EAG Recordings
3.6. Two-Choice Oviposition Assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bruce, T.J.A.; Wadhams, L.J.; Woodcock, C.M. Insect host location, a volatile situation. Trends Plant Sci. 2005, 10, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, R.L.; Kogan, M. Plant volatiles as insect attractants. Crit. Rev. Plant Sci. 1987, 5, 251–301. [Google Scholar] [CrossRef]
- Schoonhoven, L.M.; Van Loon, J.J.A.; Dicke, M. Insect-Plant Biology, 2nd ed.; Oxford University Press: New York, NY, USA, 2005. [Google Scholar]
- Zacharuk, R.Y. Ultrastructure and function of insect chemosensilla. Annu. Rev. Entomol. 1980, 25, 27–47. [Google Scholar] [CrossRef]
- Hansson, B.S.; Stensmyr, M.C. Evolution of insect olfaction. Neuron 2011, 72, 698–711. [Google Scholar] [CrossRef]
- Prelic, S.; Pal Mahadevan, V.; Venkateswaran, V.; Lavista-Llanos, S.; Hansson, B.S.; Wicher, D. Functional interaction between Drosophila olfactory sensory neurons and their support cells. Front. Cell. Neurosci. 2022, 15, 789086. [Google Scholar] [CrossRef]
- Leal, W.S. Odorant reception in insects, roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Rihani, K.; Ferveur, J.F.; Briand, L. The 40-year mystery of insect odorant-binding proteins. Biomolecules 2021, 11, 509. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.S.; Smith, D.P. Recent insights into insect olfactory receptors and odorant-binding proteins. Insects 2022, 13, 926. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb. Philos. Soc. 2018, 93, 184–200. [Google Scholar] [CrossRef]
- Zhou, J.J. Odorant-binding proteins in insects. Vitam. Horm. 2010, 83, 241–272. [Google Scholar]
- Laue, M.; Steinbrecht, R.A.; Ziegelberger, G. Immunocytochemical localization of general odorant-binding protein in olfactory sensilla of the silkmoth Antheraea polyphemus. Naturwissenschaften 1994, 81, 178–180. [Google Scholar] [CrossRef]
- Vogt, R.G.; Große-Wilde, E.; Zhou, J.J. The Lepidoptera odorant binding protein gene family, gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 2015, 62, 142–153. [Google Scholar] [CrossRef]
- Nardi, J.B.; Miller, L.A.; Walden, K.K.O.; Rovelstad, S.; Wang, L.; Frye, J.C.; Ramsdell, K.; Deem, L.S.; Robertson, H.M. Expression patterns of odorant-binding proteins in antennae of the moth Manduca sexta. Cell Tissue Res. 2003, 313, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.Z.; Liu, J.T.; Zhou, J.J.; Wang, Q.; Dong, J.Z.; Zhang, Y.J.; Li, X.C.; Li, J.; Gu, S.H. Expressional and functional comparisons of two general odorant binding proteins in Agrotis ipsilon. Insect Biochem. Mol. Biol. 2018, 98, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Wu, Z.R.; Liao, W.; Zhang, X.Q.; Pei, Y.W.; Lu, M. The binding affinity of two general odorant binding proteins in Spodoptera frugiperda to general volatiles and insecticides. Int. J. Biol. Macromol. 2023, 252, 126338. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, Y.; Wei, Z.-Q.; Hou, J.H.; Si, Y.X.; Zhang, J.; Dong, S.L.; Yan, Q. Identification and functional characterization of general odorant binding proteins in Orthaga achatina. Insects 2023, 14, 216. [Google Scholar] [CrossRef] [PubMed]
- Han, W.K.; Wei, Z.Q.; Yang, Y.L.; Liu, X.L.; Yan, Q.; Zhang, J.; Dong, S.L. Roles of GOBP1 in perception of host plant volatiles revealed by CRISPR/Cas9-mediated mutagenesis in Spodoptera litura. J. Appl. Entomol. 2023, 147, 602–610. [Google Scholar] [CrossRef]
- Jing, D.; Prabu, S.; Zhang, T.; Bai, S.; He, K.; Wang, Z. Genetic knockout and general odorant-binding/chemosensory protein interactions, Revealing the function and importance of GOBP2 in the yellow peach moth’s olfactory system. Int. J. Biol. Macromol. 2021, 193, 1659–1668. [Google Scholar] [CrossRef]
- Liu, Z.; Vidal, D.M.; Syed, Z.; Ishida, Y.; Leal, W.S. Pheromone binding to general odorant-binding proteins from the navel orange worm. J. Chem. Ecol. 2010, 36, 787–794. [Google Scholar] [CrossRef]
- Han, W.K.; Yang, Y.L.; Si, Y.X.; Wei, Z.Q.; Liu, S.R.; Liu, X.L.; Yan, Q.; Dong, S.L. Involvement of GOBP2 in the perception of a sex pheromone component in both larval and adult Spodoptera litura revealed using CRISPR/Cas9 mutagenesis. Insect Biochem. Mol. Biol. 2022, 141, 103719. [Google Scholar] [CrossRef]
- Guo, H.; Guo, P.P.; Sun, Y.L.; Huang, L.Q.; Wang, C.Z. Contribution of odorant binding proteins to olfactory detection of (Z)-11-hexadecenal in Helicoverpa armigera. Insect Biochem. Mol. Biol. 2021, 131, 103554. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.J.; Tang, J.M.; Chen, Z.Y.; Jiang, Y.S.; Wang, Z.F.; Wei, X. Analysis of genetic diversity and population structure in Sophora japonica Linn. in China with newly developed SSR markers. Plant Mol. Biol. Rep. 2019, 37, 87–97. [Google Scholar] [CrossRef]
- Yu, S.U.N.; Zuo-deng, P.E.N.G. Insights into history culture and value of Sophora japonica. J. Beijing For. Univ. Soc. Sci. 2018, 17, 23–31. [Google Scholar]
- Liu, P.; Zhang, X.; Meng, R.; Liu, C.; Li, M.; Zhang, T. Identification of chemosensory genes from the antennal transcriptome of Semiothisa cinerearia. PLoS ONE 2020, 15, e0237134. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Y.; Xu, J.W.; Li, L.L.; Wang, D.Y.; Zhang, M.L.; Yu, N.N.; Purba, E.R.; Zhang, F.; Li, X.M.; Zhang, Y.N.; et al. Analysis of chemosensory genes in Semiothisa cinerearia reveals sex-specific contributions for type-II sex pheromone chemosensation. Genomics 2020, 112, 3846–3855. [Google Scholar] [CrossRef] [PubMed]
- Yan, G. Releasing Variation and Effects on Human Health of Volatile Organic Compounds from Landscape Trees in Beijing. Ph.D. Thesis, Beijing Forestry University, Beijing, China, 2005. [Google Scholar]
- Lescop, E.; Briand, L.; Pernollet, J.C.; Guittet, E. Structural basis of the broad specificity of a general odorant-binding protein from honeybee. Biochemistry 2009, 48, 2431–2441. [Google Scholar] [CrossRef] [PubMed]
- Rojas, V.; Jiménez, H.; Palma-Millanao, R.; González-González, A.; Machuca, J.; Godoy, R.; Ceballos, R.; Mutis, A.; Venthur, H. Analysis of the grapevine moth Lobesia botrana antennal transcriptome and expression of odorant-binding and chemosensory proteins. Comp. Biochem. Phys. D Genom. Proteom. 2018, 27, 244. [Google Scholar] [CrossRef] [PubMed]
- Jing, D.; Zhang, T.; Bai, S.; He, K.; Prabu, S.; Luan, J.; Wang, Z. Sexual-biased gene expression of olfactory-related genes in the antennae of Conogethes pinicolalis (Lepidoptera, Crambidae). BMC Genom. 2020, 21, 244. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Dong, J.; Sun, Y.; Hu, Z.; Lv, Q.; Li, D. Antennal transcriptome analysis and expression profiles of putative chemosensory soluble proteins in Histia rhodope Cramer (Lepidoptera, Zygaenidae). Comp. Biochem. Phys. D Genom. Proteom. 2020, 33, 100654. [Google Scholar] [CrossRef]
- Yang, L.; Tian, X.; Gui, L.; Wang, F.; Zhang, G. Key amino acid residues involved in binding interactions between Bactrocera minax odorant-binding protein 3 (BminOBP3) and undecanol. Insects 2023, 14, 745. [Google Scholar] [CrossRef]
- Yin, J.; Zhuang, X.; Wang, Q.; Cao, Y.; Zhang, S.; Xiao, C.; Li, K. Three amino acid residues of an odorant-binding protein are involved in binding odours in Loxostege sticticalis L. Insect Mol. Biol. 2015, 24, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Conboy, N.J.A.; Mcdaniel, T.; George, D.; Ormerod, A.; Tosh, C.R. Volatile organic compounds as insect repellents and plant elicitors, An integrated pest management (IPM) strategy for glasshouse whitefly (Trialeurodes vaporariorum). J. Chem. Ecol. 2020, 46, 1090–1104. [Google Scholar] [CrossRef]
- Fan, J.; Zheng, K.; Xie, P.; Dong, Y.; Gu, Y.; Wickham, J.D. Electrophysiological and behavioral responses of Batocera horsfieldi Hope to volatiles from Pistacia chinensis Bunge. Insects 2023, 14, 911. [Google Scholar] [CrossRef] [PubMed]
- Prokopy, R.J.; Phelan, P.L.; Wright, S.E.; Minalga, A.J.; Barger, R.; Leskey, T.C. Compounds from host fruit odor attractive to adult plum curculios (Coleoptera, Curculionidae). J. Entomol. Sci. 2001, 36, 122–134. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, M.; Aikeremu, F.; Huang, H.; You, M.; Zhao, Q. Involvement of three chemosensory proteins in perception of host plant volatiles in the tea green leafhopper, Empoasca onukii. Front. Physiol. 2023, 13, 2687. [Google Scholar] [CrossRef]
- Lindmark, M.; Ganji, S.; Wallin, E.A.; Schlyter, F.; Unelius, C.R. Semiochemicals produced by fungal bark beetle symbiont Endoconidiophora rufipennis and the discovery of an anti-attractant for Ips typographus. PLoS ONE 2023, 18, e0283906. [Google Scholar] [CrossRef]
- Borrero-Echeverry, F.; Bengtsson, M.; Nakamuta, K.; Witzgall, P. Plant odor and sex pheromone are integral elements of specific mate recognition in an insect herbivore. Evolution 2018, 72, 2225–2233. [Google Scholar] [CrossRef]
- Jarrett, B.J.M.; Miller, C.W. Host plant effects on sexual selection dynamics in phytophagous insects. Annu. Rev. Entomol. 2023, 69, 41–57. [Google Scholar] [CrossRef]
- Deng, J.Y.; Wei, H.Y.; Huang, Y.P.; Du, J.W. Enhancement of attraction to sex pheromones of Spodoptera exigua by volatile compounds produced by host plants. J. Chem. Ecol. 2004, 30, 2037–2045. [Google Scholar] [CrossRef]
- Gemeno, C.; Leal, W.S.; Mori, K.; Schal, C. Behavioral and electrophysiological responses of the brownbanded cockroach, Supella longipalpa, to stereoisomers of its sex pheromone, supellapyrone. J. Chem. Ecol. 2003, 29, 1797–1811. [Google Scholar] [CrossRef]
- Deletre, E.; Chandre, F.; Williams, L.; Duménil, C.; Menut, C.; Martin, T. Electrophysiological and behavioral characterization of bioactive compounds of the Thymus vulgaris, Cymbopogon winterianus, Cuminum cyminum and Cinnamomum zeylanicum essential oils against Anopheles gambiae and prospects for their use as bednet treatments. Parasites Vectors 2015, 8, 316. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.; Busula, A.O.; Voets, M.A.; Beshir, K.B.; Caulfield, J.C.; Powers, S.J.; Verhulst, N.O.; Winskill, P.; Muwanguzi, J.; Birkett, M.A.; et al. Plasmodium-associated changes in human odor attract mosquitoes. Proc. Natl. Acad. Sci. USA 2018, 115, E4209–E4218. [Google Scholar] [CrossRef] [PubMed]
| Ligand | Source | CAS Number | Purity (%) | KD (μM) * |
|---|---|---|---|---|
| Geranyl acetone | Macklin | 3796-70-1 | ≥98% | 2.21 ± 0.17 |
| Acetophenone | Macklin | 98-86-2 | ≥99.5% | – |
| cis-3-Hexen-1-ol | Macklin | 928-96-1 | 98% | – |
| cis-2-Nonen-1-ol | TCI | 41453-56-9 | >95.0% | – |
| 2-Ethylhexan-1-ol | TCI | 104-76-7 | >99.5% | – |
| 1-Octanol | TCI | 111-87-5 | >99% | – |
| Hexanal | TCI | 66-25-1 | >98% | – |
| Nonanal | TCI | 124-19-6 | >95% | – |
| Decanal | TCI | 112-31-2 | >97% | 5.07 ± 0.27 |
| 3-Methyl-2-butenal | Macklin | 107-86-8 | 98% | – |
| 2-Hexenal | Macklin | 6728-26-3 | 98% | – |
| Octane | Macklin | 111-65-9 | >99% | – |
| cis-3-Hexenyl acetate | Macklin | 3681-71-8 | 98% | – |
| cis-3-Hexenyl n-valerate | Macklin | 35852-46-1 | 98% | 6.55 ± 0.18 |
| Ethyl acetate | Macklin | 141-78-6 | ≥99.7% | – |
| Butyl acetate | Macklin | 123-86-4 | ≥99.7% | – |
| Hexyl acetate | TCI | 142-92-7 | >99% | – |
| cis-3-Hexenyl butyrate | Macklin | 16491-36-4 | ≥98% | 14.17 ± 0.04 |
| Nonanoic acid | Macklin | 112-05-0 | >99% | – |
| Capric Acid | Macklin | 334-48-5 | >99% | – |
| 2-octeno | TCI | 111-67-1 | >95% | – |
| 1-Nonene | Macklin | 124-11-8 | 95% | 12.20 ± 0.44 |
| Dipentene | Macklin | 7705-14-8 | 95% | 14.94 ± 0.77 |
| Ocimene | Macklin | 13877-91-3 | ≥90% | – |
| α-Pinene | Macklin | 80-56-8 | 98% | 9.41 ± 0.28 |
| β-Pinene | Macklin | 127-91-3 | ≥95% | 13.19 ± 0.19 |
| Isoprene | Macklin | 78-79-5 | >99% | – |
| Ligand | Binding Energy (kcal/mol) | Hydrophobic Interactions |
|---|---|---|
| Geranyl acetone | −7.3 | Phe52, Phe55, Trp56, Leu71, Ile113, Val130, Val133, Ala134, Phe137 |
| Decanal | −5.7 | Trp56, Met24, Val27, Ala28 |
| cis-3-Hexenyl n-valerate | −6.2 | Phe31, Phe52, Trp56, Ile113, Val133, Ala134, Phe137 |
| cis-3-Hexenyl butyrate | −5.9 | Phe31, Phe52, Trp56, Ile113, Val133, Ala134, Phe137 |
| 1-Nonene | −5.3 | Val27, Phe52, Val133, Phe137, Ala28, Ile113, Phe31, Ala134 |
| Dipentene | −6.8 | Phe52, Trp56, Leu71, Phe137, Ala134, Tyr95, Val27, Ala28 |
| α-Pinene | −6.2 | Phe31, Leu71, Ile113, Val130, Val133, Ala134, Phe137 |
| β-Pinene | −6.3 | Phe31, Leu71, Ile113, Val130, Val133, Ala134, Phe137 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, J.; Wang, Z.; Yang, F.; Liu, H.; Qiao, G.; Zhang, A.; Wang, S. The Female-Biased General Odorant Binding Protein 2 of Semiothisa cinerearia Displays Binding Affinity for Biologically Active Host Plant Volatiles. Biology 2024, 13, 274. https://doi.org/10.3390/biology13040274
Tu J, Wang Z, Yang F, Liu H, Qiao G, Zhang A, Wang S. The Female-Biased General Odorant Binding Protein 2 of Semiothisa cinerearia Displays Binding Affinity for Biologically Active Host Plant Volatiles. Biology. 2024; 13(4):274. https://doi.org/10.3390/biology13040274
Chicago/Turabian StyleTu, Jingjing, Zehua Wang, Fan Yang, Han Liu, Guanghang Qiao, Aihuan Zhang, and Shanning Wang. 2024. "The Female-Biased General Odorant Binding Protein 2 of Semiothisa cinerearia Displays Binding Affinity for Biologically Active Host Plant Volatiles" Biology 13, no. 4: 274. https://doi.org/10.3390/biology13040274
APA StyleTu, J., Wang, Z., Yang, F., Liu, H., Qiao, G., Zhang, A., & Wang, S. (2024). The Female-Biased General Odorant Binding Protein 2 of Semiothisa cinerearia Displays Binding Affinity for Biologically Active Host Plant Volatiles. Biology, 13(4), 274. https://doi.org/10.3390/biology13040274






