Simple Summary
The protein arginine N-methyltransferase 5 (PRMT5) has been identified as a promising therapeutic target in various cancers. However, its role in hepatocellular carcinoma (HCC) development has not yet been investigated. This study aims to understand PRMT5′s impact on overall survival, signaling pathways, and downstream gene expression using in silico public databases and our in-house NGS data. Our results revealed an increase in PRMT5 expression in HCC compared to normal liver tissue, and this elevated expression was associated with poorer patient outcomes. Analysis of promoter CpG islands and methylation status suggested potential epigenetic mechanisms driving PRMT5 overexpression in HCC. Pathway analyses found a link between PRMT5 expression and the HIF1α pathway, with Ras-related nuclear protein (RAN) identified as a potential target of PRMT5 in HCC.
Abstract
Protein arginine N-methyltransferase 5 (PRMT5) has been identified as a potential therapeutic target for various cancer types. However, its role in regulating the hepatocellular carcinoma (HCC) transcriptome remains poorly understood. In this study, publicly available databases were employed to investigate PRMT5 expression, its correlation with overall survival, targeted pathways, and genes of interest in HCC. Additionally, we utilized in-house generated NGS data to explore PRMT5 expression in dysplastic nodules compared to hepatocellular carcinoma. Our findings revealed that PRMT5 is significantly overexpressed in HCC compared to normal liver, and elevated expression correlates with poor overall survival. To gain insights into the mechanism driving PRMT5 overexpression in HCC, we analyzed promoter CpG islands and methylation status in HCC compared to normal tissues. Pathway analysis of PRMT5 knockdown in the HCC cells revealed a connection between PRMT5 expression and genes related to the HIF1α pathway. Additionally, by filtering PRMT5-correlated genes within the HIF1α pathway and selecting up/downregulated genes in HCC patients, we identified Ras-related nuclear protein (RAN) as a target associated with overall survival. For the first time, we report that PRMT5 is implicated in the regulation of HIF1A and RAN genes, suggesting the potential prognostic utility of PRMT5 in HCC.
1. Introduction
According to the records of the International Agency for Research on Cancer (IARC), in 2020, primary liver cancer was ranked sixth in incidence and third in mortality rate. The vast majority, approximately 80%, of primary liver cancer cases are hepatocellular carcinoma (HCC) [1]. Several risk factors have been associated with HCC, including metabolic liver diseases, such as nonalcoholic fatty liver (NAFL) and nonalcoholic steatohepatitis (NASH), chronic viral infections, such as hepatitis B virus (HBV) and hepatitis C virus (HCV), alcohol abuse, inherited diseases, and Aflatoxin exposure [2]. HCC is classified into five stages, and treatment is determined based on the stage. It begins with resection and liver transplantation in the early stages, followed by targeted therapy (such as Sorafenib) in the advanced stages [3,4]. Most of the early-stage cases achieve a 5-year survival rate, and 20–50% of intermediate-stage cases attain a 3-year survival rate. Unfortunately, terminal-stage patients succumb to the disease within 6 months [5,6]. Therefore, identifying new diagnostic biomarkers, therapeutic targets, and prognostic markers is critical for diagnosing and treating the disease at its early-intermediate stages.
Recent studies have suggested that protein arginine N-methyltransferase 5 (PRMT5) could serve as a potential prognostic and therapeutic biomarker in various cancers, including breast, lung, and colorectal cancer [7,8,9]. PRMT5 functions as a catalytic enzyme that transfers methyl groups from S-adenosylmethionine (SAM) to arginine residues in multiple proteins, including histones [10]. Arginine monomethylation or symmetric dimethylation induced by PRMT5 affects cellular functions by influencing protein activity and stability, gene expression, and pre-mRNA splicing [11,12]. There is also evidence suggesting that PRMT5 plays a role in promoting cancer cell growth and proliferation [9,13]. Interestingly, PRMT5 overexpression has been positively correlated with cellular transformation in various neoplasms, including HCC [14,15,16]. As a potential molecular target, the PRMT5 inhibitor (GSK3326595) is currently in phase I/II clinical trials for acute myeloid leukemia (AML) and other cancer types [17,18]. Also, several studies have established PRMT5 as a reliable prognostic marker for cancers, including HCC [19]. Therefore, investigating PRMT5 expression at different stages of HCC could provide a hint of whether PRMT5 could also serve as a diagnostic marker of early HCC. Moreover, PRMT5 overexpression in the liver has been linked to a high-fat diet, which is a risk factor for HCC [20,21].
Previous research documents the role of PRMT5 in regulating key signaling pathways in HCC, such as the WNT signaling pathway, the ERK signaling pathway, and iron homeostasis. For instance, PRMT5 manipulates WNT signaling activity, which enhances HCC metastases. PRMT5 activates the ERK signaling pathway, which hinders the expression of B-cell translocation gene 2 (BTG2), which is responsible for G1 to S phase cell cycle arrest. On the other hand, PRMT5 exhibited a protective role in HCC via inhibiting ferritin heavy-chain-1 (FTH1) expression which reverses the iron overload process in HCC [22,23,24,25]. However, the role of PRMT5 in regulating the Hypoxia-inducible factor 1 alpha (HIF1α) signaling pathway in HCC has not been previously reported. The HIF1α pathway is involved in HCC proliferation, invasion, metastasis, angiogenesis, and drug resistance [26]. HIF1α is a transcription factor that gets activated under hypoxia condition which is associated with tumor microenvironment. It plays a crucial role in cancer cell survival by inhibiting the generation and propagation of reactive oxygen species (ROS) and by blocking ROS-mediated apoptosis [27,28,29]. HIF1α pathway inhibition demonstrates a promising therapeutic strategy for combating HCC progression and enhancing patient outcomes.
In this study, publicly available data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) were used to investigate the possible role of PRMT5 in regulating the expression of key genes in the HIF1α pathway. Furthermore, GEO data were utilized to assess the capacity of PRMT5 downstream effectors from the HIF1α pathway to serve as prognostic biomarkers. Our data proposed promoter hypermethylation as a mechanism that is involved in PRMT5 increased expression in HCC. Our data revealed an association between PRMT5 upregulation and HIF1α pathway activation, as well as increased HIF1A gene expression. Also, our data highlight the capacity of PRMT5 in regulating RAN gene expression, which is a part of the HIF1α pathway and could serve as a biomarker for HCC prognosis.
2. Materials and Methods
2.1. In Silico Analysis Using UALCAN Online Portal
The UALCAN in silico tool provides access to Level 3 RNA-seq. The UALCAN website (http://ualcan.path.uab.edu/analysis.html, accessed on 29 October 2022) was used to profile gene expression in 371 HCC patients compared to 50 normal counterparts using Cancer Genome Atlas (TCGA) level 3 RNAseq and clinical data [30,31]. In addition, UALCAN was utilized to obtain protein expression analysis via the Clinical Proteomic Tumor Analysis Consortium (CPTAC) and the International Cancer Proteogenomic Consortium (ICPC) datasets that represent High-throughput mass spectrometry data of 165 normal versus 165 HCC patients. The website conducts a Comprehensive Perl Archive Network (CPAN) to calculate the p-value using a Student t-test.
2.2. Methylation Status of Promoter CpG Islands
The genome data viewer browser allowed the visualization of biological information blotted onto the genome in a graph such as CpG islands. A genome data viewer browser provided by the National Health Institute was utilized to identify the presence of CpG islands in the promoter of the gene. The UALCAN tool provides promoter region hypermethylation analysis utilizing TCGA data obtained via Illumina Infinium HumanMethylation450 BeadChip. Promoter methylation level was evaluated in the UALCAN tool using TCGA DNA methylation data of 50 normal compared to 377 HCC cases. The website conducts CPAN to calculate the p-value using a Student t-test.
2.3. Kaplan–Meier Patient Survival Analysis
The Kaplan–Meier plotter (https://kmplot.com/analysis/, accessed on 15 December 2022) was used to analyze RNAseq data to identify overall survival (OS), relapse-free survival (RFS), progression-free survival (PFS), and disease-specific survival (DSS) in 364, 316, 370, and 362 liver cancer patients, respectively [32,33]. A upper quartile cutoff was used to generate a PRMT5 Kaplan–Meier plotter graph of OS, RFS, PFS, and DSS. A lower quartile cutoff was used to generate the RAN Kaplan-Meier plotter graph of OS, RFS, PFS, and DSS. A hazard ratio (HR) of more than 1 was considered as a bad prognosis biomarker, while an HR of less than 1 was considered as a good prognosis. An upper quartile cutoff was used to generate a MAPK3 Kaplan–Meier plotter graph of OS, RFS, PFS, and DSS. Auto select option for the best cutoff value was used to generate an FLT1 and SERPINE1 Kaplan–Meier plotter graph of OS.
2.4. QIAGEN Ingenuity Pathway Analysis (IPA)
Transcriptome data generated by Illumina Novaseq 6000 for PRMT5 knocked down JHH-7 Cell utilizing a short hairpin RNA (shRNA) were obtained from the Gene Expression Omnibus (GEO) database (GSE168745) provided by the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/geo/, accessed on 30 November 2022). Significantly altered gene lists were utilized to perform the IPA using the tool provided by Qiagen to analyze omics data.
2.5. TIMER 2.0
The Tumor Immune Estimation Resource (TIMER) is a database that represents a molecular cross-talk of tumor and tumor microenvironment. TIMER also allows the detection of gene expression correlation in multiple cancers. TIMER (https://cistrome.shinyapps.io/timer/, accessed on 9 January 2023) was used to investigate gene–gene correlations in 371 specimens of liver hepatocellular carcinoma sourced from Cancer Genome Atlas (TCGA) [34,35]. Spearman’s rho value was utilized to identify the degree of the correlation between two genes. An R-value between 0 and 0.3 was considered weak positive, 0.3 to 0.7 moderate positive, and 0.7 to 1 strong positive.
2.6. Next-Generation Sequencing (NGS)
RNA was isolated from 12 samples of dysplastic nodules and 7 samples of HCC tumor tissue. Library preparation was performed using the NEBNext® UltraTM RNA Library Prep Kit for Illumina®, and sequencing was conducted using Illumina Novaseq 6000. The data were aligned to the human reference genome sequence (ENSEMBL Homosapiens.GRCh38) using HISAT2 (hisat2-2.0.2-beta). These data were generated by our co-authors and were previously published [36]. The Bioconductor limma-voom package was utilized for RNA-seq data normalization.
2.7. Statistical Analysis
GraphPad Prism 8.4.2 was used to perform an unpaired parametric t-test to calculate the p-value. Collective data were presented as mean ± SEM. A p-value of <0.05 was considered to be statistically significant. The SPSS program was used to identify the distribution of the dataset and to draw the receiver operating characteristic (ROC) curve utilizing the GEO data set (GSE214846) to assess the gene expression clinical value as a diagnostic marker.
3. Results
3.1. PRMT5 Is Over-Expressed in HCC and Differentially Expressed in Different Disease Stages
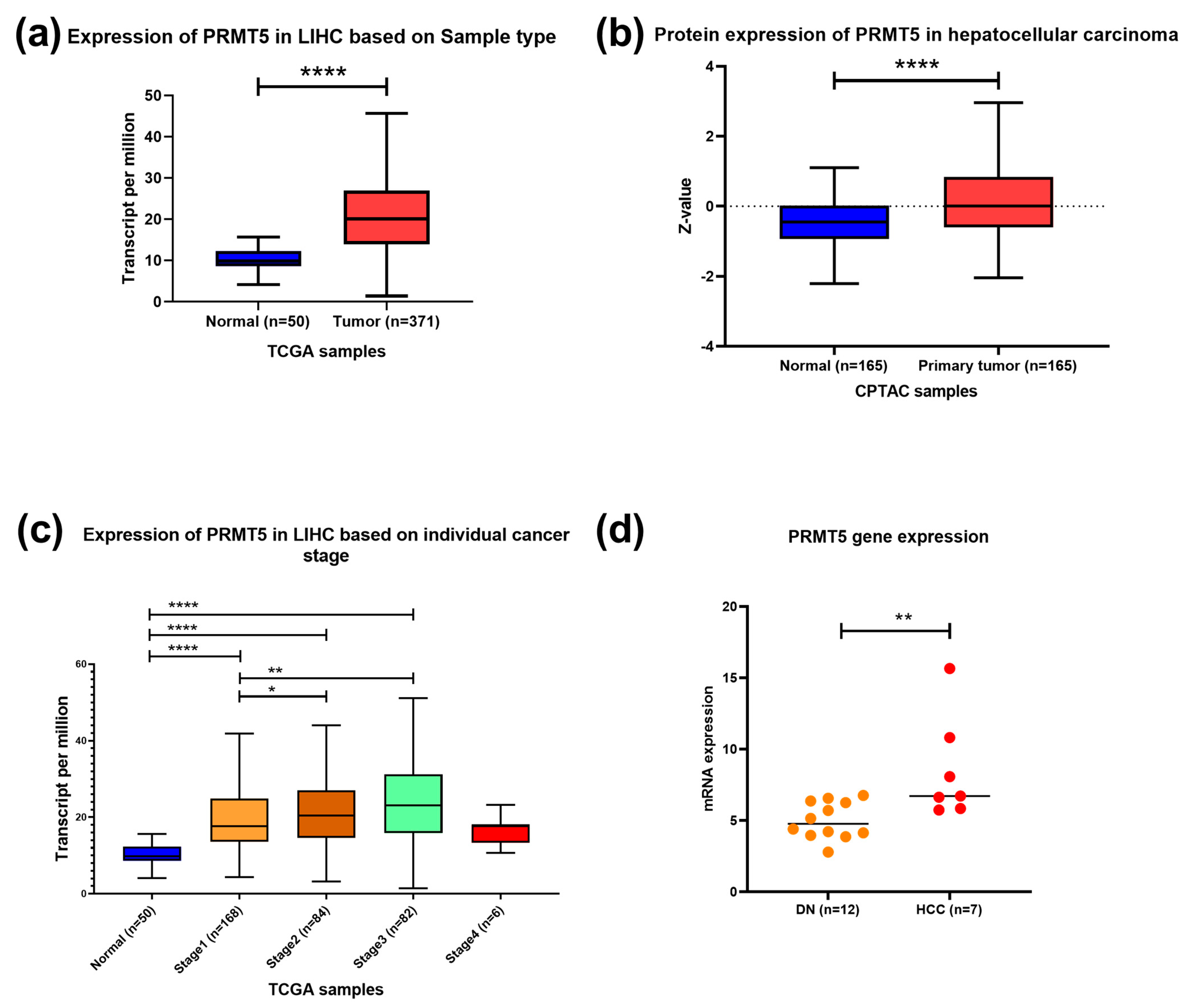
To identify PRMT5 expression in HCC, the UALCAN tool was used to analyze TCGA RNAseq, Clinical Proteomic Tumor Analysis Consortium (CPTAC), and the International Cancer Proteogenome Consortium (ICPC) data. In silico analysis showed that PRMT5 is overexpressed in HCC at the RNA and protein levels with p < 0.0001 (Figure 1a,b). PRMT5 mRNA was significantly overexpressed in all stages of HCC relative to its normal counterpart; it was very highly elevated in stages II (p < 0.05) and III (p < 0.01) relative to stage I (Figure 1c). Additionally, transcriptomic data demonstrated a significant increase in the PRMT5 expression level of HCC compared to dysplastic nodules with p < 0.01 (Figure 1d), indicating a progressive activation of PRMT5 during liver cancer development and progression.
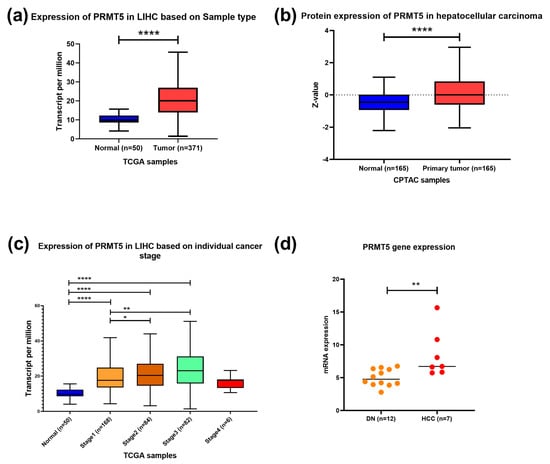
Figure 1.
PRMT5 expression in HCC. (a) PRMT5 expression in HCC clinical samples compared to normal tissue counterpart using RNA seq dataset, TCGA data, UALCAN tool. (b) PRMT5 expression in HCC clinical samples compared to normal tissue counterpart using clinical proteomic tumor analysis consortium (CPTAC) data, UALCAN tool. (c) PRMT5 expression in different stages of HCC compared to normal tissue counterpart using RNA seq dataset from TCGA data, UALCAN tool. (d) transcriptome data of PRMT5 expression in 12 samples of dysplastic nodules and 7 samples of hepatocellular carcinoma. * p < 0.05, ** p < 0.01, and **** p < 0.0001.
3.2. PRMT5 Is a Promising Disease Progression Marker for HCC
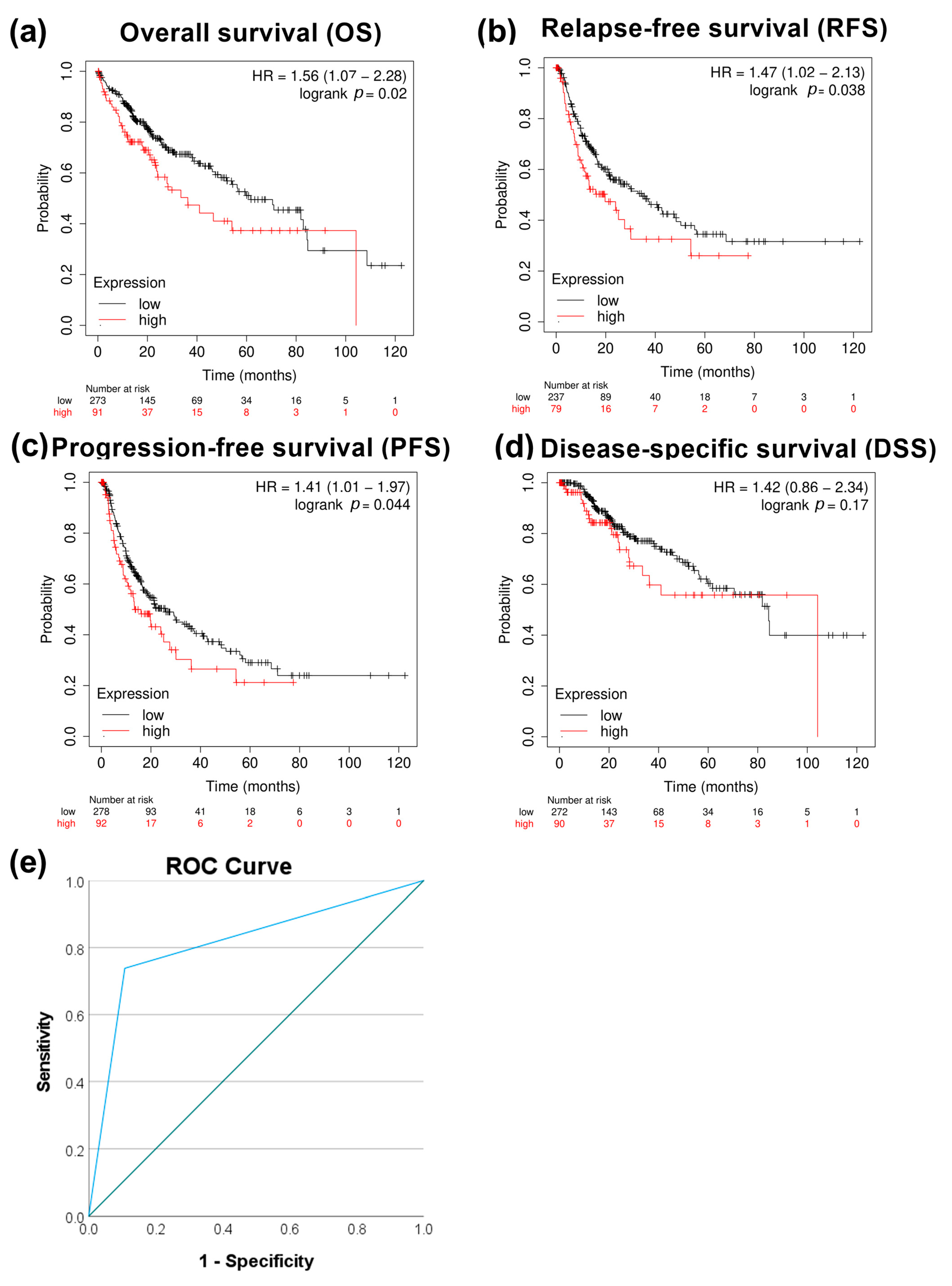
We next evaluated the correlation between PRMT5 expression and patient survival. High expression of PRMT5 significantly correlated with poor overall survival p < 0.05, relapse-free survival with p < 0.05, and progression-free survival with p < 0.05, except disease-specific survival with p > 0.05 in HCC patients (Figure 2a–d). The capacity of PRMT5 to serve as a disease progression marker ROC curve was also evaluated by comparing HCC versus normal adjacent tissues using the GSE214846 dataset. This type of analysis revealed that the area under the curve is 81.5%, sensitivity 73.8%, and specificity 89.2%, with a cutoff value of 5 (Figure 2e). This highlights the capability of PRMT5 to serve as a biomarker in HCC.
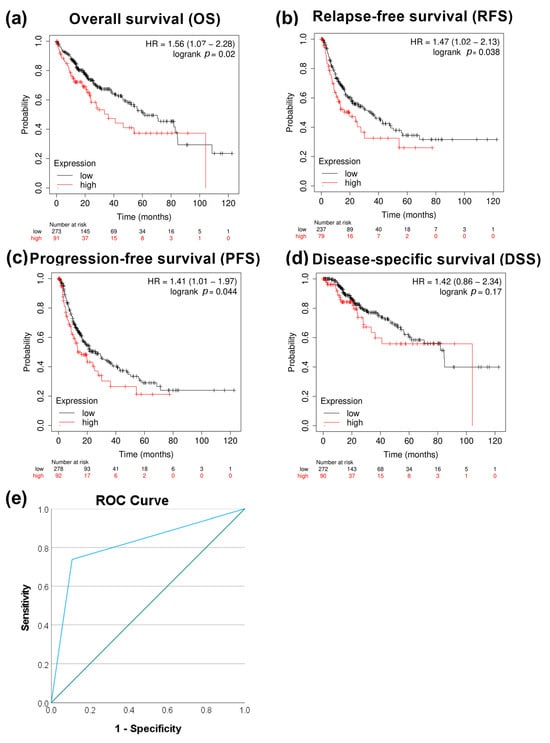
Figure 2.
Correlational analysis of PRMT5 expression versus HCC patient survival. (a) PRMT5 expression versus overall patient survival (OS). (b) PRMT5 expression versus relapse-free survival (RFS). (c) PRMT5 expression versus progression-free survival (PFS). (d) PRMT5 expression versus disease-specific survival (DSS). (e) ROC curves for PRMT5 expression at adjacent normal versus HCC tissue utilizing the GSE214846 dataset.
3.3. PRMT5 Promoter Is Hypomethylated in HCC
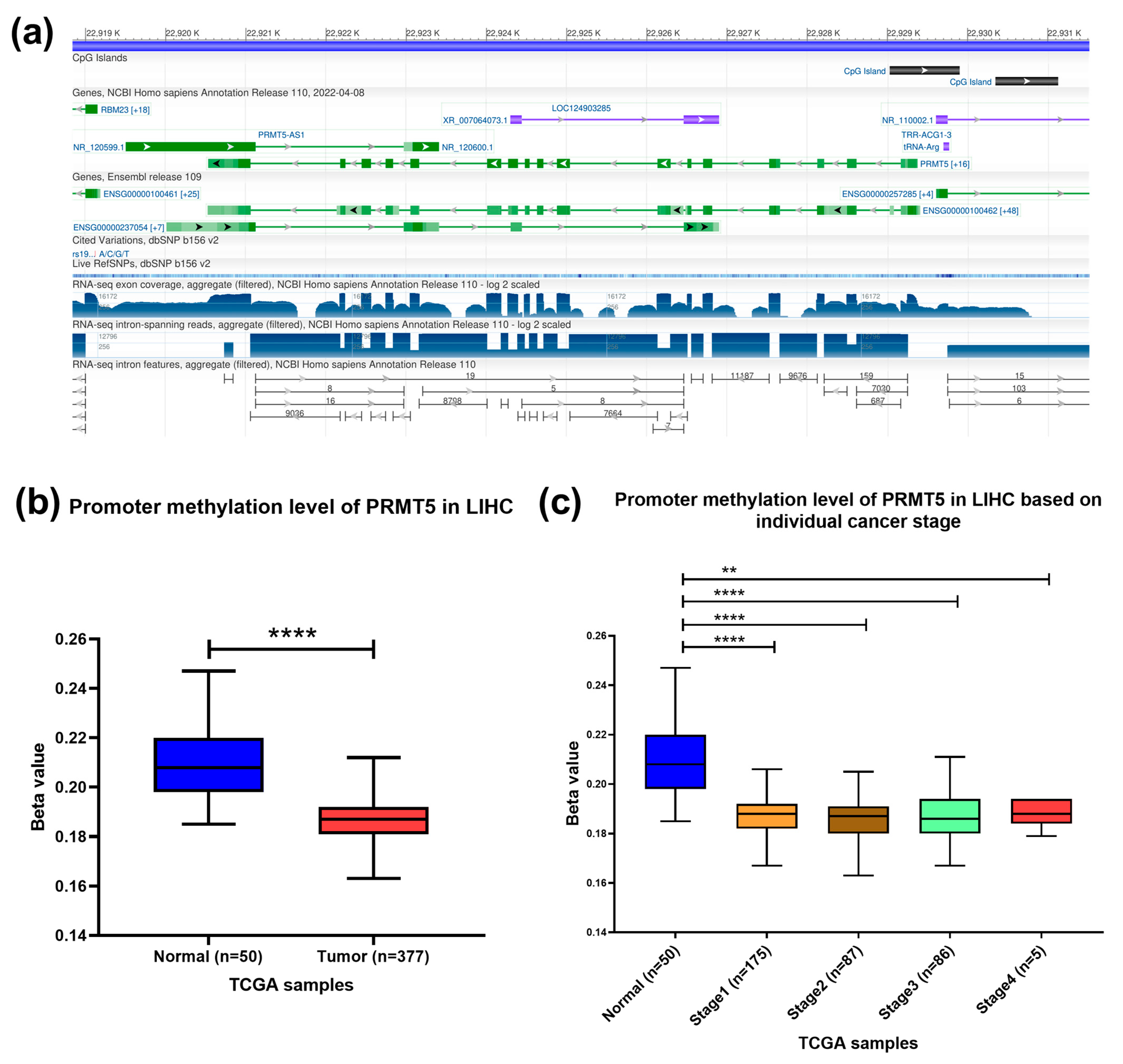
To confirm the presence of CpG islands in the promoter region of the PRMT5 gene, the Genome data viewer browser was used. An 872-nucleotide long cytosine- and guanine-rich domain present at NC_000014.9 [22,929,030–22,929,901] chromosome 14 that contains promoter region of PRMT5 gene was identified (Figure 3a). PRMT5 promoter methylation status was assessed using the publicly available tool. The results showed a significant hypomethylation at the promoter region of PRMT5 in HCC (p < 0.0001) patients compared to normal counterparts, especially in early HCC stages (p < 0.0001) (Figure 3b,c). Indicating the role of epigenetic hypomethylation of PRMT5 promoter region in PRMT5 overexpression in the early stage of HCC.
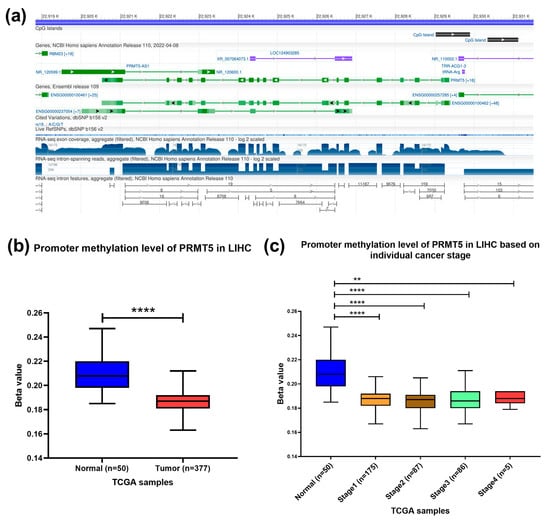
Figure 3.
Methylation level of PRMT5 promoter CpG Islands in HCC. (a) Location of CpG islands in the promoter region of PRMT5 gene. (b) PRMT5 promoter methylation level in HCC compared to normal tissues counterpart as per the TCGA database, UALCAN tool. (c) PRMT5 promoter methylation level at different stages of HCC compared to normal tissue counterpart as per the TCGA database, UALCAN tool., ** p < 0.01, and **** p < 0.0001.
3.4. PRMT5 Knockdown Manipulates the Activity of the HIF1α Pathway
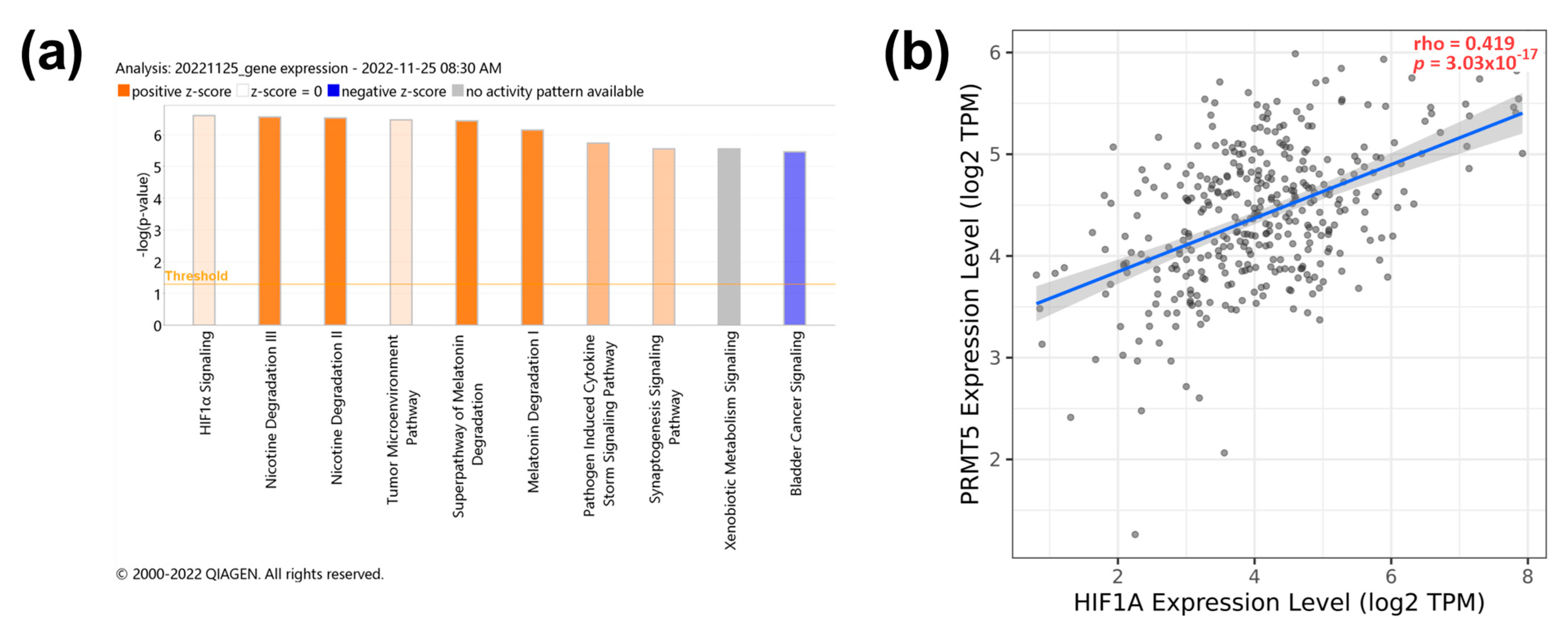
To investigate the pathways related to PRMT5 expression, transcriptomic analysis of the PRMT5-silenced JHH-7 HCC cell line was performed using IPA (−log (p-value) = 6.6). PRMT5 depletion significantly reduced the expression of genes involved in the HIF-1α pathway (Figure 4a). In that, HIF1A gene expression showed a moderate positive correlation with PRMT5 expression in HCC patients with p < 0.0001 (Figure 4b). The detailed IPA visual pathway is shown in Supplementary Figure S1, showing that PRMT5 silencing helped in the deactivation of HIF-1α via reduced expression of key genes such as MAPK3 (ERK1/2) and ribosomal S6 kinase B2 (RPS6KB2 or p70S6Kb). Also, inhibition of angiogenesis and blood vessel maturation pathways was evident as a decreased expression of the vascular endothelial growth factor receptor 1 (FLT1) and the plasminogen activator inhibitor-1 (SERPINE1) (Supplementary Figure S1). PRMT5 showed a significant (p < 0.0001) but weak positive (r < 0.3) correlation with FLT1 and SERPINE1 (Supplementary Figure S3a,c). It is worth noting that FLT1 and SERPINE1 expression were not significantly associated with overall survival according to Kaplan–Meier patient survival analysis (Supplementary Figure S3b,d). PRMT5 inhibition also reduced the expression of Ras-related nuclear protein (RAN) (Supplementary Figure S1). Collectively, these data indicate that PRMT5 enhances the expression of genes involved in tumor progression via HIF-1α pathway activation.
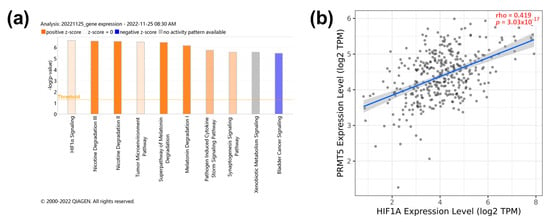
Figure 4.
PRMT5 expression positively correlates with HIF1α signaling pathway. (a) IPA pathway analysis of PRMT5 knocked down JHH-7. (b) Spearman’s rank correlation coefficient of PRMT5 versus HIF1α expression.
3.5. PRMT5 Expression Is Positively Correlated with MAPK3, and RAN Genes as a Key Regulator in HCC
To further understand the relationship between PRMT5 and the HIF1α pathway, the possibility that PRMT5 could be involved in the regulation of the key HIF1α pathway was investigated. In silico analysis was performed to address this point, with RAN and MAPK3 as the genes that were significantly co-expressed with PRMT5 in HCC patients, the genes that correlated with worse prognosis, and genes that were downregulated upon PRMT5 depletion were shortlisted (Supplementary Figure S2). The MAPK3 gene was significantly upregulated in HCC patients with early, intermediate, and late stages of the disease (p < 0.0001) (Supplementary Figure S4a,b). Also, a moderate positive correlation between PRMT5 and MAPK3 was observed in HCC (p < 0.0001) (Supplementary Figure S4c). MAPK3 overexpression was significantly associated with HCC patient overall survival (OS) (p < 0.001) and disease-specific survival (DSS) (p < 0.05). However, it is not linked to relapse-free survival (RFS) (p > 0.05) or progression-free survival (PFS) (p < 0.05) (Supplementary Figure S5a–d).
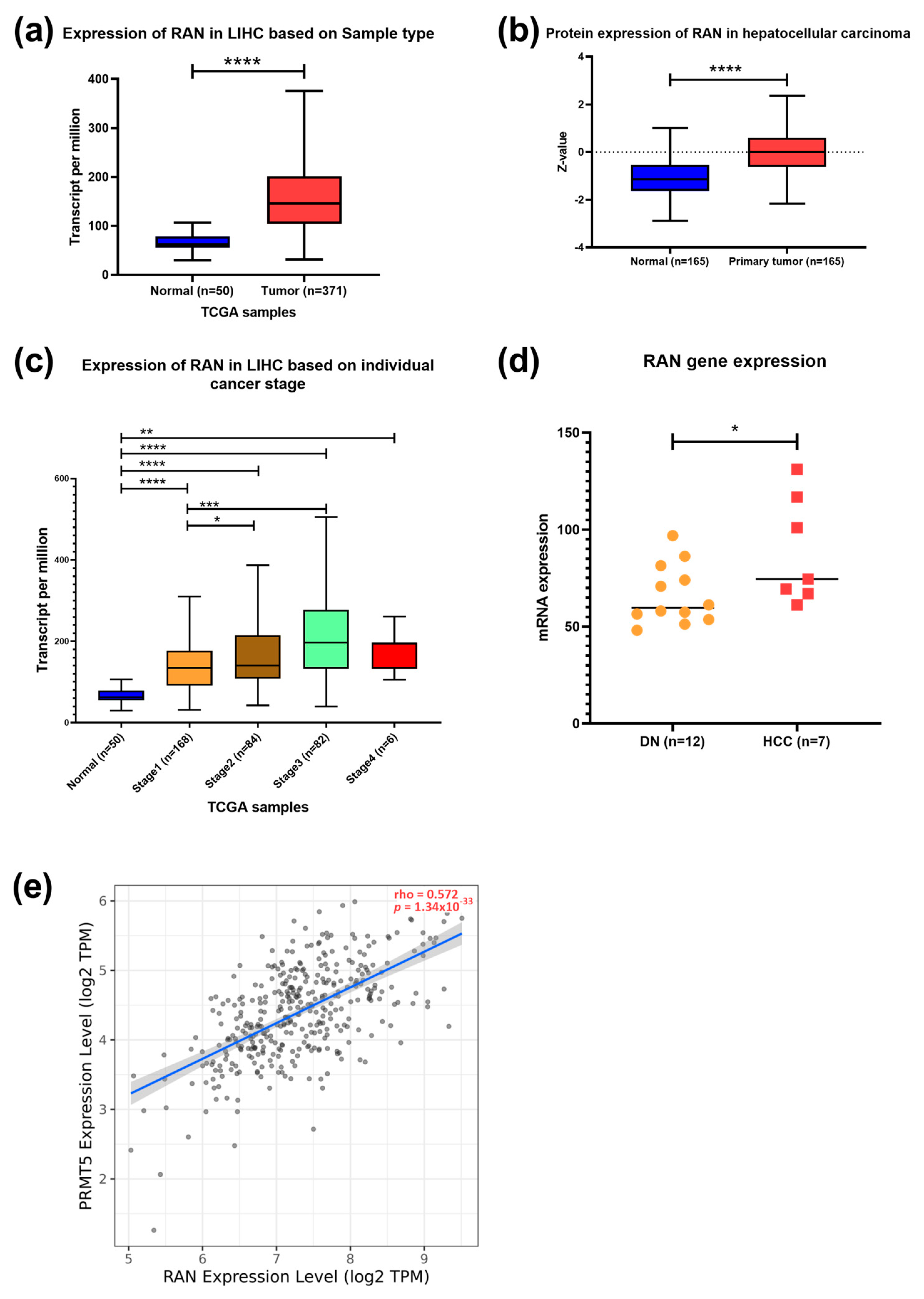
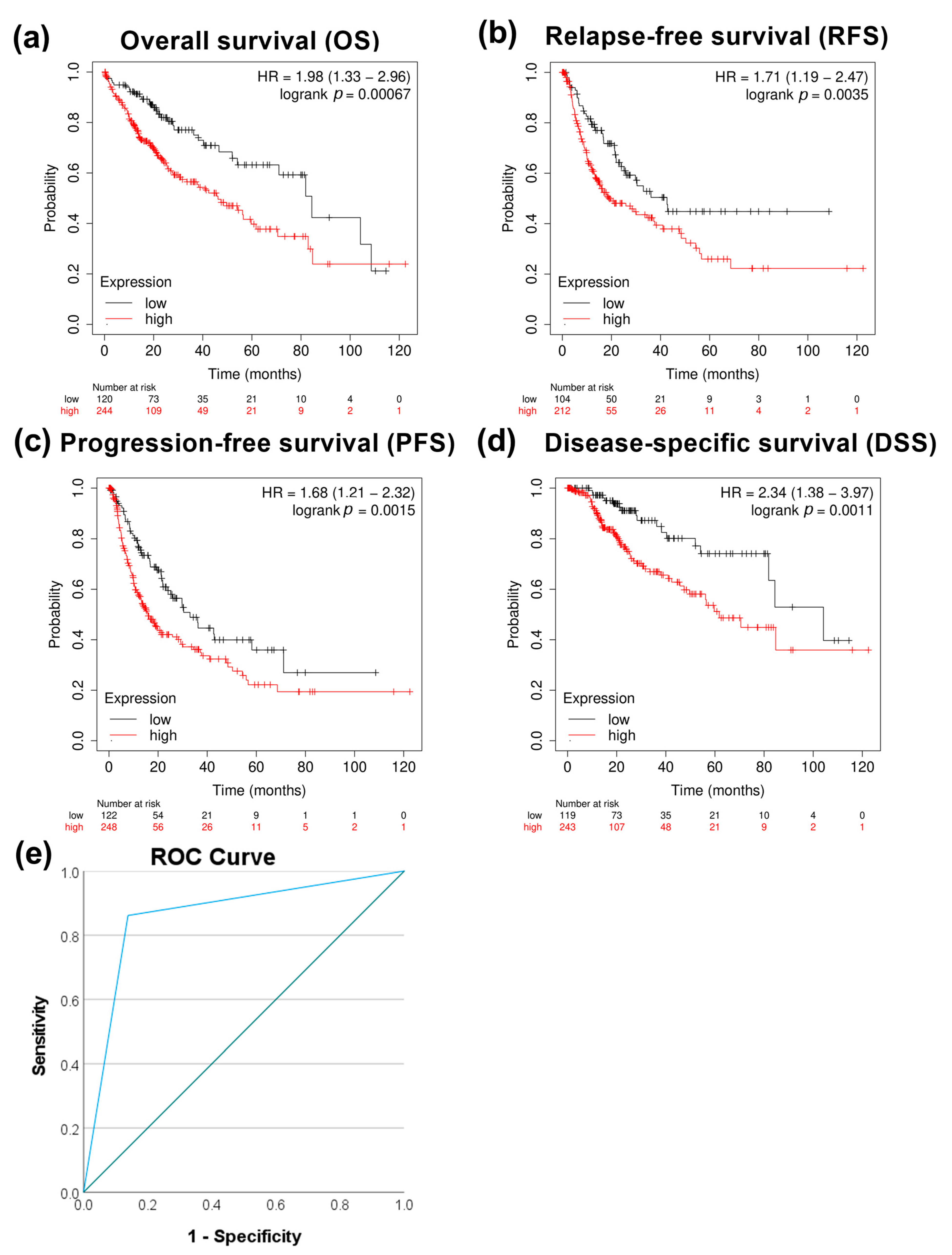
Further analysis showed increased expression of the RAS-related nuclear protein (RAN) in HCC clinical samples both at the transcriptomic and protein levels (p < 0.0001) (Figure 5a,b). TCGA data showed a higher expression of RAN in stages 2 (p < 0.05) and 3 (p < 0.001) than in stage 1 (Figure 5c). A significant upregulation of RAN gene expression in HCC tissue versus dysplastic nodule was detected using NGS data (p < 0.05) (Figure 5d). Furthermore, a moderate positive correlation between PRMT5 and RAN was detected in HCC patients (Figure 5e). To investigate the prognostic and diagnostic value of RAN gene expression, a survival curve, and a ROC curve analysis were performed. RAN overexpression was remarkably linked to poor overall survival (OS), relapse-free survival (RFS), progression-free survival (PFS), and disease-specific survival (DSS) in HCC patients (Figure 6a–d). ROC analysis of the GSE214846 dataset showed that the area under the curve is 86.2%, and both sensitivity and specificity are 86.2% with a cutoff of 65 (Figure 6e).
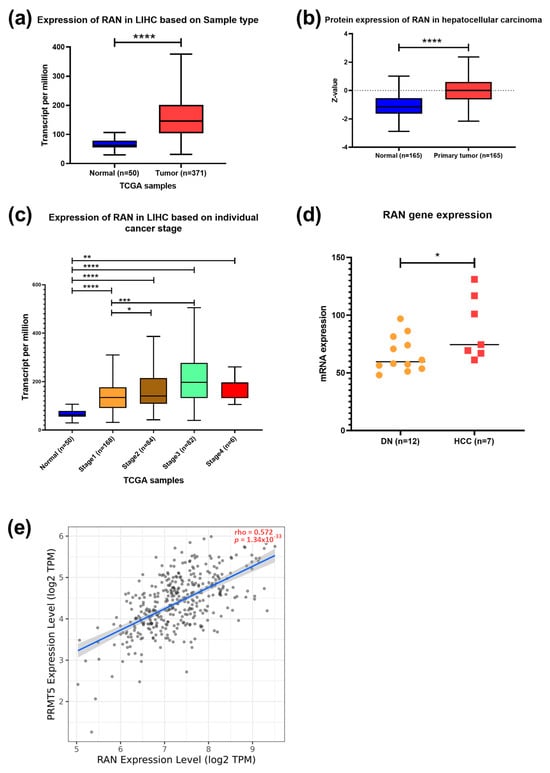
Figure 5.
Expression and correlation of RAN versus PRMT5 in HCC. (a) RAN expression in HCC compared to normal tissue counterpart. (b) RAN expression in different stages of HCC compared to normal tissue counterpart. (c) RAN expression in different stages of HCC compared to normal tissue counterpart. (d) Transcriptomic data of RAN expression in 12 samples of dysplastic nodules and 7 samples of hepatocellular carcinoma. (e) Spearman’s rank correlation coefficient of PRMT5 expression and RAN. Data in a–c were generated using RNA seq datasets, TCGA database in UALCAN tool; * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001.
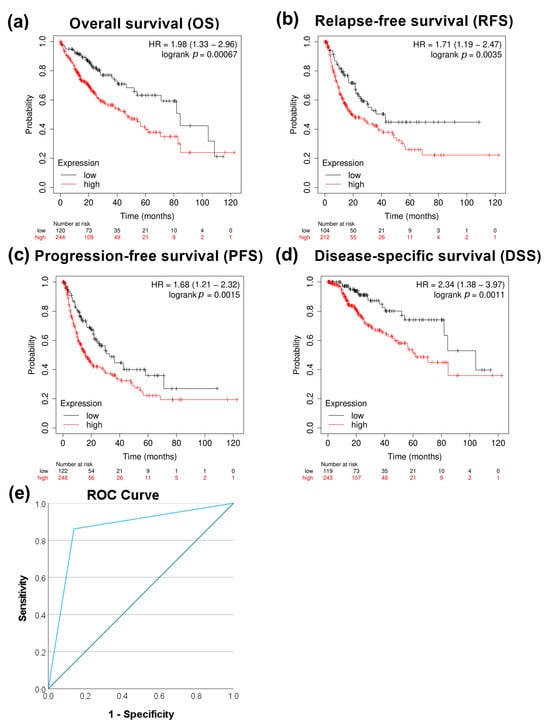
Figure 6.
Correlational analysis of RAN expression versus HCC patient survival. (a) RAN expression versus overall survival (OS). (b) RAN expression versus relapse-free survival (RFS). (c) RAN expression versus progression-free survival (PFS). (d) RAN expression versus disease-specific survival (DSS). (e) ROC curves for RAN expression at adjacent normal versus HCC tissue utilizing the GSE214846 dataset.
4. Discussion
Our analysis of the data has unveiled a notable pattern of PRMT5 overexpression, specifically in hepatocellular carcinoma (HCC). This overexpression is not uniform but exhibits a gradual increase as the disease progresses through various stages. HCC’s progression is well-documented to follow a stepwise sequence, initiating from regenerative nodules within the liver parenchyma and gradually evolving into dysplastic nodules (DNs). These DNs are known to possess cellular abnormalities that indicate a higher risk of developing into fully formed HCC lesions. Our findings highlighted the distinct elevation of PRMT5 levels in HCC compared to dysplastic nodules, indicating a significant role for PRMT5 in the step-by-step advancement of HCC. This observation underscores the potential of PRMT5 as a key player in the intricate process of HCC development. The gradual increase in PRMT5 expression across different stages of HCC progression suggests a dynamic involvement of PRMT5 in driving the malignant transformation of liver cells. Moreover, understanding this progressive upregulation of PRMT5 in HCC sheds light on the molecular mechanisms underlying the transition from pre-neoplastic lesions to fully established HCC. This insight is crucial for identifying potential therapeutic targets aimed at disrupting the pathways influenced by PRMT5, thereby impeding or reversing the progression of HCC. Further investigations into the specific interactions and downstream effects of PRMT5 in HCC development are warranted to fully comprehend its role and therapeutic implications in combating this aggressive form of cancer.
While previous studies have explored the role of PRMT5 in HCC, transcriptional activation of PRMT5 in HCC has not been examined. Multiple transcription factors have been implicated in PRMT5 overexpression in different cancer subtypes, including nuclear transcription factor Y (NF-Y) in prostate cancer, nuclear factor kappa B (NF-κB) in diffuse large B-cell lymphoma, and the fused MLL-1 protein in acute myeloid leukemia (AML) [37,38,39]. Additionally, epigenetic alterations, such as N-alpha-acetyltransferase 40 (NAA40)-induced acetylation, have been reported to contribute to PRMT5 overexpression in colorectal cancer [40]. DNA methylation, a heritable epigenetic alteration, occurs due to DNA methyltransferase (DNMTs) activity adding a methyl group to cytosine [41]. DNA methylation plays a crucial role in gene expression modulation by hindering the binding of transcription factors to DNA [42]. Our study suggests DNA hypomethylation as a potential cause of PRMT5 overexpression in the early stage of HCC. However, other mechanisms could influence PRMT5 expression in later stages, such as histone modifications and transcription factors. Indeed, identifying PRMT5 promoter CpG islands hypermethylation in HCC cell lines in vitro is crucial in addition to validation in patient biopsy samples.
The HIF1α pathway is a critical signaling pathway involved in the development and progression of HCC [43,44]. Under hypoxic conditions, HIF1α stabilizes and translocates to the nucleus, where it induces the expression of genes involved in glucose metabolism, angiogenesis, and cell proliferation [45]. HIF1α is often overexpressed in HCC, promoting tumor growth, metastasis, and drug resistance, with increased expression associated with poor patient outcomes [26]. Our data shed light on a positive relationship between PRMT5 and HIF1α expression as well as HIF1α pathway activity. Even though some of the previous work proposed a protective role of PRMT5 in HCC, our data indicated inhibition of HIF1α signaling upon targeting PRMT5. Therefore, we emphasize the previous data that identify PRMT5 as a promising therapeutic target in HCC [22,23,24,25]. Indeed, testing the therapeutic capacity of PRMT5 inhibitors against HCC via clinical trials is pivotal.
Furthermore, we identified the role of PRMT5 in regulating the expression of potential biomarker genes involved in the HIF1α pathway. Our results revealed a correlation between PRMT5 expression and two such biomarkers, MAPK3 and RAN. Previous studies discussed PRMT5′s role in modulating the activity of ERK in glioblastoma neurospheres and lung cancer, with ERK being encoded by the MAPK3 gene [46,47]. Our study also showed a positive correlation between PRMT5 and MAPK3 gene expression in HCC patients and the JHH-7 HCC cell line.
RAN is a small GTP-binding protein crucial for RNA and protein transport through the nuclear membrane [48]. It plays a role in microtubule polymerization and mitotic spindle formation, thus impacting the cell cycle [49]. RAN has been implicated in tumor progression and metastasis in various cancer subtypes, such as breast and pancreatic cancer, where it affects proliferation and apoptosis [49,50]. RAN has also been considered a potential therapeutic target in certain cancers [51]. Its expression in cancer has been linked to epigenetic regulation, such as long noncoding RNA LINC00858 in gastric cancer and microRNAs (MiR-802) in colorectal cancer [52,53]. Furthermore, RAN expression has been correlated with promoter hypomethylation in HCC and suggested as a potential prognostic marker [54]. Our data supported these findings, indicating RAN as a prognostic biomarker. This study establishes a positive correlation between PRMT5 and the RAN gene through co-expression analysis in HCC patients and investigation of RAN expression upon PRMT5 knockdown in the HCC cell line, suggesting a regulatory effect of PRMT5 on RAN. It is worth investigating whether the PRMT5 regulatory effect is due to epigenetic modulation, where it is capable of inducing histone symmetric demethylation, or due to alternative splicing. Chromatin immunoprecipitation assay targeting PRMT5 unique histone modifications such as H4R3me2s and H3R8me2s and RNAseq-based alternative splicing analysis could expand the knowledge of PRMT5-induced expression regulation of RAN [9]. We propose RAN and PRMT5 as promising disease progression biomarkers for HCC, with higher specificity (>80%) compared to alpha-fetoprotein (AFP), which has a histological specificity of 70.4% [55]. Immunohistochemistry screening of RAN and PRMT5 expression in HCC tissue is warranted, along with further investigations involving a larger sample size population. Additionally, we illustrate the significant difference in RAN expression between DN and HCC and report variable expression across different disease stages. Indeed, expanding the experiment into a larger sample size would allow further generalization of our hypothesis. RAN translocate HIF1α into the cytoplasm, contributing to cancer progression in mitochondria [27,28,29]. Understanding the mechanism of action in which RAN contributes to HCC pathogenesis will strengthen the knowledge and pave the way for further research in this field. Therefore, PRMT5 and RAN could be a promising diagnostic marker for early-stage HCC and allow early detection of the disease, which is correlated with better patient outcomes.
5. Conclusions
In conclusion, the data presented in this study strongly suggested a positive correlation between PRMT5 expression and various pro-cancer pathways, notably the HIF1α pathway in HCC. The intricate analysis conducted revealed compelling evidence indicating that PRMT5 may play a role in activating the HIF1α pathway, potentially through the MAPK3 (ERK1/2) signaling cascade. This activation mechanism could significantly contribute to the enhanced growth and metastatic potential observed in HCC cells with elevated PRMT5 expression levels. Furthermore, our findings point towards a directly proportional relationship between PRMT5 and the RAN gene, a pivotal component within the HIF1α pathway. The identification of RAN as a potential diagnostic biomarker underscores the clinical relevance of understanding PRMT5′s involvement in HCC pathogenesis. However, it is essential to acknowledge that our study represents a stepping stone in unraveling the complex molecular interactions underlying the PRMT5-HIF1α pathway axis in HCC. Further in-depth research is warranted to fully elucidate the precise mechanisms through which PRMT5 activates the HIF1α pathway and to decipher the functional consequences of this activation. These insights are crucial for developing targeted therapeutic strategies aimed at disrupting the PRMT5-driven pathways implicated in HCC progression, ultimately improving patient outcomes and treatment efficacy in combating this aggressive form of liver cancer.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biology13040216/s1. Supplementary Figure S1: PRMT5 expression positively correlates with HIF1α signaling pathway. IPA visual diagram of the HIF1α pathway illustrates the location and predicted changes in the pathway molecules in the HHJ-7 cell line upon PRMT5 knockdown. Differentially expressed genes are marked purple. Green color refers to decrease expression while red color refers to increased expression. Molecular activity variations are highlighted orange for activation and blue for inhibition. Supplementary Figure S2: filtration criteria of HIF1α signaling pathway genes controlled by PRMT5 expression to identify potential biomarkers. Supplementary Figure S3: Correlation of angiogenesis related genes expression vs. PRMT5 expression and patient survival in HCC. (a) Spearman’s rank correlation coefficient of PRMT5 versus FLT1 expression (b) FLT1 expression versus overall patient survival (OS) (c) Spearman’s rank correlation coefficient of PRMT5 versus SERPINE1 expression (d) SERPINE1 expression versus overall patient survival (OS). Supplementary Figure S4: Expression and correlation of MAPK3 vs. PRMT5 in HCC. (a) MAPK3 expression in HCC compared to normal tissue counterpart. (b) MAPK3 expression in different stages of HCC compared to normal tissue counterpart. (c) Spearman’s rank correlation coefficient of PRMT5 expression and MAPK3. Supplementary Figure S5: Correlational analysis of MAPK3 expression versus HCC patient survival. (a) MAPK3 expression versus overall patient survival (OS). (b) MAPK3 expression versus relapse-free survival (RFS). (c) MAPK3 expression versus progression-free survival (PFS). (d) MAPK3 expression versus disease-specific survival (DSS).
Author Contributions
Conceptualization, J.S.M. and W.A.; methodology, W.A.; formal analysis, W.A., D.C. and F.S.S.-A.; investigation, W.A. and D.C.; data curation, W.A., B.A.Z. and D.C.; writing—original draft preparation, J.S.M. and W.A.; writing—review and editing, J.S.M., M.H. and J.-U.M.; supervision, J.S.M. and J.-U.M. All authors have read and agreed to the published version of the manuscript.
Funding
J.S.M. is funded by the King Hussein Award from the King Hussein Cancer Foundation, Jordan (2022-KHA-001), and the Research Institute of Medical and Health Sciences, University of Sharjah (Project no. 22010901118).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data in this study were available in the public database. These data can be obtained from the GEO database (GSE168745, and GSE214846).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Tunissiolli, N.M.; Castanhole-Nunes, M.M.U.; Biselli-Chicote, P.M.; Pavarino, É.C.; da Silva, R.F.; da Silva, R.d.C.M.A.; Goloni-Bertollo, E.M. Hepatocellular Carcinoma: A Comprehensive Review of Biomarkers, Clinical Aspects, and Therapy. Asian Pac. J. Cancer Prev. 2017, 18, 863–872. [Google Scholar] [PubMed]
- Suresh, D.; Srinivas, A.N.; Kumar, D.P. Etiology of Hepatocellular Carcinoma: Special Focus on Fatty Liver Disease. Front. Oncol. 2020, 10, 601710. [Google Scholar] [CrossRef] [PubMed]
- Berliner, L.; Lemke, H.U.; vanSonnenberg, E.; Ashamalla, H.; Mattes, M.D.; Dosik, D.; Hazin, H.; Shah, S.; Mohanty, S.; Verma, S.; et al. Model-guided therapy for hepatocellular carcinoma: A role for information technology in predictive, preventive and personalized medicine. EPMA J. 2014, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Fuster, J.; Bruix, J. Prognosis of hepatocellular carcinoma. Hepato-Gastroenterol. 2002, 49, 7–11. [Google Scholar]
- Chen, V.L.; Xu, D.; Wicha, M.S.; Lok, A.S.; Parikh, N.D. Utility of Liquid Biopsy Analysis in Detection of Hepatocellular Carcinoma, Determination of Prognosis, and Disease Monitoring: A Systematic Review. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2020, 18, 2879–2902.e9. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.W.; Cho, Y.; Bae, G.U.; Kim, S.N.; Kim, Y.K. Protein arginine methyltransferases: Promising targets for cancer therapy. Exp. Mol. Med. 2021, 53, 788–808. [Google Scholar] [CrossRef] [PubMed]
- Lattouf, H.; Poulard, C.; Le Romancer, M. PRMT5 prognostic value in cancer. Oncotarget 2019, 10, 3151–3153. [Google Scholar] [CrossRef] [PubMed]
- Abumustafa, W.; Zamer, B.A.; Khalil, B.A.; Hamad, M.; Maghazachi, A.A.; Muhammad, J.S. Protein arginine N-methyltransferase 5 in colorectal carcinoma: Insights into mechanisms of pathogenesis and therapeutic strategies. Biomed. Pharmacother. 2022, 145, 112368. [Google Scholar] [CrossRef] [PubMed]
- Tewary, S.K.; Zheng, Y.G.; Ho, M.-C. Protein arginine methyltransferases: Insights into the enzyme structure and mechanism at the atomic level. Cell Mol. Life Sci. 2019, 76, 2917–2932. [Google Scholar] [CrossRef] [PubMed]
- Litzler, L.C.; Zahn, A.; Meli, A.P.; Hébert, S.; Patenaude, A.-M.; Methot, S.P.; Sprumont, A.; Bois, T.; Kitamura, D.; Costantino, S.; et al. PRMT5 is essential for B cell development and germinal center dynamics. Nat. Commun. 2019, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Ronai, Z.A. PRMT5 function and targeting in cancer. Cell Stress 2020, 4, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-R.; Li, H.-N.; Zhang, L.-J.; Zhang, C.; He, J.-G. Protein Arginine Methyltransferase 5 Promotes Esophageal Squamous Cell Carcinoma Proliferation and Metastasis via LKB1/AMPK/mTOR Signaling Pathway. Front. Bioeng. Biotechnol. 2021, 9, 645375. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Hu, Q.; Xu, J.; Ji, S.; Dai, W.; Liu, W.; Xu, W.; Sun, Q.; Zhang, Z.; Ni, Q.; et al. PRMT5 enhances tumorigenicity and glycolysis in pancreatic cancer via the FBW7/cMyc axis. Cell Commun. Signal. 2019, 17, 30. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.Y.; Lee, J.S.; Park, E.R.; Shen, Y.N.; Kim, M.Y.; Shin, H.J.; Joo, H.Y.; Cho, E.H.; Moon, S.M.; Shin, U.S.; et al. Protein arginine methyltransferase 5 is implicated in the aggressiveness of human hepatocellular carcinoma and controls the invasive activity of cancer cells. Oncol. Rep. 2018, 40, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Vishwanath, S.N.; Erdjument-Bromage, H.; Tempst, P.; Sif, S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 2004, 24, 9630–9645. [Google Scholar] [CrossRef] [PubMed]
- An Open-Label, Dose Escalation Study to Investigate the Safety, Pharmacokinetics, Pharmacodynamics and Clinical Activity of GSK3326595 in Participants with Solid Tumors and Non-Hodgkin’s Lymphoma. Available online: https://ClinicalTrials.gov/show/NCT02783300 (accessed on 15 November 2022).
- Study to Investigate the Safety and Clinical Activity of GSK3326595 and Other Agents to Treat Myelodysplastic Syndrome (MDS) and Acute Myeloid Leukemia (AML). Available online: https://ClinicalTrials.gov/show/NCT03614728 (accessed on 1 October 2022).
- Lei, Y.; Han, P.; Tian, D. Protein arginine methyltransferases and hepatocellular carcinoma: A review. Transl. Oncol. 2021, 14, 101194. [Google Scholar] [CrossRef] [PubMed]
- George, E.S.; Sood, S.; Broughton, A.; Cogan, G.; Hickey, M.; Chan, W.S.; Sudan, S.; Nicoll, A.J. The Association between Diet and Hepatocellular Carcinoma: A Systematic Review. Nutrients 2021, 13, 172. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Liu, J.; Zhang, X.O.; Sibley, K.; Najjar, S.M.; Lee, M.M.; Wu, Q. Inhibition of protein arginine methyltransferase 5 enhances hepatic mitochondrial biogenesis. J. Biol. Chem. 2018, 293, 10884–10894. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhu, Y.; Zhou, Z.; Xu, J.; Jin, S.; Xu, K.; Zhang, H.; Sun, Q.; Wang, J.; Xu, J. PRMT5 promotes cell proliferation by inhibiting BTG2 expression via the ERK signaling pathway in hepatocellular carcinoma. Cancer Med. 2018, 7, 869–882. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Peng, Y.; Hu, J.; Zhan, H.; Yang, L.; Gao, Q.; Jia, H.; Luo, R.; Dai, Z.; Tang, Z.; et al. Metadherin-PRMT5 complex enhances the metastasis of hepatocellular carcinoma through the WNT-β-catenin signaling pathway. Carcinogenesis 2020, 41, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Dong, S.; Li, Z.; Lu, L.; Zhang, S.; Chen, X.; Cen, X.; Wu, Y. Targeting protein arginine methyltransferase 5 inhibits human hepatocellular carcinoma growth via the downregulation of beta-catenin. J. Transl. Med. 2015, 13, 349. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, J.S.; Bajbouj, K.; Shafarin, J.; Hamad, M. Estrogen-induced epigenetic silencing of FTH1 and TFRC genes reduces liver cancer cell growth and survival. Epigenetics 2020, 15, 1302–1318. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xiao, Z.; Yang, L.; Gao, Y.; Zhu, Q.; Hu, L.; Huang, D.; Xu, Q. Hypoxia-inducible factors in hepatocellular carcinoma (Review). Oncol. Rep. 2020, 43, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Li, T.; Li, X.; Zhang, L.; Sun, L.; He, X.; Zhong, X.; Jia, D.; Song, L.; Semenza, G.L.; et al. HIF-1-mediated suppression of acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 2014, 8, 1930–1942. [Google Scholar] [CrossRef]
- Bao, X.; Zhang, J.; Huang, G.; Yan, J.; Xu, C.; Dou, Z.; Sun, C.; Zhang, H. The crosstalk between HIFs and mitochondrial dysfunctions in cancer development. Cell Death Dis. 2021, 12, 215. [Google Scholar] [CrossRef]
- Li, H.S.; Zhou, Y.N.; Li, L.; Li, S.F.; Long, D.; Chen, X.L.; Zhang, J.B.; Feng, L.; Li, Y.P. HIF-1α protects against oxidative stress by directly targeting mitochondria. Redox Biol. 2019, 25, 101109. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. GeroScience 2023, 45, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Czauderna, C.; Poplawski, A.; O’Rourke, C.J.; Castven, D.; Pérez-Aguilar, B.; Becker, D.; Heilmann-Heimbach, S.; Odenthal, M.; Amer, W.; Schmiel, M.; et al. Epigenetic modifications precede molecular alterations and drive human hepatocarcinogenesis. JCI Insight 2021, 6, e146196. [Google Scholar] [CrossRef]
- Zhang, H.T.; Zhang, D.; Zha, Z.G.; Hu, C.D. Transcriptional activation of PRMT5 by NF-Y is required for cell growth and negatively regulated by the PKC/c-Fos signaling in prostate cancer cells. Biochim. Biophys. Acta 2014, 1839, 1330–1340. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Guo, H.; Bates, P.D.; Zhang, S.; Zhang, H.; Nomie, K.J.; Li, Y.; Lu, L.; Seibold, K.R.; Wang, F.; et al. PRMT5 is upregulated by B-cell receptor signaling and forms a positive-feedback loop with PI3K/AKT in lymphoma cells. Leukemia 2019, 33, 2898–2911. [Google Scholar] [CrossRef] [PubMed]
- Serio, J.; Ropa, J.; Chen, W.; Mysliwski, M.; Saha, N.; Chen, L.; Wang, J.; Miao, H.; Cierpicki, T.; Grembecka, J.; et al. The PAF complex regulation of Prmt5 facilitates the progression and maintenance of MLL fusion leukemia. Oncogene 2018, 37, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Demetriadou, C.; Pavlou, D.; Mpekris, F.; Achilleos, C.; Stylianopoulos, T.; Zaravinos, A.; Papageorgis, P.; Kirmizis, A. NAA40 contributes to colorectal cancer growth by controlling PRMT5 expression. Cell Death Dis. 2019, 10, 019–1487. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Li, Y.; Robertson, K.D. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, J.; Yuan, H.; Li, X.; Li, W. Hypoxia-inducible factor-1α: A critical target for inhibiting the metastasis of hepatocellular carcinoma. Oncol. Lett. 2022, 24, 284. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Wu, J. Hypoxia inducible factor in hepatocellular carcinoma: A therapeutic target. World J. Gastroenterol. 2015, 21, 12171–12178. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.; Zadeh, L.R.; Baradaran, B.; Molavi, O.; Ghesmati, Z.; Sabzichi, M.; Ramezani, F. Up-down regulation of HIF-1α in cancer progression. Gene 2021, 798, 145796. [Google Scholar] [CrossRef] [PubMed]
- Banasavadi-Siddegowda, Y.K.; Russell, L.; Frair, E.; Karkhanis, V.A.; Relation, T.; Yoo, J.Y.; Zhang, J.; Sif, S.; Imitola, J.; Baiocchi, R.; et al. PRMT5-PTEN molecular pathway regulates senescence and self-renewal of primary glioblastoma neurosphere cells. Oncogene 2017, 36, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Jing, P.; Zhao, N.; Ye, M.; Zhang, Y.; Zhang, Z.; Sun, J.; Wang, Z.; Zhang, J.; Gu, Z. Protein arginine methyltransferase 5 promotes lung cancer metastasis via the epigenetic regulation of miR-99 family/FGFR3 signaling. Cancer Lett. 2018, 427, 38–48. [Google Scholar] [CrossRef] [PubMed]
- de Boor, S.; Knyphausen, P.; Kuhlmann, N.; Wroblowski, S.; Brenig, J.; Scislowski, L.; Baldus, L.; Nolte, H.; Krüger, M.; Lammers, M. Small GTP-binding protein Ran is regulated by posttranslational lysine acetylation. Proc. Natl. Acad. Sci. USA 2015, 112, E3679–E3688. [Google Scholar] [CrossRef] [PubMed]
- Boudhraa, Z.; Carmona, E.; Provencher, D.; Mes-Masson, A.M. Ran GTPase: A Key Player in Tumor Progression and Metastasis. Front. Cell Dev. Biol. 2020, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Lu, Y.; Zhao, X.; Sun, Y.; Shi, Y.; Fan, H.; Liu, C.; Zhou, J.; Nie, Y.; Wu, K.; et al. Ran GTPase protein promotes human pancreatic cancer proliferation by deregulating the expression of Survivin and cell cycle proteins. Biochem. Biophys. Res. Commun. 2013, 440, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Yuen, H.F.; Gunasekharan, V.K.; Chan, K.K.; Zhang, S.D.; Platt-Higgins, A.; Gately, K.; O’Byrne, K.; Fennell, D.A.; Johnston, P.G.; Rudland, P.S.; et al. RanGTPase: A candidate for Myc-mediated cancer progression. J. Natl. Cancer Inst. 2013, 105, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Meng, Q.; Bai, L.; Wang, R.; Sun, Y.; Li, J.; Fan, J.; Tian, T. LINC00858 stabilizes RAN expression and promotes metastasis of gastric cancer. Biol. Direct 2022, 17, 41. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Liu, L.; Xu, L.; Wang, H.; Hua, Q.; He, P. MiR-802 Suppresses Colorectal Cancer Cell Viability, Migration and Invasion by Targeting RAN. Cancer Manag. Res. 2020, 12, 2291–2300. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-H.; Wang, J.; Zhang, Y.; Fan, Y.-C.; Wang, K. Prognostic potential of the small GTPase Ran and its methylation in hepatocellular carcinoma. Hepatobiliary Pancreat. Dis. Int. 2022, 21, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.L.; Mo, F.; Johnson, P.J.; Siu, D.Y.; Chan, M.H.; Lau, W.Y.; Lai, P.B.; Lam, C.W.; Yeo, W.; Yu, S.C. Performance of serum α-fetoprotein levels in the diagnosis of hepatocellular carcinoma in patients with a hepatic mass. HPB Off. J. Int. Hepato Pancreato Biliary Assoc. 2014, 16, 366–372. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).