Simple Summary
The role of human papillomavirus (HPV) in cervical carcinogenesis is widely documented; however, with an increasing number of scientific publications on the molecular and cellular mechanisms activated by the virus and, specifically, by high-risk HPVs (HR-HPVs) that are involved in the development of uterine cervical cancer (CaCU) and its precursor lesions, we consider it is important to present a review of scientific articles that address ten of the mechanisms associated with at least seven of the fourteen hallmarks of cancer recently proposed. Understanding the mechanisms activated by HR-HPVs in the context of the distinctive physiological capabilities of cancer will allow the identification of clinically relevant biomarkers to improve the diagnosis and treatment of CaCU.
Abstract
Human papillomaviruses (HPVs) and, specifically, high-risk HPVs (HR-HPVs) are identified as necessary factors in the development of cancer of the lower genital tract, with CaCU standing out as the most prevalent tumor. This review summarizes ten mechanisms activated by HR-HPVs during cervical carcinogenesis, which are broadly associated with at least seven of the fourteen distinctive physiological capacities of cancer in the newly established model by Hanahan in 2022. These mechanisms involve infection by human papillomavirus, cellular tropism, genetic predisposition to uterine cervical cancer (CaCU), viral load, viral physical state, regulation of epigenetic mechanisms, loss of function of the E2 protein, deregulated expression of E6/E7 oncogenes, regulation of host cell protein function, and acquisition of the mesenchymal phenotype.
Keywords:
HPV; uterine cervical cancer; viral load; viral physical state; integration; methylation; metastasis 1. Introduction
According to data published by the International Agency for Research on Cancer of the World Health Organization (IARC-WHO; Globocan 2020), worldwide, uterine cervical cancer (CaCU) is the fourth most common cancer and the third cause of death in women. In Latin America, it is not only the third most common cancer, but also the third cause of death in the female population [1,2,3].
Seventeen years have passed since the first marketing of vaccines against the human papillomavirus (HPV) was authorized. However, a recent report from the WHO indicates that, to date, only 60% of the member states of the organization have introduced the HPV vaccine in their national vaccination schedule and that until 2021, only 13% of girls in the world had completed the planned vaccination schedule [4]. Therefore, CaCU continues to be a global public health problem, with a particularly high burden in low- and middle-income countries (LMICs), such as Mexico and Ecuador, where the incidence and mortality rate of CaCU occupy an alarming second place [1].
For this reason, in the current review, we present an outline of a series of distinctive molecular multi-step mechanisms that are involved in the carcinogenic process of the cervix and that could be considered as molecular targets for the timely treatment of neoplasms caused by HPV.
2. Human Papillomavirus Infection
HPVs are small icosahedral viruses, approximately 50 to 60 nm in diameter, non-enveloped, containing a circular double-stranded DNA genome (between 7000 and 8000 bp) (see Figure 1), infecting mucosal and skin epithelia in a specific manner and inducing cell proliferation [5,6,7].
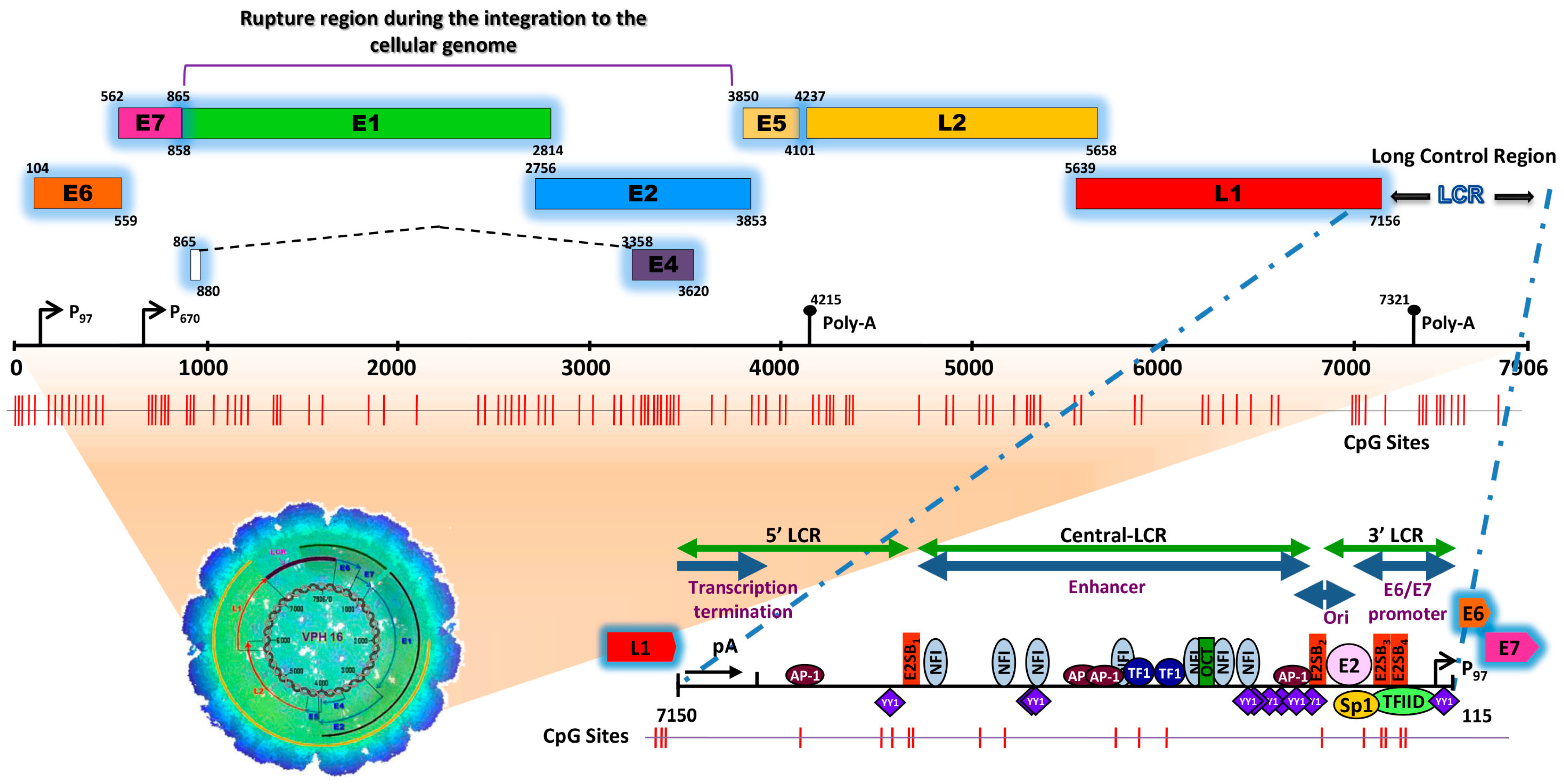
Figure 1.
Schematic representation of the structure of the HPV type 16 (HPV16) genome and its long control region (LCR) as a representative model of genital HPVs. The red vertical lines indicate the position of the 112 CpG sites along the viral genome. The bottom of the schematic illustrates the segments into which the LCR is divided as well as the cellular transcription factors that bind to it [8,9,10,11,12,13,14,15]. To illustrate the genomic structure of HPV16, the latest update of the genomic sequence was used, with NCBI Reference Sequence NC_001526, as well as PISMA software for the localization of each of the CpGs sites [16] and Vector NTI® Express Designer Software v1.5.1 (Thermo Fisher Scientific Inc., Waltham, MA, USA) for the identification of the ORFs of each of the HPV16 genes.
The HPV genome is organized similarly to chromatin [17] and is divided into three functional regions. The first is a “non-coding upstream regulatory region”, also known as the long control region (LCR) or upper regulatory region (URR). This region contains the p97 core promoter along with cis-enhancer elements that include binding sites for the viral proteins E1 (E1BS) and E2 (E2BS)—required for the commencement of HPV replication—and binding sites for several cellular transcription factors, including Sp1, YY1, TEF-10, AP1, Oct-1, NF1, KRF-1 and glucocorticoid response elements (GREs), required for the initiation of transcription [8,9,10,11,12,13,14,15,18]. The second is called the “early (E) region” and consists of the open reading frames (ORFs) for E1, E2, E4, E5, E6 and E7, where the E1, E2 and E4 proteins are mainly associated with replication, transcription, and viral integration. The E5 protein regulates cell proliferation and apoptosis and facilitates the activity of E6 and E7, while E6 and E7 act as oncoproteins and are associated with cancer development and progression [19,20,21]. The third region is known as the “late region (L)”, comprises 40% of the viral genome and includes the ORFs L1 and L2 that encode the viral capsid proteins [22].
In 1983, Harald Zur Hausen and his working group established, for the first time, the relationship between HPV and CaCU [23], but it was not until 1995 that the IARC-WHO evaluated and considered HPV as a biological agent with carcinogenic risk for humans [24,25]. Currently, there are 229 different types of HPV [26,27], classified by the IARC-WHO into three groups according to their oncogenic potential. Group 1, referred to as ‘carcinogenic or oncogenic’ (also called high-risk or HR-HPV), includes types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 59. Of these types, HPV 16 and 18 are considered to be the most important for their association with CaCU. Group 2, referred to as intermediate-risk, is subdivided into Group 2A, called ‘probably carcinogenic’, which includes only HPV 68, and Group 2B called ‘possibly carcinogenic’, which includes types 26, 53, 66, 67, 69, 70, 73 and 82. Group 3, called ‘not classifiable as carcinogenic (low risk)’, includes types 6, 11, 40, 42, 53, 54 and 57 [28,29,30,31,32].
To date, only viral types in Group 1 (HPVs or HR-HPVs) have been associated with the development of both CaCU and other types of cancer, including anogenital cancers (penis, vulva, and vagina) and cancers of the head and neck [28,33,34,35]. In addition to this, a recent study showed that almost one in three men worldwide are infected with at least one type of genital HPV and around one in five men are infected with one or more types of HR-HPV (14). This indicates that men frequently harbor genital HPV infections, emphasizing the importance of incorporating men in efforts to control HPV infection and reduce the incidence of HPV-related diseases in both men and women [36].
3. Cellular Tropism
The uterine cervix is divided into three regions, i.e., the exocervix (also called ectocervix), the endocervix and the squamocolumnar junction (SCJ) or transformation zone (considered a misnomer for a benign process, since the term “transformation” is currently used in oncology to refer to malignant neoplastic transformation). The ectocervix is composed of a non-keratinized stratified squamous epithelium and contains four phenotypically distinct cell populations: epithelial stem or stem-like cells, located in the basal and parabasal layer, and differentiated cells, located in the intermediate and superficial layers. The endocervix is lined by a single layer of mucinous columnar cells (also referred to as columnar epithelium or glandular epithelium). The SCJ is the transition area between the ectocervix and the endocervix and consists of endocervical squamous metaplasia cells, which include endocervical reserve cells (a specialized type of tissue stem cell) and possibly cuboidal cells located, more precisely, in the squamocolumnar junction, which have the capacity to divide and renew [37,38,39,40,41].
John Doorbar [42] extensively described the cellular tropism of HPV. His findings allowed us to establish that in the case of non-keratinized stratified squamous epithelium, the presence of a micro-wound is required that allows infectious virions to access the basal layer and specifically infect stem-like cells. Once infected, the stem-like cells form a reservoir of infection, and in these cells, the viral genome is maintained in an episomal state with a low copy number; as the cells divide, they produce daughter cells that are pushed towards the epithelial surface, giving rise to transient productive infections possibly progressing to high-grade neoplasia or squamous cell carcinoma [42,43,44]. Conversely, it is suggested that HPV can also infect mucinous columnar cells, reserve cells and cuboidal cells located in the SCJ and endocervix, where infection of these cell types is associated with different patterns of disease progression and the development of adenocarcinoma [45,46,47,48].
Importantly, most research models of HPV-associated cervical carcinogenesis focus on the non-keratinized stratified squamous epithelium, while the columnar epithelium of the endocervix and the metaplastic epithelium (which contains reserve cells and cuboidal cells) of the SCJ have received less attention. Evidence of this difference in research focus is that the mechanism by which HR-HPV infects stem-like cells is widely known. Specifically, these cells are characterized by expressing α6β4 integrin receptors, the epidermal growth factor receptor (EGFR), the keratinocyte growth factor receptor (KGFR) and heterotetrameric annexin A2/S100A10 (A2t) receptors, which are necessary for the entry of virions into the cell [49,50,51]. On the other hand, in the case of the epithelia of the endocervical region and the SCJ, it is only known that reserve cells that have a CK17/p63 phenotype are easily accessible targets for HPV infection [52,53,54,55].
4. Genetic Predisposition to Cervical Cancer
Several genome-wide association studies (GWASs) in different populations have provided evidence that there is a certain genetic susceptibility associated with the development of CaCU. A GWAS study of the British population identified certain single-nucleotide polymorphisms (SNPs) in the PAX8, CLPTM1L and HLA genes, with the SNPs rs10175462 in PAX8, rs27069 in CLPTM1L and rs9272050 in HLA-DQA1 being strongly associated with the risk of developing CaCU [56]. Another GWAS study of the Saudi population determined that the SNPs T10C in the GFB1 gene and G399A in the XRCC1 gene were associated with a 1.5-fold increase in the risk of developing CaCU [57]. Some studies reported that functional SNPs in codon 72 of TP53 and SNP609 in the NQO1 gene are associated with the risk of developing CaCU [58]. Finally, the homozygous CC genotype in the SNP rs4646903 of the CYP1A1 gene—which participates in genetic repair mechanisms—and the CT heterozygous genotype in the SNP rs1801133 of the MTHFR gene—which participates in cellular detoxification—are not only associated with the development of CaCU and high-grade dysplasia, but may also contribute to disease progression [59].
5. Viral Load
Initially, the detection of HR-HPV viral load was used as an additional test to relate the viral copy number to an active infectious process and reduce false-negative results in HPV diagnostic assays [60]. Other studies correlated the viral load of HR-HPV with the age of the patient, histological severity, multiple viral types, the area of the cervical lesion and the sampling method (endocervical and exocervical) [61,62]. The viral load has also been proposed as a significant marker of progression towards precancerous lesions, that is, as the viral load increases, the risk of cervical lesions increases. The risk is further enhanced if the HPV genotype is high-risk, if the viral infection is persistent during the cervical disease, and if recurrent infections are contracted with different HPV genotypes [63,64,65,66]. For example, recent studies reported that a high viral load of HR-HPVs, specifically HPV16, is significantly related to a higher risk of developing CIN2+, suggesting that viral load could be a relevant biomarker to identify women with high susceptibility to developing precancerous lesions in the uterine cervix [67,68,69].
6. Viral Physical State
At the outset of viral infection in the stem-like cells of the ectocervix, the HPV genome persists as a naked nucleic acid (also called an episome), and it depends on the host cell to enable replication. This occurs in the nucleus, with the genome replicating as an extrachromosomal element each time the cell divides [70]. As infected cells differentiate and move towards the surface of the epithelium, high levels of viral DNA are replicated, packaged into virions, and released from the surface of the epithelium as virus-laden squamous cells [47], thus completing the viral life cycle.
Conversely, it was reported that during the infectious phase, HPV can remain in its episomal form, integrate into the genome of the host cell or even be present in a coexisting state (episomal/integrated) [71,72,73,74]. In 1987, Awady and collaborators analyzed the integration of HPV16 in the SiHa cell line and reported, for the first time, the deletion of 251 nucleotides of the viral sequence within the ORFs E2 and E4 (viral integration site), and a deletion in chromosome 13 of 4.8 kb of the cellular genomic sequence (integration site in the cellular genome) [75]. Another study on invasive CaCU samples carried out by Kalantari and collaborators reported that the HPV16 genome was integrated between the E1 and E2 regions and that the integration site in the cellular genome was located in the chromosomal regions 1q25, 3q28, 6p25, 11p13 and 18q22 [76]. In Figure 1 illustrates the region of breakage and integration in the HPV16 genome.
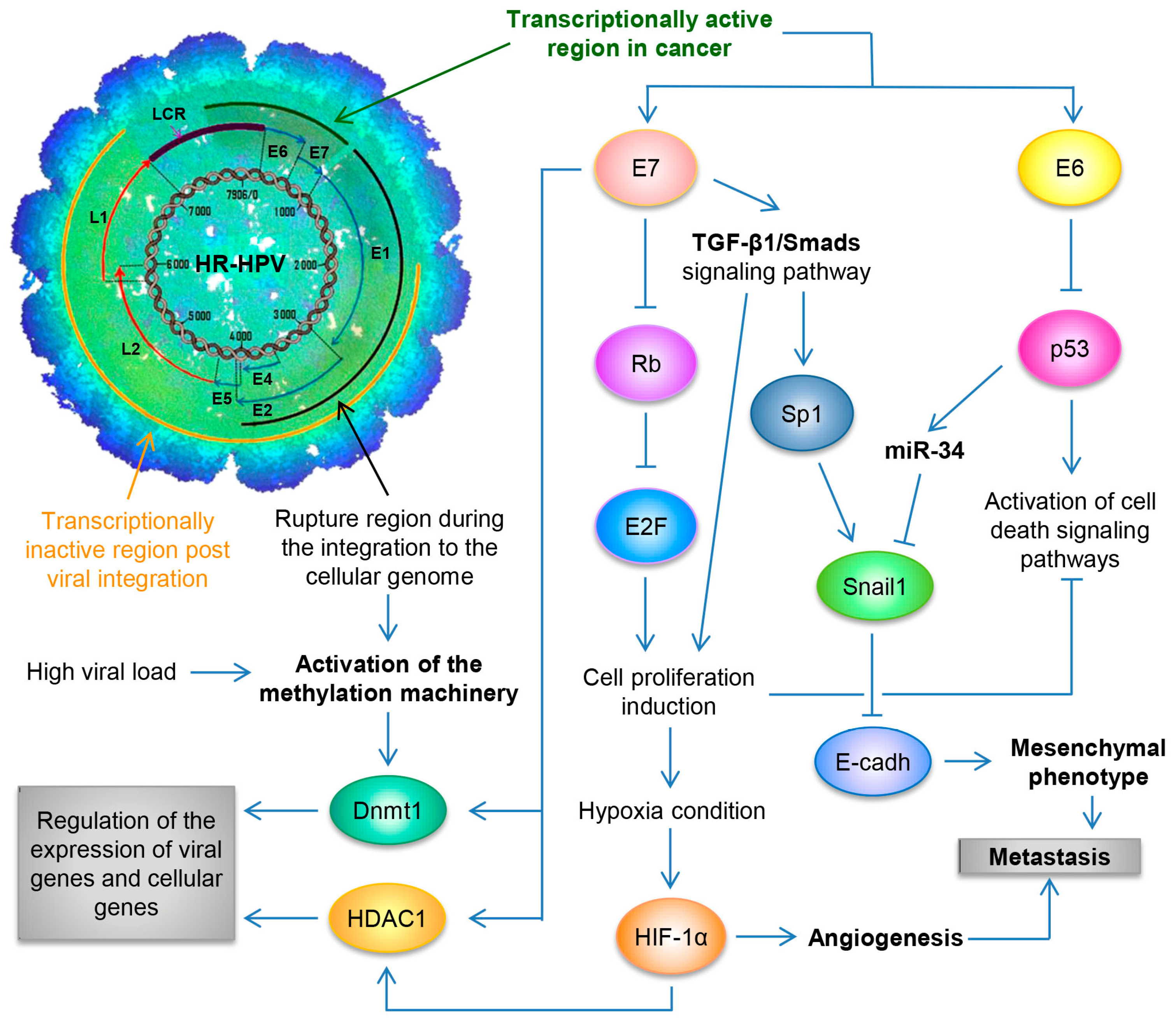
By utilizing the Capture-HPV NGS method using tumor biopsies of patients with CaCU, it was determined that HR-HPVs are inserted into intact and repeated regions of the cellular genome, specifically in MYC, NUDT15, MED4, ITM2B, RB1 loci, LPAR6, KLF5, KLF12, PIBF1, RB1, AKT3, SST, ID1, LPP, AFF3, BCL6, CCAT1, CCAT2, RAB11A, RAB22A, MAST4 and MAP2, among others [77]. By considering these findings coupled with results from women with normal cytology, those positive for HR-HPV and those with an integrated viral physical status [78,79], we can hypothesize that the integration of the viral genome at the start of infection in the target cell, will not only lead to genomic instability but also induce the development of a malignant neoplastic process (see Figure 2).
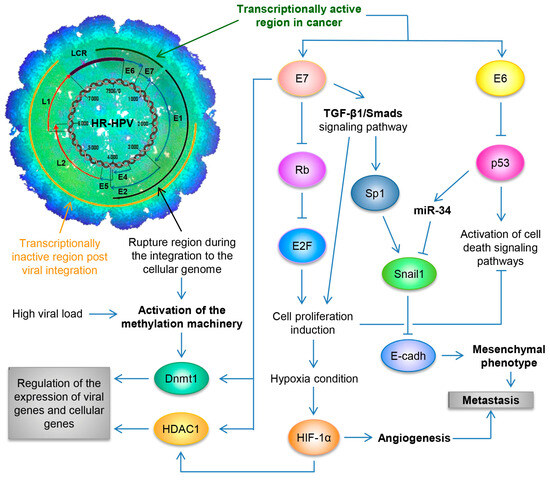
Figure 2.
Schematic representation of the molecular mechanisms induced by HPV during the carcinogenic process of the uterine cervix. Both the viral load and the integration phenomenon induce the activation of the methylation machinery, which results in the regulation of the expression of viral genes and cellular genes. Loss of E2 function, either by methylation of the E2SB regions or by deletion of the viral genome during the integration phenomenon, causes the deregulated expression of the E6/E7 oncoproteins, which will consequently induce uncontrolled cell proliferation, evasion of cell death, activation of the angiogenic process and the acquisition of the mesenchymal or metastatic phenotype.
7. Initiation of Epigenetic Mechanisms
In general, it is understood that tumor-associated DNA viruses are organized into nucleosomes to regulate the expression of their genes through histone modifications, particularly, histone acetylation. Methylation of the viral genome occurs during infection as a cellular defense mechanism against the entry of foreign genomes [80,81]. In this section, we highlight the most relevant epigenetic mechanisms induced by HPV during carcinogenesis.
7.1. Activation of the Cellular Methylation Machinery
The first studies referring to HPV DNA methylation were carried out in the LCR of the viral genome using different techniques. One of these involved methylation-specific PCR (MSP), which made it possible to report different methylation states (hypomethylated, hemimethylated and hypermethylated) depending on the amplification specificity of the primers [82,83,84,85,86,87]. Another comprised bisulfite sequencing PCR (BSP), which allowed reporting of the methylation patterns or methylation frequencies (%) of each of the CpG sites of the LCR [88,89,90,91,92,93,94].
Based on different publications alluding to the methylation of the LCR of HR-HPVs and given the premise that the methylation machinery is activated as a defense mechanism against foreign genomes, the question is raised as to how HR-HPVs activate the cellular methylation machinery. Based on previous studies, it can be inferred that HPV activates the methylation machinery through two physical mechanisms. The first occurs during HPV infection, when the entry of the viral particles into the target cell activates the methylation of the viral DNA via DNA methyltransferase 1 (Dnmt1). Following the differentiation of the host cell, the viral LCR is hypomethylated to regulate the expression of viral genes during the normal viral life cycle in the non-keratinized stratified squamous epithelium [95,96,97,98]. The second mechanism involves the integration of the viral genome, which activates the cellular methylation machinery again. However, during this process, methylation occurs only in regions where the viral genome is integrated into tandems, and the distal viral genomes are transcriptionally active and hypomethylated [94,99,100,101].
Findings published by Fernández et al. [102] on the DNA methylomes of HR-HPVs suggest that the viral load and the integration of the viral genome could play an important role in inducing different methylation patterns as the disease evolves. For example, in this study, the HeLa (derived from an adenocarcinoma that contains between 10 and 50 integrated copies of HPV18 per cell), SiHa (derived from a grade II cervical squamous cell carcinoma containing from 1 to 2 integrated copies of HPV16 per cell), and Ca Ski (derived from a cervical squamous cell carcinoma that contains between 500 and 600 integrated copies of HPV16 per cell) cell lines were used [102,103,104], and it was found that the HPV18 genome in HeLa cells is mostly demethylated, with site-specific methylation only in the E2 and L1 regions, while in SiHa cells, the HPV16 genome is demethylated in the LCR, E6, E7 and E1 regions, with methylation in the E2/E4, E5, L2 and L1 regions. Interestingly, in Ca Ski cells, it was found that the majority of HPV16 genomes are hypermethylated, and only a few are hypomethylated, suggesting that the latter are those which are transcriptionally active [102].
7.2. Histone Rearrangement
Favre and collaborators were the first to describe that the HPV genome is associated with the canonical histones H2A, H2B, H3 and H4 [17]. It was reported that the E2, E6 and E7 proteins of HR-HPV have the capacity not only to bind to the CBP/p300 coactivator complex and inhibit its histone acetyltransferase (HAT) activity, but also to block the ability of p300 to activate p53-responsive promoter elements. This results in the deregulation of cellular signaling, which decreases genome stability and favors the cellular transformation process [105,106,107,108].
8. Loss of E2 Protein Function
It is generally understood that the oncogenic HPV E2 protein is a negative regulator of the expression of the E6 and E7 oncogenes [72]. It is understood that the loss of E2 function can occur in two ways: the first is through the integration process, where breaks in the E1/E2 regions lead to the functional loss of the E2 gene [72,76,109,110]; and the second involves the methylation of the CpG sites located in the E2BSs of the HPV LCR, specifically, E2BS1, E2BS3 and E2BS4, which results in the activation of the p97 promoter and the subsequent loss of the repressive function of the E2 protein on the transcription of E7/E6 [96,111,112,113,114]. Therefore, the loss of E2 function could be considered a key step in carcinogenesis.
9. Deregulated Expression of the E6/E7 Oncogenes
Various studies reported that the loss of E2 function—either due to the phenomenon of viral genome integration or due to the methylation of the E2BSs in the HPV LCR—is associated with the overexpression or the aberrant expression of E6 and E7 [102,115,116,117]. However, these same studies mentioned that there was no significant difference when comparing the expression levels of the E6/E7 oncogenes in samples that expressed E2 and contained HPV genomes in a purely episomal state or in a coexisting state, with those in samples that contained HPV genomes in a purely integrated state, without E2 expression. This indicates that methylation at specific sites of the E2BSs in the LCR plays an important role not only in the loss of E2 function in those samples harboring transcriptionally active E2 genes, but also in regulating the expression level of E6/E7. This suggests that the overexpression of the E6/E7 oncogenes can be favored only in cases where the following criteria are met: (1) there is a high number of viral genomes in the episomal state with intact E2 genes and with site-specific methylation in E2BS-I and -II; (2) there is a low or moderate viral load, and the viral genomes are integrated at distal sites in a single copy and probably under the control of strong promoter regions in the host cell genome [102,117].
10. Regulation of Host Cell Protein Function
It was reported that the E6 oncoprotein of HR-HPV can evade cell death by apoptosis through two pathways. The first is through the proteasomal degradation of p53 via its association with the ubiquitin ligase UBE3a (E6AP) [118,119,120]. The second is through the interaction of E6 with hADA3—a protein that functions as a coactivator of p53-mediated transactivation for a variety of target promoters—where E6 induces the degradation of hADA3, thus inactivating the function of p53 and overriding the arrest of p14ARF-induced cell growth, despite the presence of normal levels of p53 [121,122]. Conversely, it is widely accepted that the E7 oncoprotein of HR-HPVs plays two main roles to induce the transforming and proliferative process in cells. Firstly, it binds with members of the retinoblastoma protein (pRb) family, such as p107 and p130 [123], which promotes the transcriptional activity of E2F transcription factors, thus regulating cell cycle entry and the progression from the G1 phase to the S phase of the cell cycle [124]. Secondly, it destabilizes pRb via degradation through the ubiquitin–proteasome pathway, leading to oncogenic transformation [125].
Table 1 exemplifies host cell proteins that interact with HPV proteins and their effect on different cellular mechanisms.

Table 1.
Host cell proteins that interact with HPV proteins.
11. Acquisition of the Mesenchymal Phenotype
An established feature of solid tumors which are not associated with oncogenic viruses is the acquisition of a mesenchymal phenotype. This is characterized by the overexpression of N-cadherin, vimentin, fibronectin, Twist, FOX C2, SOX 10, MMP-2, MMP-3, MMP-9, Snail and Slug (currently designated as Snai1 and Snai2, respectively, by the HUGO Gene Nomenclature Committee) and a decrease in the expression of E-cadherin (currently designated as CDH1 by the HUGO Gene Nomenclature Committee), desmoplakin, cytokeratin and occludin [203,204]. Nevertheless, Hellner and collaborators [205] reported that both E6 and E7 induced the expression of N-cadherin and that the expression of E7 in primary human foreskin keratinocytes (HFK) induced elevated levels of vimentin and fibronectin, as well as reduced levels of CDH1, while the levels of the regulators Twist and Snai1 remained unchanged. Another study performed in a NIKS cell model demonstrated that HPV E7 not only induced the expression of Dnmt1 but also was associated with the suppression of CDH1 expression. However, despite the expression of Dnmt1, no methylation of the CDH1 promoter region was observed, nor was any alteration observed in the expression of the negative regulators of CDH1 (Snai1/Snai2) [206]. Furthermore, a study utilizing Madin–Darby canine kidney (MDCK) cells proposed that both E6 and E7 of HPV16 could play an important role in the epithelial–mesenchymal transition (EMT) process by inducing the expression of the transcriptional factors Snai2, Twist, ZEB1 and ZEB2 and reducing CDH1 expression [207].
It is widely accepted that a common characteristic of tumors associated with oncogenic viruses—such as Epstein–Barr virus (EBV), human papillomaviruses (HPV) and hepatitis B and C viruses (HBV, HCV)—when acquiring the mesenchymal phenotype, is the suppression of CDH1 expression [208]. The interaction of viral oncoproteins with Dnmt1 plays an important role in suppressing the expression of CDH1 via methylation of its promoter region [209]. However, since reports showed that HPV may or may not methylate the CDH1 promoter region—despite inducing Dnmt1 overexpression and promoting its activity [206,210,211,212]—and given the fact that HPV does not significantly alter the expression of negative regulators of CDH1 [203,205,206], the question arises as to how HPV participates in regulating CDH1 expression and in inducing a mesenchymal phenotype.
Following the premise that both E6 and E7 of HPV have the ability to induce the expression of, bind to and stimulate the methyltransferase activity of Dnmt1 [185,189] and that E7 interacts with Mi2β, as well as with HDAC1 and HDAC2, to modulate the expression of cellular genes and viral genes by chromatin rearrangement [157,213], and knowing that the cell lines HeLa (adenocarcinoma of the cervix), SiHa (squamous cell carcinoma) and Ca Ski (squamous cell carcinoma of the cervix) are representative of the most common types of CaCU with HR-HPV infection and have different viral load and different epithelial origin, our working group previously reported that HR-HPVs can induce a mesenchymal phenotype by negatively regulating the expression of CDH1 through different pathways in which E7, Snai1 and epigenetic mechanisms are involved [214]. For example, in HeLa cells, it was found that E7 suppresses the expression of CDH1 via total methylation of the CDH1 promoter region and overexpression of Snai1, most likely forming an E7/Snai1/Dnmt1 repressive complex. Similarly, in SiHa cells, methylation of 17.65% of the CDH1 promoter region was observed, with significant expression of Snai1, which gave rise to a slight expression of CDH1; this suggests that CDH1 may be regulated by a complex consisting of E7/Snai1/HDAC1. Conversely, the Ca Ski cell line did not exhibit a mesenchymal phenotype, since it showed a high level of expression of CDH1, without methylation of its promoter region, and low levels of expression of Snai1 and Snai2 [214]. Our results demonstrated that HR-HPVs can regulate the expression of TEM markers in different ways, most likely depending on the infected epithelium and the viral load.
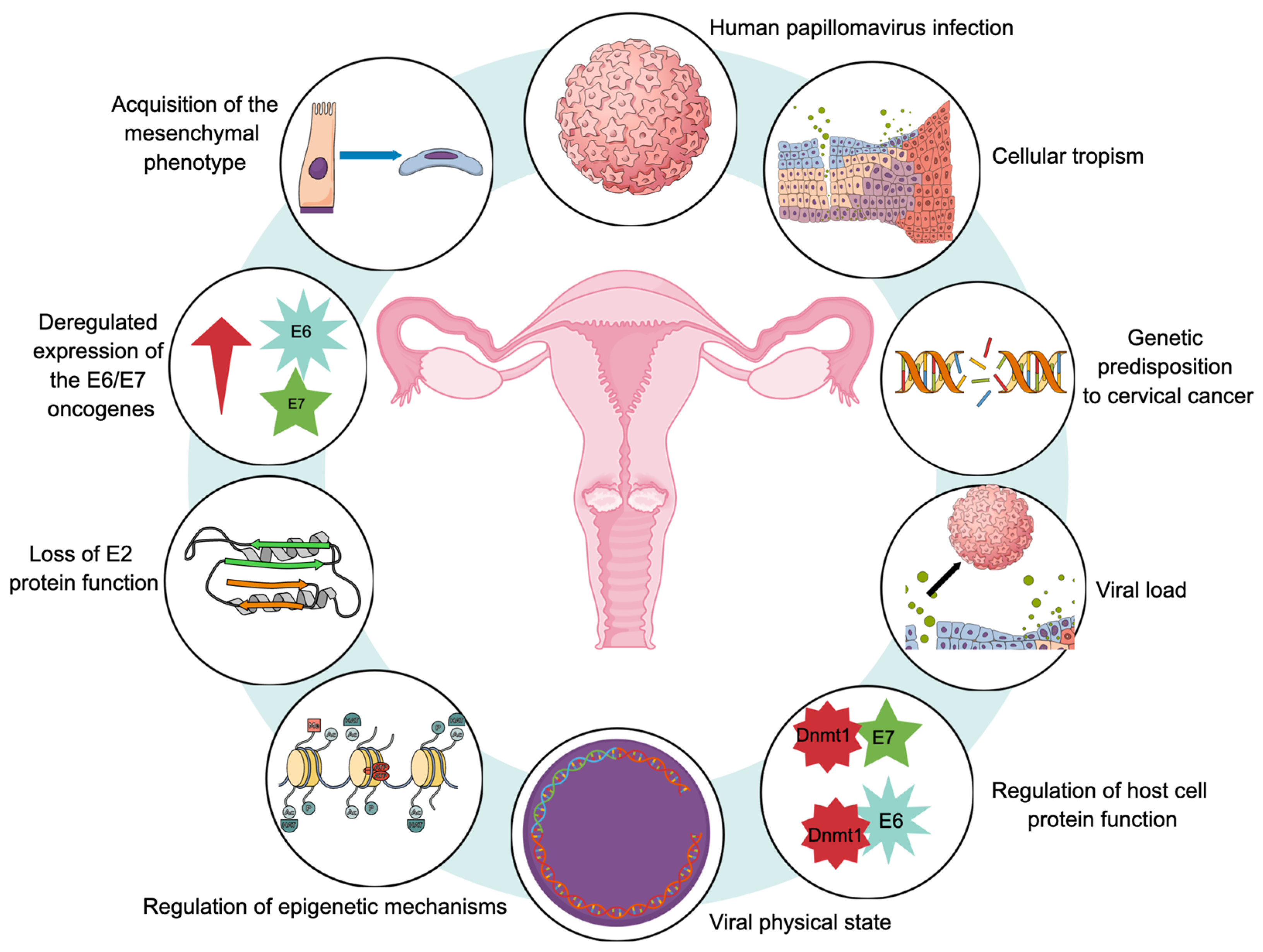
Figure 3 provides a representative diagram of the main molecular hallmarks that have been reported during the carcinogenic process of CaCU.
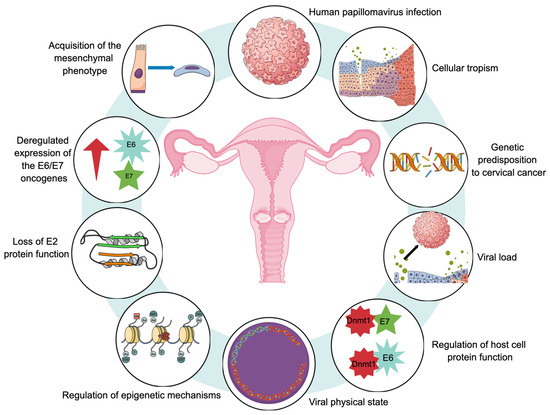
Figure 3.
Proposed molecular hallmarks for cervical carcinogenesis. All of these markers would play an important role in the development of uterine cervical cancer.
12. Conclusions
HR-HPVs, through their oncoproteins E6 and E7, are responsible for the cellular changes linked to the development of CaCU. During the process of viral genome replication—whether as an episome, integrated or coexisting in the episomal and integrated states—modifications are generated in the host cell machinery, which induce genomic instability and the development of the carcinogenic process. In this review, we described ten mechanisms activated by HR-HPVs during cervical carcinogenesis, which are broadly associated with at least seven of the fourteen distinctive physiological capacities of cancer in a newly established model [215]. Specifically, the mechanisms involved are among those that promote epigenetic modifications, instability in the host cell genome, sustained proliferative signaling, replicative immortality, resistance to cell death, evasion of the immune response and activation of invasion and metastasis. Therefore, improved understanding of the viral oncogenic mechanisms will allow us to develop new tools for the early diagnosis of cervical lesions and to identify other therapeutic targets with a focus on the early phases of cervical malignancy.
Author Contributions
P.R.-C., V.A.-V. and J.G.O.T. wrote the manuscript and participated in scientific discussions. B.V.C. and J.O.S. provided scientific direction and led scientific discussions. G.D.B.-O. provided the concept design and scientific direction, led scientific discussions and contributed to the editing and drafting of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This review was funded by Universidad de Cuenca.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank the Vice-Rectorate of Research of University of Cuenca for the support in the publication of this article.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- WHO. International Agency for Research on Cancer. 2022. Available online: https://gco.iarc.fr/today/home (accessed on 4 August 2023).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Singh, D.; Vignat, J.; Lorenzoni, V.; Eslahi, M.; Ginsburg, O.; Lauby-Secretan, B.; Arbyn, M.; Basu, P.; Bray, F.; Vaccarella, S. Global estimates of incidence and mortality of cervical cancer in 2020: A baseline analysis of the WHO Global Cervical Cancer Elimination Initiative. Lancet Glob. Health 2023, 11, e197–e206. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO HPV Vaccine Global Market Study. 2022. Available online: https://www.who.int/publications/m/item/who-hpv-vaccine-global-market-study-april-2022 (accessed on 7 August 2023).
- McMurray, H.R.; Nguyen, D.; Westbrook, T.F.; McAnce, D.J. Biology of human papillomaviruses. Int. J. Exp. Pathol. 2001, 82, 15–33. [Google Scholar] [CrossRef]
- WHO. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. In Human Papillomaviruses; WHO: Lyon, France, 1995; Volume 64. Available online: https://www.ncbi.nlm.nih.gov/books/NBK424408 (accessed on 10 August 2023).
- zur Hausen, H.; de Villiers, E.M. Human papillomaviruses. Annu. Rev. Microbiol. 1994, 48, 427–447. [Google Scholar] [CrossRef]
- Apt, D.; Watts, R.M.; Suske, G.; Bernard, H.U. High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter. Virology 1996, 224, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Butz, K.; Hoppe-Seyler, F. Transcriptional control of human papillomavirus (HPV) oncogene expression: Composition of the HPV type 18 upstream regulatory region. J. Virol. 1993, 67, 6476–6486. [Google Scholar] [CrossRef] [PubMed]
- Gloss, B.; Yeo-Gloss, M.; Meisterenst, M.; Rogge, L.; Winnacker, E.L.; Bernard, H.U. Clusters of nuclear factor I binding sites identify enhancers of several papillomaviruses but alone are not sufficient for enhancer function. Nucleic Acids Res. 1989, 17, 3519–3533. [Google Scholar] [CrossRef]
- Hoppe-Seyler, F.; Butz, K.; zur Hausen, H. Repression of the human papillomavirus type 18 enhancer by the cellular transcription factor Oct-1. J. Virol. 1991, 65, 5613–5618. [Google Scholar] [CrossRef]
- Kanaya, T.; Kyo, S.; Laimins, L.A. The 5′ region of the human papillomavirus type 31 upstream regulatory region acts as an enhancer which augments viral early expression through the action of YY1. Virology 1997, 237, 159–169. [Google Scholar] [CrossRef]
- Kyo, S.; Klumpp, D.J.; Inoue, M.; Kanaya, T.; Laimins, L.A. Expression of AP1 during cellular differentiation determines human papillomavirus E6/E7 expression in stratified epithelial cells. J. Gen. Virol. 1997, 78, 401–411. [Google Scholar] [CrossRef]
- O’Connor, M.; Chan, S.-Y.; Bernard, H.-U. Transcription Factor Binding Sites in the Long Control Region of Genital HPVs. In Human Papillomaviruses; 1995 compendium, part III-A; Los Alamos National Laboratory: Los Alamos, NM, USA, 1995; pp. 21–40. Available online: https://pave.niaid.nih.gov/lanl-archives/compendium/95PDF/3/oconnor.pdf (accessed on 19 August 2023).
- Sailaja, G.; Watts, R.M.; Bernard, H.U. Many different papillomaviruses have low transcriptional activity in spite of strong epithelial specific enhancers. J. Gen. Virol. 1999, 80, 1715–1724. [Google Scholar] [CrossRef][Green Version]
- Alcantara-Silva, R.; Alvarado-Hermida, M.; Diaz-Contreras, G.; Sanchez-Barrios, M.; Carrera, S.; Galvan, S.C. PISMA: A Visual Representation of Motif Distribution in DNA Sequences. Bioinform. Biol. Insights 2017, 11, 1177932217700907. [Google Scholar] [CrossRef]
- Favre, M.; Breitburd, F.; Croissant, O.; Orth, G. Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J. Virol. 1977, 21, 1205–1209. [Google Scholar] [CrossRef]
- Parker, J.N.; Zhao, W.; Askins, K.J.; Broker, T.R.; Chow, L.T. Mutational analyses of differentiation-dependent human papillomavirus type 18 enhancer elements in epithelial raft cultures of neonatal foreskin keratinocytes. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 1997, 8, 751–762. [Google Scholar]
- Bhattacharjee, R.; Das, S.S.; Biswal, S.S.; Nath, A.; Das, D.; Basu, A.; Malik, S.; Kumar, L.; Kar, S.; Singh, S.K.; et al. Mechanistic role of HPV-associated early proteins in cervical cancer: Molecular pathways and targeted therapeutic strategies. Crit. Rev. Oncol./Hematol. 2022, 174, 103675. [Google Scholar] [CrossRef] [PubMed]
- Harari, A.; Chen, Z.; Burk, R.D. Human papillomavirus genomics: Past, present and future. Curr. Probl. Dermatol. 2014, 45, 1–18. [Google Scholar] [CrossRef]
- Nelson, C.W.; Mirabello, L. Human papillomavirus genomics: Understanding carcinogenicity. Tumour Virus Res. 2023, 15, 200258. [Google Scholar] [CrossRef]
- Hafkamp, H.C.; Manni, J.J.; Speel, E.J. Role of human papillomavirus in the development of head and neck squamous cell carcinomas. Acta Oto-Laryngol. 2004, 124, 520–526. [Google Scholar] [CrossRef]
- Durst, M.; Gissmann, L.; Ikenberg, H.; zur Hausen, H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl. Acad. Sci. USA 1983, 80, 3812–3815. [Google Scholar] [CrossRef] [PubMed]
- WHO. Human Papillomaviruses. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; WHO: Lyon, France, 2007; Volume 90, pp. 1–636. Available online: https://www.ncbi.nlm.nih.gov/books/NBK321760 (accessed on 17 January 2024).
- zur Hausen, H. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology 1991, 184, 9–13. [Google Scholar] [CrossRef]
- Arroyo Muhr, L.S.; Eklund, C.; Dillner, J. Misclassifications in human papillomavirus databases. Virology 2021, 558, 57–66. [Google Scholar] [CrossRef]
- PaVE: The PapillomaVirus Episteme. National Institute of Allergy and Infectious Diseases. 2023. Available online: https://pave.niaid.nih.gov/search/search_database (accessed on 3 September 2023).
- Bosch, F.X.; Lorincz, A.; Munoz, N.; Meijer, C.J.; Shah, K.V. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 2002, 55, 244–265. [Google Scholar] [CrossRef]
- Clifford, G.M.; Howell-Jones, R.; Franceschi, S. Judging the carcinogenicity of human papillomavirus types by single/multiple infection ratio in cervical cancer. Int. J. Cancer 2011, 129, 1792–1794. [Google Scholar] [CrossRef]
- Moeinzadeh, M.; Kheirkhah, B.; Amini, K.; Pouryasin, A. Classification and identification of human papillomavirus based on its prevalence and development of cervical lesion among Iranian women. BioImpacts BI 2020, 10, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Schiffman, M.; Clifford, G.; Buonaguro, F.M. Classification of weakly carcinogenic human papillomavirus types: Addressing the limits of epidemiology at the borderline. Infect. Agents Cancer 2009, 4, 8. [Google Scholar] [CrossRef]
- Schiffman, M.; Herrero, R.; Desalle, R.; Hildesheim, A.; Wacholder, S.; Rodriguez, A.C.; Bratti, M.C.; Sherman, M.E.; Morales, J.; Guillen, D.; et al. The carcinogenicity of human papillomavirus types reflects viral evolution. Virology 2005, 337, 76–84. [Google Scholar] [CrossRef]
- Alhamlan, F.S.; Alfageeh, M.B.; Al Mushait, M.A.; Al-Badawi, I.A.; Al-Ahdal, M.N. Human Papillomavirus-Associated Cancers. Adv. Exp. Med. Biol. 2021, 1313, 1–14. [Google Scholar] [CrossRef]
- de Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef] [PubMed]
- WHO. Human Papillomavirus and Related Diseases Report, World. ICO/IARC Information Centre on HPV and Cancer. 2023. Available online: https://hpvcentre.net/statistics/reports/XWX.pdf (accessed on 5 September 2023).
- Bruni, L.; Albero, G.; Rowley, J.; Alemany, L.; Arbyn, M.; Giuliano, A.R.; Markowitz, L.E.; Broutet, N.; Taylor, M. Global and regional estimates of genital human papillomavirus prevalence among men: A systematic review and meta-analysis. Lancet Glob. Health 2023, 11, e1345–e1362. [Google Scholar] [CrossRef]
- Fadare, O.; Roma, A.A. Normal Anatomy of the Uterine Cervix. In Atlas of Uterine Pathology. Atlas of Anatomic Pathology; Springer: Cham, Switzerland, 2019; pp. 193–196. [Google Scholar] [CrossRef]
- Herfs, M.; Vargas, S.O.; Yamamoto, Y.; Howitt, B.E.; Nucci, M.R.; Hornick, J.L.; McKeon, F.D.; Xian, W.; Crum, C.P. A novel blueprint for ‘top down’ differentiation defines the cervical squamocolumnar junction during development, reproductive life, and neoplasia. J. Pathol. 2013, 229, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Kenemans, P.; Davina, J.H.M.; de Haan, R.W.; Hafez, E.S.E. The Cervix. In Atlas of Human Reproduction: By Scanning Electron Microscopy; Springer: Dordrecht, The Netherlands, 1982; pp. 45–54. [Google Scholar] [CrossRef]
- Martens, J.E.; Smedts, F.M.; Ploeger, D.; Helmerhorst, T.J.; Ramaekers, F.C.; Arends, J.W.; Hopman, A.H. Distribution pattern and marker profile show two subpopulations of reserve cells in the endocervical canal. Int. J. Gynecol. Pathol. 2009, 28, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Murall, C.L.; Jackson, R.; Zehbe, I.; Boulle, N.; Segondy, M.; Alizon, S. Epithelial stratification shapes infection dynamics. PLoS Comput. Biol. 2019, 15, e1006646. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The biology and life-cycle of human papillomaviruses. Vaccine 2012, 30 (Suppl. 5), F55–F70. [Google Scholar] [CrossRef]
- Griffin, H.; Soneji, Y.; Van Baars, R.; Arora, R.; Jenkins, D.; van de Sandt, M.; Wu, Z.; Quint, W.; Jach, R.; Okon, K.; et al. Stratification of HPV-induced cervical pathology using the virally encoded molecular marker E4 in combination with p16 or MCM. Mod. Pathol. 2015, 28, 977–993. [Google Scholar] [CrossRef] [PubMed]
- Stanley, M.A. Epithelial cell responses to infection with human papillomavirus. Clin. Microbiol. Rev. 2012, 25, 215–222. [Google Scholar] [CrossRef]
- Chow, L.T.; Broker, T.R.; Steinberg, B.M. The natural history of human papillomavirus infections of the mucosal epithelia. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2010, 118, 422–449. [Google Scholar] [CrossRef]
- Doorbar, J. Papillomavirus life cycle organization and biomarker selection. Dis. Markers 2007, 23, 297–313. [Google Scholar] [CrossRef]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25 (Suppl. 1), 2–23. [Google Scholar] [CrossRef]
- Egawa, N.; Egawa, K.; Griffin, H.; Doorbar, J. Human Papillomaviruses; Epithelial Tropisms, and the Development of Neoplasia. Viruses 2015, 7, 3863–3890. [Google Scholar] [CrossRef]
- Kines, R.C.; Schiller, J.T. Harnessing Human Papillomavirus’ Natural Tropism to Target Tumors. Viruses 2022, 14, 1656. [Google Scholar] [CrossRef]
- Ozbun, M.A.; Campos, S.K. The long and winding road: Human papillomavirus entry and subcellular trafficking. Curr. Opin. Virol. 2021, 50, 76–86. [Google Scholar] [CrossRef]
- Raff, A.B.; Woodham, A.W.; Raff, L.M.; Skeate, J.G.; Yan, L.; Da Silva, D.M.; Schelhaas, M.; Kast, W.M. The evolving field of human papillomavirus receptor research: A review of binding and entry. J. Virol. 2013, 87, 6062–6072. [Google Scholar] [CrossRef] [PubMed]
- Herfs, M.; Yamamoto, Y.; Laury, A.; Wang, X.; Nucci, M.R.; McLaughlin-Drubin, M.E.; Munger, K.; Feldman, S.; McKeon, F.D.; Xian, W.; et al. A discrete population of squamocolumnar junction cells implicated in the pathogenesis of cervical cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 10516–10521. [Google Scholar] [CrossRef]
- Martens, J.E.; Arends, J.; Linden, P.J.Q.V.D.; Boer, B.A.G.D.; Helmerhorst, T.J.M. Cytokeratin 17 and p63 are Markers of the HPV Target Cell, the Cervical Stem Cell. Anticancer Res. 2004, 24, 771–776. [Google Scholar] [PubMed]
- Regauer, S.; Reich, O. CK17 and p16 expression patterns distinguish (atypical) immature squamous metaplasia from high-grade cervical intraepithelial neoplasia (CIN III). Histopathology 2007, 50, 629–635. [Google Scholar] [CrossRef]
- Regauer, S.; Reich, O. The origin of Human Papillomavirus (HPV)—Induced cervical squamous cancer. Curr. Opin. Virol. 2021, 51, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Bowden, S.J.; Bodinier, B.; Kalliala, I.; Zuber, V.; Vuckovic, D.; Doulgeraki, T.; Whitaker, M.D.; Wielscher, M.; Cartwright, R.; Tsilidis, K.K.; et al. Genetic variation in cervical preinvasive and invasive disease: A genome-wide association study. Lancet Oncol. 2021, 22, 548–557. [Google Scholar] [CrossRef]
- Al-Harbi, N.M.; Bin Judia, S.S.; Mishra, K.N.; Shoukri, M.M.; Alsbeih, G.A. Genetic Predisposition to Cervical Cancer and the Association With XRCC1 and TGFB1 Polymorphisms. Int. J. Gynecol. Cancer 2017, 27, 1949–1956. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Z.; Ma, D.; Huettner, P.C.; Massad, L.S.; Nguyen, L.; Borecki, I.; Rader, J.S. TP53, MDM2, NQO1, and susceptibility to cervical cancer. Cancer Epidemiol. Biomark. Prev. 2010, 19, 755–761. [Google Scholar] [CrossRef]
- von Keyserling, H.; Bergmann, T.; Schuetz, M.; Schiller, U.; Stanke, J.; Hoffmann, C.; Schneider, A.; Lehrach, H.; Dahl, A.; Kaufmann, A.M. Analysis of 4 single-nucleotide polymorphisms in relation to cervical dysplasia and cancer development using a high-throughput ligation-detection reaction procedure. Int. J. Gynecol. Cancer 2011, 21, 1664–1671. [Google Scholar] [CrossRef]
- Schmitt, M.; Depuydt, C.; Benoy, I.; Bogers, J.; Antoine, J.; Pawlita, M.; Arbyn, M. Viral load of high-risk human papillomaviruses as reliable clinical predictor for the presence of cervical lesions. Cancer Epidemiol. Biomark. Prev. 2013, 22, 406–414. [Google Scholar] [CrossRef]
- Lu, X.; Wang, T.; Zhang, Y.; Liu, Y. Analysis of influencing factors of viral load in patients with high-risk human papillomavirus. Virol. J. 2021, 18, 6. [Google Scholar] [CrossRef]
- Veitia, D.; Liuzzi, J.; Avila, M.; Rodriguez, I.; Toro, F.; Correnti, M. Association of viral load and physical status of HPV-16 with survival of patients with head and neck cancer. Ecancermedicalscience 2020, 14, 1082. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shi, X.; Liu, J.; Zhang, L. Correlation between human papillomavirus viral load and cervical lesions classification: A review of current research. Front. Med. 2023, 10, 1111269. [Google Scholar] [CrossRef] [PubMed]
- Hortlund, M.; van Mol, T.; Van de Pol, F.; Bogers, J.; Dillner, J. Human papillomavirus load and genotype analysis improves the prediction of invasive cervical cancer. Int. J. Cancer 2021, 149, 684–691. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, C.; Pan, J.; Sun, C.; Zhou, H.; Meng, Y. Significance of the viral load of high-risk HPV in the diagnosis and prediction of cervical lesions: A retrospective study. BMC Women’s Health 2021, 21, 353. [Google Scholar] [CrossRef] [PubMed]
- Ylitalo, N.; Sorensen, P.; Josefsson, A.M.; Magnusson, P.K.; Andersen, P.K.; Ponten, J.; Adami, H.O.; Gyllensten, U.B.; Melbye, M. Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: A nested case-control study. Lancet 2000, 355, 2194–2198. [Google Scholar] [CrossRef] [PubMed]
- Baumann, A.; Henriques, J.; Selmani, Z.; Meurisse, A.; Lepiller, Q.; Vernerey, D.; Valmary-Degano, S.; Paget-Bailly, S.; Riethmuller, D.; Ramanah, R.; et al. HPV16 Load Is a Potential Biomarker to Predict Risk of High-Grade Cervical Lesions in High-Risk HPV-Infected Women: A Large Longitudinal French Hospital-Based Cohort Study. Cancers 2021, 13, 4149. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhao, S.; Hu, S.; Zhao, K.; Zhang, Q.; Zhang, X.; Pan, Q.; Zhao, F. Role of Human Papillomavirus DNA Load in Predicting the Long-term Risk of Cervical Cancer: A 15-Year Prospective Cohort Study in China. J. Infect. Dis. 2019, 219, 215–222. [Google Scholar] [CrossRef]
- Tao, X.; Austin, R.M.; Yu, T.; Zhong, F.; Zhou, X.; Cong, Q.; Sui, L.; Zhao, C. Risk stratification for cervical neoplasia using extended high-risk HPV genotyping in women with ASC-US cytology: A large retrospective study from China. Cancer Cytopathol. 2022, 130, 248–258. [Google Scholar] [CrossRef]
- Boshart, M.; Gissmann, L.; Ikenberg, H.; Kleinheinz, A.; Scheurlen, W.; zur Hausen, H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 1984, 3, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Oyervides-Munoz, M.A.; Perez-Maya, A.A.; Rodriguez-Gutierrez, H.F.; Gomez-Macias, G.S.; Fajardo-Ramirez, O.R.; Trevino, V.; Barrera-Saldana, H.A.; Garza-Rodriguez, M.L. Understanding the HPV integration and its progression to cervical cancer. Infect. Genet. Evol. 2018, 61, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Hwang, E.S.; Park, S.N.; Ahn, H.K.; Um, S.J.; Kim, C.J.; Kim, S.J.; Namkoong, S.E. Physical status and expression of HPV genes in cervical cancers. Gynecol. Oncol. 1997, 65, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Kulmala, S.M.; Syrjanen, S.M.; Gyllensten, U.B.; Shabalova, I.P.; Petrovichev, N.; Tosi, P.; Syrjanen, K.J.; Johansson, B.C. Early integration of high copy HPV16 detectable in women with normal and low grade cervical cytology and histology. J. Clin. Pathol. 2006, 59, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Matsukura, T.; Koi, S.; Sugase, M. Both episomal and integrated forms of human papillomavirus type 16 are involved in invasive cervical cancers. Virology 1989, 172, 63–72. [Google Scholar] [CrossRef] [PubMed]
- el Awady, M.K.; Kaplan, J.B.; O’Brien, S.J.; Burk, R.D. Molecular analysis of integrated human papillomavirus 16 sequences in the cervical cancer cell line SiHa. Virology 1987, 159, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Kalantari, M.; Blennow, E.; Hagmar, B.; Johansson, B. Physical state of HPV16 and chromosomal mapping of the integrated form in cervical carcinomas. Diagn. Mol. Pathol. 2001, 10, 46–54. [Google Scholar] [CrossRef]
- Holmes, A.; Lameiras, S.; Jeannot, E.; Marie, Y.; Castera, L.; Sastre-Garau, X.; Nicolas, A. Mechanistic signatures of HPV insertions in cervical carcinomas. NPJ Genom. Med. 2016, 1, 16004. [Google Scholar] [CrossRef]
- Canadas, M.P.; Darwich, L.; Sirera, G.; Cirigliano, V.; Bofill, M.; Clotet, B.; Videla, S. New molecular method for the detection of human papillomavirus type 16 integration. Clin. Microbiol. Infect. 2010, 16, 836–842. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, C.; Gao, W.; Wang, L.; Pan, Y.; Gao, Y.; Lu, Z.; Ke, Y. Genome-wide profiling of the human papillomavirus DNA integration in cervical intraepithelial neoplasia and normal cervical epithelium by HPV capture technology. Sci. Rep. 2016, 6, 35427. [Google Scholar] [CrossRef]
- Fischer, N. Infection-induced epigenetic changes and their impact on the pathogenesis of diseases. Semin. Immunopathol. 2020, 42, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Milavetz, B.I.; Balakrishnan, L. Viral epigenetics. Methods Mol. Biol. 2015, 1238, 569–596. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Choi, Y.D.; Lee, J.S.; Lee, J.H.; Nam, J.H.; Choi, C.; Kweon, S.S.; Fackler, M.J.; Sukumar, S. Quantitative assessment of DNA methylation for the detection of cervical neoplasia in liquid-based cytology specimens. Virchows Arch. 2010, 457, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Choi, Y.D.; Lee, J.S.; Lee, J.H.; Nam, J.H.; Choi, C. Assessment of DNA methylation for the detection of cervical neoplasia in liquid-based cytology specimens. Gynecol. Oncol. 2010, 116, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Turan, T.; Kalantari, M.; Cuschieri, K.; Cubie, H.A.; Skomedal, H.; Bernard, H.U. High-throughput detection of human papillomavirus-18 L1 gene methylation, a candidate biomarker for the progression of cervical neoplasia. Virology 2007, 361, 185–193. [Google Scholar] [CrossRef]
- Turan, T.; Kalantari, M.; Calleja-Macias, I.E.; Cubie, H.A.; Cuschieri, K.; Villa, L.L.; Skomedal, H.; Barrera-Saldana, H.A.; Bernard, H.U. Methylation of the human papillomavirus-18 L1 gene: A biomarker of neoplastic progression? Virology 2006, 349, 175–183. [Google Scholar] [CrossRef]
- Badal, V.; Chuang, L.S.; Tan, E.H.; Badal, S.; Villa, L.L.; Wheeler, C.M.; Li, B.F.; Bernard, H.U. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: Genomic hypomethylation correlates with carcinogenic progression. J. Virol. 2003, 77, 6227–6234. [Google Scholar] [CrossRef]
- Robert, M.F.; Morin, S.; Beaulieu, N.; Gauthier, F.; Chute, I.C.; Barsalou, A.; MacLeod, A.R. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 2003, 33, 61–65. [Google Scholar] [CrossRef]
- Torres-Rojas, F.I.; Alarcon-Romero, L.D.C.; Leyva-Vazquez, M.A.; Ortiz-Ortiz, J.; Mendoza-Catalan, M.A.; Hernandez-Sotelo, D.; Del Moral-Hernandez, O.; Rodriguez-Ruiz, H.A.; Leyva-Illades, D.; Flores-Alfaro, E.; et al. Methylation of the L1 gene and integration of human papillomavirus 16 and 18 in cervical carcinoma and premalignant lesions. Oncol. Lett. 2018, 15, 2278–2286. [Google Scholar] [CrossRef] [PubMed]
- Wentzensen, N.; Sun, C.; Ghosh, A.; Kinney, W.; Mirabello, L.; Wacholder, S.; Shaber, R.; LaMere, B.; Clarke, M.; Lorincz, A.T.; et al. Methylation of HPV18, HPV31, and HPV45 genomes and cervical intraepithelial neoplasia grade 3. J. Natl. Cancer Inst. 2012, 104, 1738–1749. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.A.; Wentzensen, N.; Mirabello, L.; Ghosh, A.; Wacholder, S.; Harari, A.; Lorincz, A.; Schiffman, M.; Burk, R.D. Human papillomavirus DNA methylation as a potential biomarker for cervical cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 2125–2137. [Google Scholar] [CrossRef] [PubMed]
- Kalantari, M.; Villa, L.L.; Calleja-Macias, I.E.; Bernard, H.U. Human papillomavirus-16 and -18 in penile carcinomas: DNA methylation, chromosomal recombination and genomic variation. Int. J. Cancer 2008, 123, 1832–1840. [Google Scholar] [CrossRef] [PubMed]
- Balderas-Loaeza, A.; Anaya-Saavedra, G.; Ramirez-Amador, V.A.; Guido-Jimenez, M.C.; Kalantari, M.; Calleja-Macias, I.E.; Bernard, H.U.; Garcia-Carranca, A. Human papillomavirus-16 DNA methylation patterns support a causal association of the virus with oral squamous cell carcinomas. Int. J. Cancer 2007, 120, 2165–2169. [Google Scholar] [CrossRef] [PubMed]
- Kalantari, M.; Garcia-Carranca, A.; Morales-Vazquez, C.D.; Zuna, R.; Montiel, D.P.; Calleja-Macias, I.E.; Johansson, B.; Andersson, S.; Bernard, H.U. Laser capture microdissection of cervical human papillomavirus infections: Copy number of the virus in cancerous and normal tissue and heterogeneous DNA methylation. Virology 2009, 390, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Kalantari, M.; Calleja-Macias, I.E.; Tewari, D.; Hagmar, B.; Lie, K.; Barrera-Saldana, H.A.; Wiley, D.J.; Bernard, H.U. Conserved methylation patterns of human papillomavirus type 16 DNA in asymptomatic infection and cervical neoplasia. J. Virol. 2004, 78, 12762–12772. [Google Scholar] [CrossRef]
- Fertey, J.; Hagmann, J.; Ruscheweyh, H.J.; Munk, C.; Kjaer, S.; Huson, D.; Haedicke-Jarboui, J.; Stubenrauch, F.; Iftner, T. Methylation of CpG 5962 in L1 of the human papillomavirus 16 genome as a potential predictive marker for viral persistence: A prospective large cohort study using cervical swab samples. Cancer Med. 2020, 9, 1058–1068. [Google Scholar] [CrossRef]
- Vinokurova, S.; von Knebel Doeberitz, M. Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS ONE 2011, 6, e24451. [Google Scholar] [CrossRef]
- Badal, S.; Badal, V.; Calleja-Macias, I.E.; Kalantari, M.; Chuang, L.S.; Li, B.F.; Bernard, H.U. The human papillomavirus-18 genome is efficiently targeted by cellular DNA methylation. Virology 2004, 324, 483–492. [Google Scholar] [CrossRef]
- Kim, K.; Garner-Hamrick, P.A.; Fisher, C.; Lee, D.; Lambert, P.F. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J. Virol. 2003, 77, 12450–12459. [Google Scholar] [CrossRef] [PubMed]
- Burnett, T.S.; Gallimore, P.H. Introduction of cloned human papillomavirus 1a DNA into rat fibroblasts: Integration, de novo methylation and absence of cellular morphological transformation. J. Gen. Virol. 1985, 66, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Van Tine, B.A.; Kappes, J.C.; Banerjee, N.S.; Knops, J.; Lai, L.; Steenbergen, R.D.; Meijer, C.L.; Snijders, P.J.; Chatis, P.; Broker, T.R.; et al. Clonal selection for transcriptionally active viral oncogenes during progression to cancer. J. Virol. 2004, 78, 11172–11186. [Google Scholar] [CrossRef]
- Kalantari, M.; Lee, D.; Calleja-Macias, I.E.; Lambert, P.F.; Bernard, H.U. Effects of cellular differentiation, chromosomal integration and 5-aza-2′-deoxycytidine treatment on human papillomavirus-16 DNA methylation in cultured cell lines. Virology 2008, 374, 292–303. [Google Scholar] [CrossRef]
- Fernandez, A.F.; Rosales, C.; Lopez-Nieva, P.; Grana, O.; Ballestar, E.; Ropero, S.; Espada, J.; Melo, S.A.; Lujambio, A.; Fraga, M.F.; et al. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 2009, 19, 438–451. [Google Scholar] [CrossRef]
- Meissner, J.D. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J. Gen. Virol. 1999, 80, 1725–1733. [Google Scholar] [CrossRef]
- Diao, M.K.; Liu, C.Y.; Liu, H.W.; Li, J.T.; Li, F.; Mehryar, M.M.; Wang, Y.J.; Zhan, S.B.; Zhou, Y.B.; Zhong, R.G.; et al. Integrated HPV genomes tend to integrate in gene desert areas in the CaSki, HeLa, and SiHa cervical cancer cell lines. Life Sci. 2015, 127, 46–52. [Google Scholar] [CrossRef]
- Bernat, A.; Avvakumov, N.; Mymryk, J.S.; Banks, L. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene 2003, 22, 7871–7881. [Google Scholar] [CrossRef]
- Zimmermann, H.; Degenkolbe, R.; Bernard, H.U.; O’Connor, M.J. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 1999, 73, 6209–6219. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Huang, S.M.; Baglia, L.A.; McCance, D.J. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999, 18, 5061–5072. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lee, B.; Kim, J.; Kim, D.W.; Choe, J. cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J. Biol. Chem. 2000, 275, 7045–7051. [Google Scholar] [CrossRef] [PubMed]
- Cricca, M.; Venturoli, S.; Leo, E.; Costa, S.; Musiani, M.; Zerbini, M. Disruption of HPV 16 E1 and E2 genes in precancerous cervical lesions. J. Virol. Methods 2009, 158, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Arias-Pulido, H.; Peyton, C.L.; Joste, N.E.; Vargas, H.; Wheeler, C.M. Human papillomavirus type 16 integration in cervical carcinoma in situ and in invasive cervical cancer. J. Clin. Microbiol. 2006, 44, 1755–1762. [Google Scholar] [CrossRef] [PubMed]
- Jacquin, E.; Baraquin, A.; Ramanah, R.; Carcopino, X.; Morel, A.; Valmary-Degano, S.; Bravo, I.G.; de Sanjose, S.; Riethmuller, D.; Mougin, C.; et al. Methylation of human papillomavirus Type 16 CpG sites at E2-binding site 1 (E2BS1), E2BS2, and the Sp1-binding site in cervical cancer samples as determined by high-resolution melting analysis-PCR. J. Clin. Microbiol. 2013, 51, 3207–3215. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, B.; Sengupta, S. CpG methylation of HPV 16 LCR at E2 binding site proximal to P97 is associated with cervical cancer in presence of intact E2. Virology 2006, 354, 280–285. [Google Scholar] [CrossRef]
- Thain, A.; Jenkins, O.; Clarke, A.R.; Gaston, K. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J. Virol. 1996, 70, 7233–7235. [Google Scholar] [CrossRef]
- Reuschenbach, M.; Huebbers, C.U.; Prigge, E.S.; Bermejo, J.L.; Kalteis, M.S.; Preuss, S.F.; Seuthe, I.M.; Kolligs, J.; Speel, E.J.; Olthof, N.; et al. Methylation status of HPV16 E2-binding sites classifies subtypes of HPV-associated oropharyngeal cancers. Cancer 2015, 121, 1966–1976. [Google Scholar] [CrossRef] [PubMed]
- Zorzi, M.; Del Mistro, A.; Giorgi Rossi, P.; Laurino, L.; Battagello, J.; Lorio, M.; Solda, M.; Martinotti Gabellotti, E.; Maran, M.; Dal Cin, A.; et al. Risk of CIN2 or more severe lesions after negative HPV-mRNA E6/E7 overexpression assay and after negative HPV-DNA test: Concurrent cohorts with a 5-year follow-up. Int. J. Cancer 2020, 146, 3114–3123. [Google Scholar] [CrossRef]
- Giorgi Rossi, P.; Bisanzi, S.; Allia, E.; Mongia, A.; Carozzi, F.; Gillio-Tos, A.; De Marco, L.; Ronco, G.; Gustinucci, D.; Del Mistro, A.; et al. Determinants of Viral Oncogene E6-E7 mRNA Overexpression in a Population-Based Large Sample of Women Infected by High-Risk Human Papillomavirus Types. J. Clin. Microbiol. 2017, 55, 1056–1065. [Google Scholar] [CrossRef]
- Das Ghosh, D.; Bhattacharjee, B.; Sen, S.; Premi, L.; Mukhopadhyay, I.; Chowdhury, R.R.; Roy, S.; Sengupta, S. Some novel insights on HPV16 related cervical cancer pathogenesis based on analyses of LCR methylation, viral load, E7 and E2/E4 expressions. PLoS ONE 2012, 7, e44678. [Google Scholar] [CrossRef]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991, 10, 4129–4135. [Google Scholar] [CrossRef]
- Scheffner, M.; Huibregtse, J.M.; Vierstra, R.D.; Howley, P.M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993, 75, 495–505. [Google Scholar] [CrossRef]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell. Biol. 1993, 13, 4918–4927. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Zhao, Y.; Meng, G.; Zeng, M.; Srinivasan, S.; Delmolino, L.M.; Gao, Q.; Dimri, G.; Weber, G.F.; Wazer, D.E.; et al. Human papillomavirus oncoprotein E6 inactivates the transcriptional coactivator human ADA3. Mol. Cell. Biol. 2002, 22, 5801–5812. [Google Scholar] [CrossRef] [PubMed]
- Shamanin, V.A.; Sekaric, P.; Androphy, E.J. hAda3 degradation by papillomavirus type 16 E6 correlates with abrogation of the p14ARF-p53 pathway and efficient immortalization of human mammary epithelial cells. J. Virol. 2008, 82, 3912–3920. [Google Scholar] [CrossRef] [PubMed]
- Munger, K.; Werness, B.A.; Dyson, N.; Phelps, W.C.; Harlow, E.; Howley, P.M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989, 8, 4099–4105. [Google Scholar] [CrossRef] [PubMed]
- Dyson, N.; Howley, P.M.; Munger, K.; Harlow, E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989, 243, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Boyer, S.N.; Wazer, D.E.; Band, V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996, 56, 4620–4624. [Google Scholar] [PubMed]
- Hwang, E.S.; Nottoli, T.; Dimaio, D. The HPV16 E5 protein: Expression, detection, and stable complex formation with transmembrane proteins in COS cells. Virology 1995, 211, 227–233. [Google Scholar] [CrossRef]
- Chen, S.L.; Lin, S.T.; Tsai, T.C.; Hsiao, W.C.; Tsao, Y.P. ErbB4 (JM-b/CYT-1)-induced expression and phosphorylation of c-Jun is abrogated by human papillomavirus type 16 E5 protein. Oncogene 2007, 26, 42–53. [Google Scholar] [CrossRef]
- Regan, J.A.; Laimins, L.A. Bap31 is a novel target of the human papillomavirus E5 protein. J. Virol. 2008, 82, 10042–10051. [Google Scholar] [CrossRef]
- Kotnik Halavaty, K.; Regan, J.; Mehta, K.; Laimins, L. Human papillomavirus E5 oncoproteins bind the A4 endoplasmic reticulum protein to regulate proliferative ability upon differentiation. Virology 2014, 452–453, 223–230. [Google Scholar] [CrossRef]
- Webb Strickland, S.; Brimer, N.; Lyons, C.; Vande Pol, S.B. Human Papillomavirus E6 interaction with cellular PDZ domain proteins modulates YAP nuclear localization. Virology 2018, 516, 127–138. [Google Scholar] [CrossRef]
- Jing, M.; Bohl, J.; Brimer, N.; Kinter, M.; Vande Pol, S.B. Degradation of tyrosine phosphatase PTPN3 (PTPH1) by association with oncogenic human papillomavirus E6 proteins. J. Virol. 2007, 81, 2231–2239. [Google Scholar] [CrossRef]
- Hoover, A.C.; Strand, G.L.; Nowicki, P.N.; Anderson, M.E.; Vermeer, P.D.; Klingelhutz, A.J.; Bossler, A.D.; Pottala, J.V.; Hendriks, W.J.; Lee, J.H. Impaired PTPN13 phosphatase activity in spontaneous or HPV-induced squamous cell carcinomas potentiates oncogene signaling through the MAP kinase pathway. Oncogene 2009, 28, 3960–3970. [Google Scholar] [CrossRef] [PubMed]
- Spanos, W.C.; Hoover, A.; Harris, G.F.; Wu, S.; Strand, G.L.; Anderson, M.E.; Klingelhutz, A.J.; Hendriks, W.; Bossler, A.D.; Lee, J.H. The PDZ binding motif of human papillomavirus type 16 E6 induces PTPN13 loss, which allows anchorage-independent growth and synergizes with ras for invasive growth. J. Virol. 2008, 82, 2493–2500. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Glaunsinger, B.; Mantovani, F.; Banks, L.; Javier, R.T. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 2000, 74, 9680–9693. [Google Scholar] [CrossRef]
- Bonilla-Delgado, J.; Bulut, G.; Liu, X.; Cortes-Malagon, E.M.; Schlegel, R.; Flores-Maldonado, C.; Contreras, R.G.; Chung, S.H.; Lambert, P.F.; Uren, A.; et al. The E6 oncoprotein from HPV16 enhances the canonical Wnt/beta-catenin pathway in skin epidermis in vivo. Mol. Cancer Res. MCR 2012, 10, 250–258. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Mo, D.; Liu, H.; Veena, M.S.; Srivatsan, E.S.; Massoumi, R.; Rettig, M.B. Inactivation of the CYLD deubiquitinase by HPV E6 mediates hypoxia-induced NF-kappaB activation. Cancer Cell 2008, 14, 394–407. [Google Scholar] [CrossRef]
- Thomas, M.; Myers, M.P.; Massimi, P.; Guarnaccia, C.; Banks, L. Analysis of Multiple HPV E6 PDZ Interactions Defines Type-Specific PDZ Fingerprints That Predict Oncogenic Potential. PLoS Pathog. 2016, 12, e1005766. [Google Scholar] [CrossRef]
- Lee, S.S.; Weiss, R.S.; Javier, R.T. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 1997, 94, 6670–6675. [Google Scholar] [CrossRef]
- Kiyono, T.; Hiraiwa, A.; Fujita, M.; Hayashi, Y.; Akiyama, T.; Ishibashi, M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 1997, 94, 11612–11616. [Google Scholar] [CrossRef]
- Habig, M.; Smola, H.; Dole, V.S.; Derynck, R.; Pfister, H.; Smola-Hess, S. E7 proteins from high- and low-risk human papillomaviruses bind to TGF-beta-regulated Smad proteins and inhibit their transcriptional activity. Arch. Virol. 2006, 151, 1961–1972. [Google Scholar] [CrossRef]
- Lee, D.K.; Kim, B.C.; Kim, I.Y.; Cho, E.A.; Satterwhite, D.J.; Kim, S.J. The human papilloma virus E7 oncoprotein inhibits transforming growth factor-beta signaling by blocking binding of the Smad complex to its target sequence. J. Biol. Chem. 2002, 277, 38557–38564. [Google Scholar] [CrossRef]
- Yun, H.Y.; Kim, M.W.; Lee, H.S.; Kim, W.; Shin, J.H.; Kim, H.; Shin, H.C.; Park, H.; Oh, B.H.; Kim, W.K.; et al. Structural basis for recognition of the tumor suppressor protein PTPN14 by the oncoprotein E7 of human papillomavirus. PLoS Biol. 2019, 17, e3000367. [Google Scholar] [CrossRef]
- Hatterschide, J.; Bohidar, A.E.; Grace, M.; Nulton, T.J.; Kim, H.W.; Windle, B.; Morgan, I.M.; Munger, K.; White, E.A. PTPN14 degradation by high-risk human papillomavirus E7 limits keratinocyte differentiation and contributes to HPV-mediated oncogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 7033–7042. [Google Scholar] [CrossRef]
- Szalmas, A.; Tomaic, V.; Basukala, O.; Massimi, P.; Mittal, S.; Konya, J.; Banks, L. The PTPN14 Tumor Suppressor Is a Degradation Target of Human Papillomavirus E7. J. Virol. 2017, 91, e00057-17. [Google Scholar] [CrossRef]
- White, E.A.; Munger, K.; Howley, P.M. High-Risk Human Papillomavirus E7 Proteins Target PTPN14 for Degradation. mBio 2016, 7, E01530-16. [Google Scholar] [CrossRef]
- Jones, D.L.; Alani, R.M.; Munger, K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997, 11, 2101–2111. [Google Scholar] [CrossRef]
- Funk, J.O.; Waga, S.; Harry, J.B.; Espling, E.; Stillman, B.; Galloway, D.A. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997, 11, 2090–2100. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Ferril, S.; Snider, A.; Barbosa, M. In-vivo analysis of hpv e7 protein association with prb, p107 and p130. Int. J. Oncol. 1995, 6, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chen, W.; Roman, A. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc. Natl. Acad. Sci. USA 2006, 103, 437–442. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin-Drubin, M.E.; Huh, K.W.; Munger, K. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J. Virol. 2008, 82, 8695–8705. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.G.; Lee, D.; Kim, J.; Seo, T.; Choe, J. Human papillomavirus type 16 E7 binds to E2F1 and activates E2F1-driven transcription in a retinoblastoma protein-independent manner. J. Biol. Chem. 2002, 277, 2923–2930. [Google Scholar] [CrossRef] [PubMed]
- Dyson, N.; Guida, P.; Munger, K.; Harlow, E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J. Virol. 1992, 66, 6893–6902. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, M.C.; Ruesch, M.N.; Laimins, L.A. Human papillomavirus E7 oncoproteins bind a single form of cyclin E in a complex with cdk2 and p107. Virology 1996, 215, 73–82. [Google Scholar] [CrossRef]
- Nguyen, C.L.; Munger, K. Direct association of the HPV16 E7 oncoprotein with cyclin A/CDK2 and cyclin E/CDK2 complexes. Virology 2008, 380, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lim, C.; Seo, T.; Kwon, H.; Min, H.; Choe, J. The viral oncogene human papillomavirus E7 deregulates transcriptional silencing by Brm-related gene 1 via molecular interactions. J. Biol. Chem. 2002, 277, 48842–48848. [Google Scholar] [CrossRef]
- Pang, C.L.; Toh, S.Y.; He, P.; Teissier, S.; Ben Khalifa, Y.; Xue, Y.; Thierry, F. A functional interaction of E7 with B-Myb-MuvB complex promotes acute cooperative transcriptional activation of both S- and M-phase genes. (129 c). Oncogene 2014, 33, 4039–4049. [Google Scholar] [CrossRef]
- Brehm, A.; Nielsen, S.J.; Miska, E.A.; McCance, D.J.; Reid, J.L.; Bannister, A.J.; Kouzarides, T. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999, 18, 2449–2458. [Google Scholar] [CrossRef]
- Ashrafi, G.H.; Haghshenas, M.; Marchetti, B.; Campo, M.S. E5 protein of human papillomavirus 16 downregulates HLA class I and interacts with the heavy chain via its first hydrophobic domain. Int. J. Cancer 2006, 119, 2105–2112. [Google Scholar] [CrossRef]
- Gruener, M.; Bravo, I.G.; Momburg, F.; Alonso, A.; Tomakidi, P. The E5 protein of the human papillomavirus type 16 down-regulates HLA-I surface expression in calnexin-expressing but not in calnexin-deficient cells. Virol. J. 2007, 4, 116. [Google Scholar] [CrossRef]
- Lo Cigno, I.; Calati, F.; Borgogna, C.; Zevini, A.; Albertini, S.; Martuscelli, L.; De Andrea, M.; Hiscott, J.; Landolfo, S.; Gariglio, M. Human Papillomavirus E7 Oncoprotein Subverts Host Innate Immunity via SUV39H1-Mediated Epigenetic Silencing of Immune Sensor Genes. J. Virol. 2020, 94, e01812-19. [Google Scholar] [CrossRef]
- Li, S.; Labrecque, S.; Gauzzi, M.C.; Cuddihy, A.R.; Wong, A.H.; Pellegrini, S.; Matlashewski, G.J.; Koromilas, A.E. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 1999, 18, 5727–5737. [Google Scholar] [CrossRef]
- Chiang, C.; Pauli, E.K.; Biryukov, J.; Feister, K.F.; Meng, M.; White, E.A.; Munger, K.; Howley, P.M.; Meyers, C.; Gack, M.U. The Human Papillomavirus E6 Oncoprotein Targets USP15 and TRIM25 To Suppress RIG-I-Mediated Innate Immune Signaling. J. Virol. 2018, 92, e01737-17. [Google Scholar] [CrossRef]
- Ronco, L.V.; Karpova, A.Y.; Vidal, M.; Howley, P.M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998, 12, 2061–2072. [Google Scholar] [CrossRef]
- Vambutas, A.; DeVoti, J.; Pinn, W.; Steinberg, B.M.; Bonagura, V.R. Interaction of human papillomavirus type 11 E7 protein with TAP-1 results in the reduction of ATP-dependent peptide transport. Clin. Immunol. 2001, 101, 94–99. [Google Scholar] [CrossRef]
- Lau, L.; Gray, E.E.; Brunette, R.L.; Stetson, D.B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 2015, 350, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Donnelly, C.R.; Gong, W.; Heath, B.R.; Hao, Y.; Donnelly, L.A.; Moghbeli, T.; Tan, Y.S.; Lin, X.; Bellile, E.; et al. HPV16 drives cancer immune escape via NLRX1-mediated degradation of STING. J. Clin. Investig. 2020, 130, 1635–1652. [Google Scholar] [CrossRef] [PubMed]
- Barnard, P.; McMillan, N.A. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-alpha. Virology 1999, 259, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Kim, E.J.; Kwon, H.J.; Hwang, E.S.; Namkoong, S.E.; Um, S.J. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J. Biol. Chem. 2000, 275, 6764–6769. [Google Scholar] [CrossRef] [PubMed]
- Spitkovsky, D.; Hehner, S.P.; Hofmann, T.G.; Moller, A.; Schmitz, M.L. The human papillomavirus oncoprotein E7 attenuates NF-kappa B activation by targeting the Ikappa B kinase complex. J. Biol. Chem. 2002, 277, 25576–25582. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.J.; Kalinina, A.; Wang, J.; Nakayama, K.; Nakayama, K.I.; Bagchi, S. The papillomavirus E7 oncoprotein is ubiquitinated by UbcH7 and Cullin 1- and Skp2-containing E3 ligase. J. Virol. 2004, 78, 5338–5346. [Google Scholar] [CrossRef]
- Brimer, N.; Lyons, C.; Vande Pol, S.B. Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology 2007, 358, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, J.; Zhang, L.; Fu, H.; Li, J.; Tian, T.; Zuo, J.; Lv, W.; Ma, X. BCCIPbeta facilitates p53 ubiquitination via binding with E6 protein in high-risk HPV positive head and neck squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2020, 529, 685–691. [Google Scholar] [CrossRef]
- White, E.A.; Sowa, M.E.; Tan, M.J.; Jeudy, S.; Hayes, S.D.; Santha, S.; Munger, K.; Harper, J.W.; Howley, P.M. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc. Natl. Acad. Sci. USA 2012, 109, E260–E267. [Google Scholar] [CrossRef] [PubMed]
- Huh, K.; Zhou, X.; Hayakawa, H.; Cho, J.Y.; Libermann, T.A.; Jin, J.; Harper, J.W.; Munger, K. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J. Virol. 2007, 81, 9737–9747. [Google Scholar] [CrossRef] [PubMed]
- Darnell, G.A.; Schroder, W.A.; Antalis, T.M.; Lambley, E.; Major, L.; Gardner, J.; Birrell, G.; Cid-Arregui, A.; Suhrbier, A. Human papillomavirus E7 requires the protease calpain to degrade the retinoblastoma protein. J. Biol. Chem. 2007, 282, 37492–37500. [Google Scholar] [CrossRef]
- Crook, T.; Tidy, J.A.; Vousden, K.H. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 1991, 67, 547–556. [Google Scholar] [CrossRef]
- Werness, B.A.; Levine, A.J.; Howley, P.M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 1990, 248, 76–79. [Google Scholar] [CrossRef]
- Lechner, M.S.; Laimins, L.A. Inhibition of p53 DNA binding by human papillomavirus E6 proteins. J. Virol. 1994, 68, 4262–4273. [Google Scholar] [CrossRef]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136. [Google Scholar] [CrossRef]
- Iftner, T.; Elbel, M.; Schopp, B.; Hiller, T.; Loizou, J.I.; Caldecott, K.W.; Stubenrauch, F. Interference of papillomavirus E6 protein with single-strand break repair by interaction with XRCC1. EMBO J. 2002, 21, 4741–4748. [Google Scholar] [CrossRef]
- Srivenugopal, K.S.; Ali-Osman, F. The DNA repair protein, O(6)-methylguanine-DNA methyltransferase is a proteolytic target for the E6 human papillomavirus oncoprotein. Oncogene 2002, 21, 5940–5945. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, S.; Meng, Q.; Ma, Y.; Katiyar, P.; Schlegel, R.; Rosen, E.M. BRCA1 interaction with human papillomavirus oncoproteins. J. Biol. Chem. 2005, 280, 33165–33177. [Google Scholar] [CrossRef]
- Yim, E.K.; Lee, K.H.; Myeong, J.; Tong, S.Y.; Um, S.J.; Park, J.S. Novel interaction between HPV E6 and BARD1 (BRCA1-associated ring domain 1) and its biologic roles. DNA Cell Biol. 2007, 26, 753–761. [Google Scholar] [CrossRef]
- Hsu, C.H.; Peng, K.L.; Jhang, H.C.; Lin, C.H.; Wu, S.Y.; Chiang, C.M.; Lee, S.C.; Yu, W.C.; Juan, L.J. The HPV E6 oncoprotein targets histone methyltransferases for modulating specific gene transcription. Oncogene 2012, 31, 2335–2349. [Google Scholar] [CrossRef] [PubMed]
- Au Yeung, C.L.; Tsang, W.P.; Tsang, T.Y.; Co, N.N.; Yau, P.L.; Kwok, T.T. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol. Rep. 2010, 24, 1599–1604. [Google Scholar] [CrossRef]
- Baldwin, A.; Huh, K.W.; Munger, K. Human papillomavirus E7 oncoprotein dysregulates steroid receptor coactivator 1 localization and function. J. Virol. 2006, 80, 6669–6677. [Google Scholar] [CrossRef] [PubMed]
- Avvakumov, N.; Torchia, J.; Mymryk, J.S. Interaction of the HPV E7 proteins with the pCAF acetyltransferase. Oncogene 2003, 22, 3833–3841. [Google Scholar] [CrossRef]
- Huang, S.M.; McCance, D.J. Down regulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p300 and P/CAF. J. Virol. 2002, 76, 8710–8721. [Google Scholar] [CrossRef] [PubMed]
- Burgers, W.A.; Blanchon, L.; Pradhan, S.; de Launoit, Y.; Kouzarides, T.; Fuks, F. Viral oncoproteins target the DNA methyltransferases. Oncogene 2007, 26, 1650–1655. [Google Scholar] [CrossRef] [PubMed]
- Filippova, M.; Song, H.; Connolly, J.L.; Dermody, T.S.; Duerksen-Hughes, P.J. The human papillomavirus 16 E6 protein binds to tumor necrosis factor (TNF) R1 and protects cells from TNF-induced apoptosis. J. Biol. Chem. 2002, 277, 21730–21739. [Google Scholar] [CrossRef]
- Filippova, M.; Parkhurst, L.; Duerksen-Hughes, P.J. The human papillomavirus 16 E6 protein binds to Fas-associated death domain and protects cells from Fas-triggered apoptosis. J. Biol. Chem. 2004, 279, 25729–25744. [Google Scholar] [CrossRef]
- Filippova, M.; Johnson, M.M.; Bautista, M.; Filippov, V.; Fodor, N.; Tungteakkhun, S.S.; Williams, K.; Duerksen-Hughes, P.J. The large and small isoforms of human papillomavirus type 16 E6 bind to and differentially affect procaspase 8 stability and activity. J. Virol. 2007, 81, 4116–4129. [Google Scholar] [CrossRef]
- Thomas, M.; Banks, L. Human papillomavirus (HPV) E6 interactions with Bak are conserved amongst E6 proteins from high and low risk HPV types. J. Gen. Virol. 1999, 80, 1513–1517. [Google Scholar] [CrossRef] [PubMed]
- Shimada, M.; Yamashita, A.; Saito, M.; Ichino, M.; Kinjo, T.; Mizuki, N.; Klinman, D.M.; Okuda, K. The human papillomavirus E6 protein targets apoptosis-inducing factor (AIF) for degradation. Sci. Rep. 2020, 10, 14195. [Google Scholar] [CrossRef] [PubMed]
- Severino, A.; Abbruzzese, C.; Manente, L.; Valderas, A.A.; Mattarocci, S.; Federico, A.; Starace, G.; Chersi, A.; Mileo, A.M.; Paggi, M.G. Human papillomavirus-16 E7 interacts with Siva-1 and modulates apoptosis in HaCaT human immortalized keratinocytes. J. Cell. Physiol. 2007, 212, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Mannhardt, B.; Weinzimer, S.A.; Wagner, M.; Fiedler, M.; Cohen, P.; Jansen-Durr, P.; Zwerschke, W. Human papillomavirus type 16 E7 oncoprotein binds and inactivates growth-inhibitory insulin-like growth factor binding protein 3. Mol. Cell. Biol. 2000, 20, 6483–6495. [Google Scholar] [CrossRef] [PubMed]
- Mileo, A.M.; Abbruzzese, C.; Mattarocci, S.; Bellacchio, E.; Pisano, P.; Federico, A.; Maresca, V.; Picardo, M.; Giorgi, A.; Maras, B.; et al. Human papillomavirus-16 E7 interacts with glutathione S-transferase P1 and enhances its role in cell survival. PLoS ONE 2009, 4, e7254. [Google Scholar] [CrossRef] [PubMed]
- Katzenellenbogen, R.A.; Egelkrout, E.M.; Vliet-Gregg, P.; Gewin, L.C.; Gafken, P.R.; Galloway, D.A. NFX1-123 and poly(A) binding proteins synergistically augment activation of telomerase in human papillomavirus type 16 E6-expressing cells. J. Virol. 2007, 81, 3786–3796. [Google Scholar] [CrossRef]
- Gewin, L.; Myers, H.; Kiyono, T.; Galloway, D.A. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004, 18, 2269–2282. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Dakic, A.; Zhang, Y.; Dai, Y.; Chen, R.; Schlegel, R. HPV E6 protein interacts physically and functionally with the cellular telomerase complex. Proc. Natl. Acad. Sci. USA 2009, 106, 18780–18785. [Google Scholar] [CrossRef]
- Veldman, T.; Liu, X.; Yuan, H.; Schlegel, R. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc. Natl. Acad. Sci. USA 2003, 100, 8211–8216. [Google Scholar] [CrossRef]
- Scarth, J.A.; Patterson, M.R.; Morgan, E.L.; Macdonald, A. The human papillomavirus oncoproteins: A review of the host pathways targeted on the road to transformation. J. Gen. Virol. 2021, 102, 001540. [Google Scholar] [CrossRef]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Ribatti, D. Epithelial-mesenchymal transition in morphogenesis, cancer progression and angiogenesis. Exp. Cell Res. 2017, 353, 1–5. [Google Scholar] [CrossRef]
- Hellner, K.; Mar, J.; Fang, F.; Quackenbush, J.; Munger, K. HPV16 E7 oncogene expression in normal human epithelial cells causes molecular changes indicative of an epithelial to mesenchymal transition. Virology 2009, 391, 57–63. [Google Scholar] [CrossRef]
- Laurson, J.; Khan, S.; Chung, R.; Cross, K.; Raj, K. Epigenetic repression of E-cadherin by human papillomavirus 16 E7 protein. Carcinogenesis 2010, 31, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.S.; Kato, I.; Kim, H.R. A novel function of HPV16-E6/E7 in epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2013, 435, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Bode, A.M.; Dong, Z.; Cao, Y. The epithelial-mesenchymal transition (EMT) is regulated by oncoviruses in cancer. FASEB J. 2016, 30, 3001–3010. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, S.A.; Marra, M.A. Oncogenic Viruses and the Epigenome: How Viruses Hijack Epigenetic Mechanisms to Drive Cancer. Int. J. Mol. Sci. 2023, 24, 9543. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, Z.J.; Jolly, C.; Androphy, E.J.; Mercer, A.; Matthews, C.M.; Hibma, M.H. Transcriptional repression of E-cadherin by human papillomavirus type 16 E6. PLoS ONE 2012, 7, e48954. [Google Scholar] [CrossRef]
- Liu, G. CDH1 promoter methylation in patients with cervical carcinoma: A systematic meta-analysis with trial sequential analysis. Future Oncol 2018, 14, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Holubekova, V.; Mendelova, A.; Grendar, M.; Mersakova, S.; Kapustova, I.; Jasek, K.; Vanochova, A.; Danko, J.; Lasabova, Z. Methylation pattern of CDH1 promoter and its association with CDH1 gene expression in cytological cervical specimens. Oncol. Lett. 2016, 12, 2613–2621. [Google Scholar] [CrossRef] [PubMed]
- Durzynska, J.; Lesniewicz, K.; Poreba, E. Human papillomaviruses in epigenetic regulations. Mutat. Res. Rev. Mutat. Res. 2017, 772, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Rosendo-Chalma, P.; Antonio-Vejar, V.; Bigoni-Ordonez, G.D.; Patino-Morales, C.C.; Cano-Garcia, A.; Garcia-Carranca, A. CDH1 and SNAI1 are regulated by E7 from human papillomavirus types 16 and 18. Int. J. Oncol. 2020, 57, 301–313. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).