Microalgae Inoculation Significantly Shapes the Structure, Alters the Assembly Process, and Enhances the Stability of Bacterial Communities in Shrimp-Rearing Water
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Microalgal Culture
2.2. Experimental Design
2.3. Environmental and Bacterial Sample Collection
2.4. Bacterial Illumina HiSeq Sequencing and Bioinformatic Analysis
2.5. Statistical Analysis
3. Results
3.1. Variations in Microalgal and Nutrient Factors
3.2. Dynamics of Bacterial Community Composition and Diversity
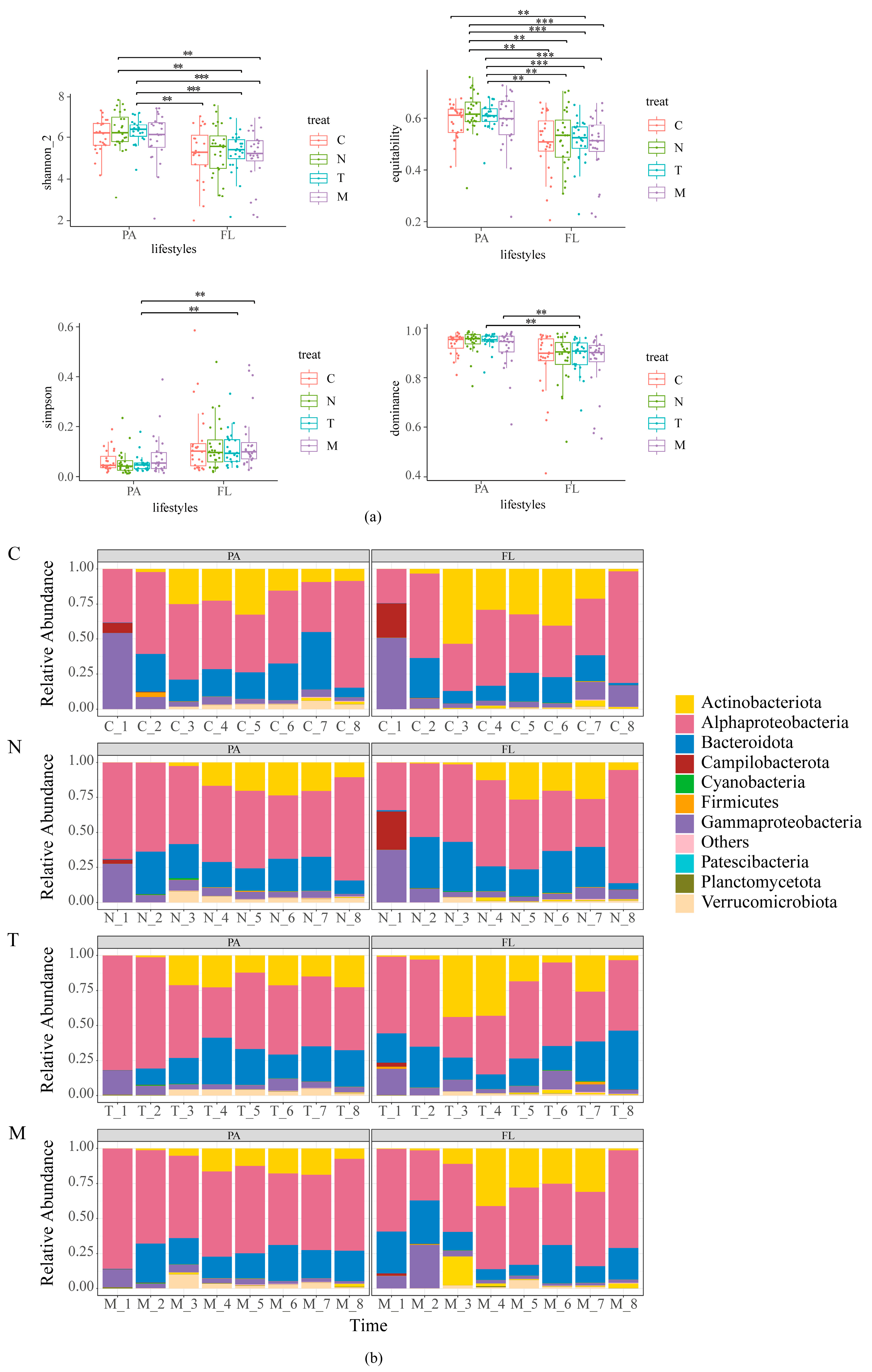
3.2.1. Alpha Diversity and Taxonomic Composition
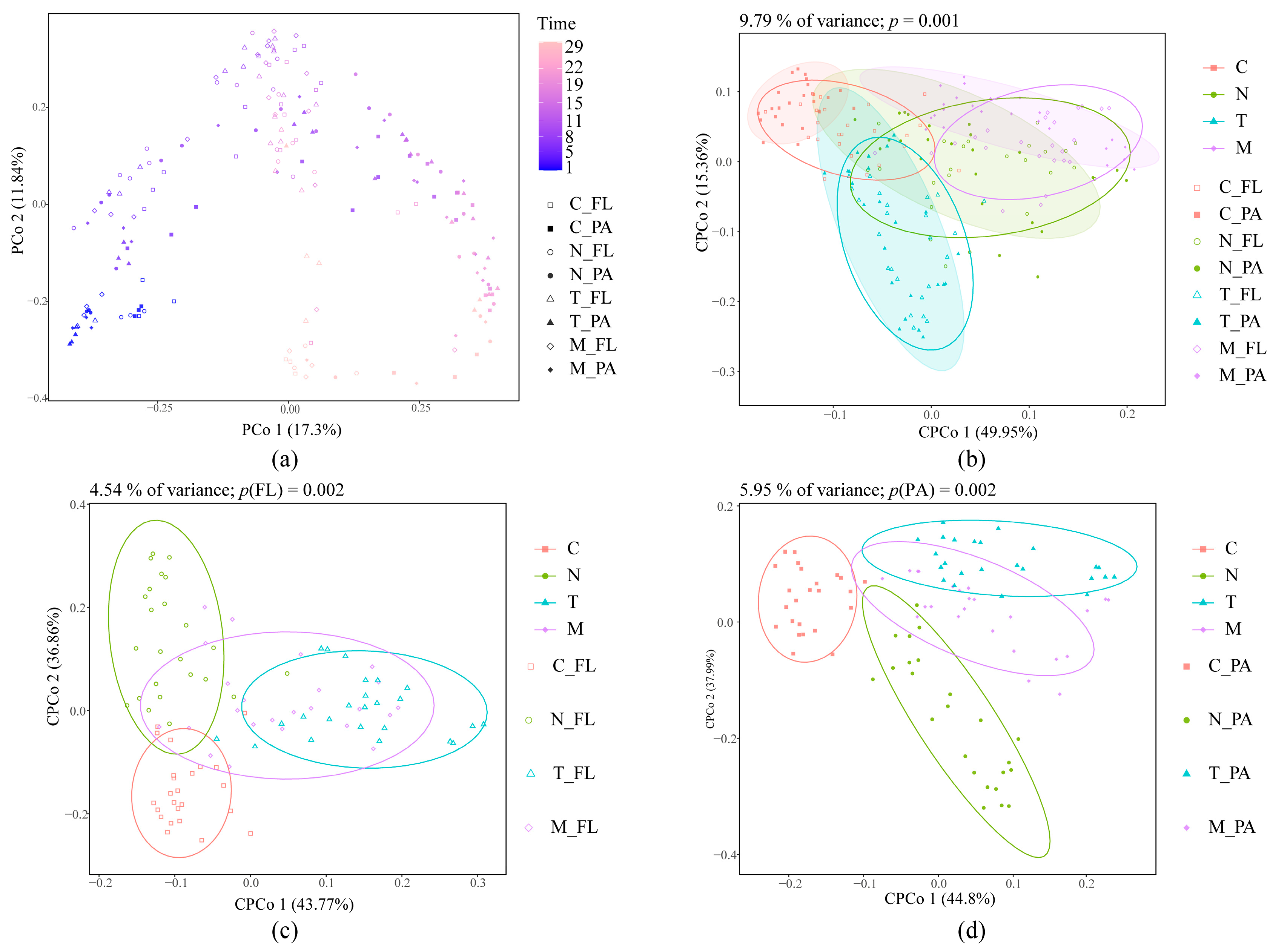
3.2.2. Beta Diversity
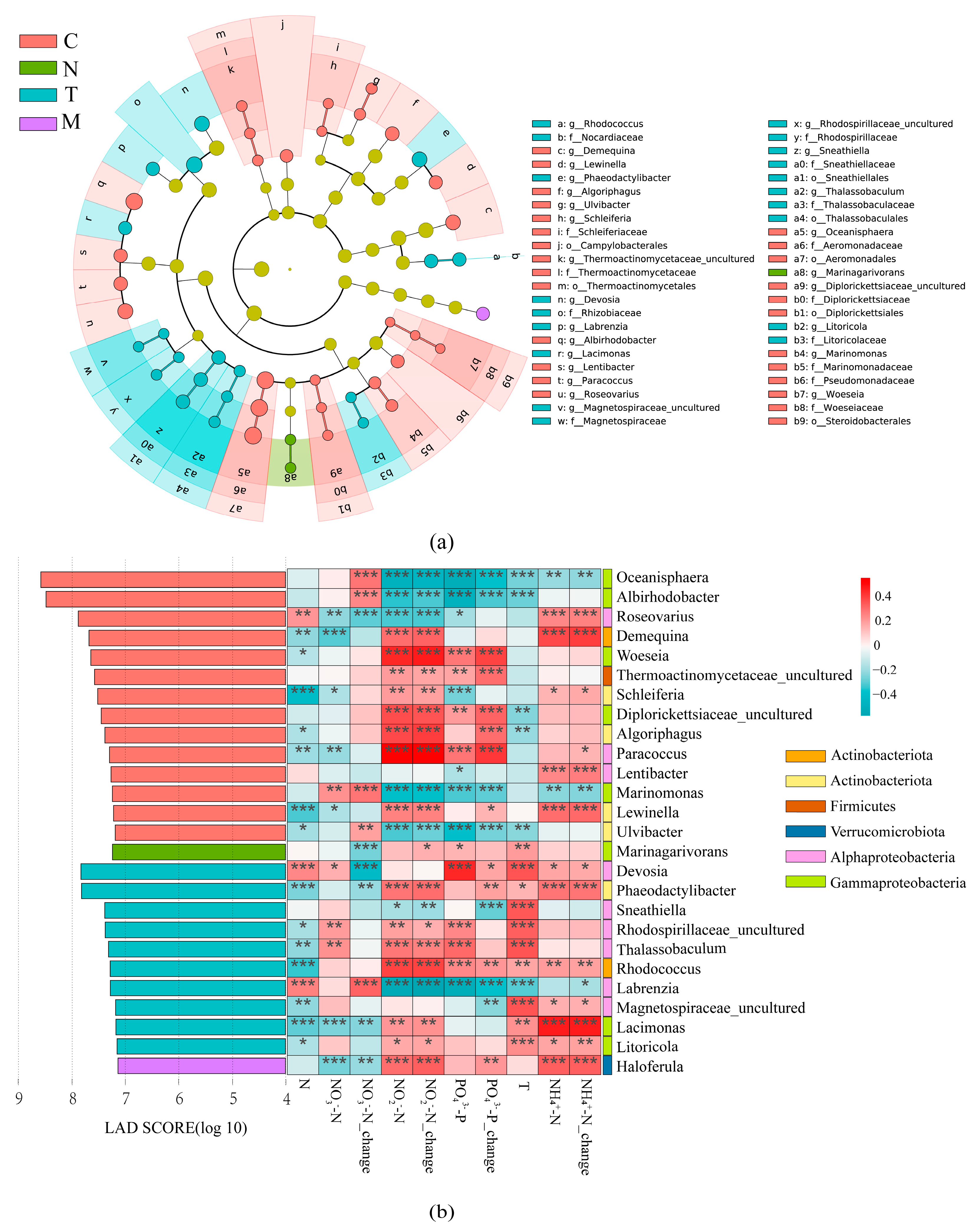
3.3. Differences in Species among Treatments
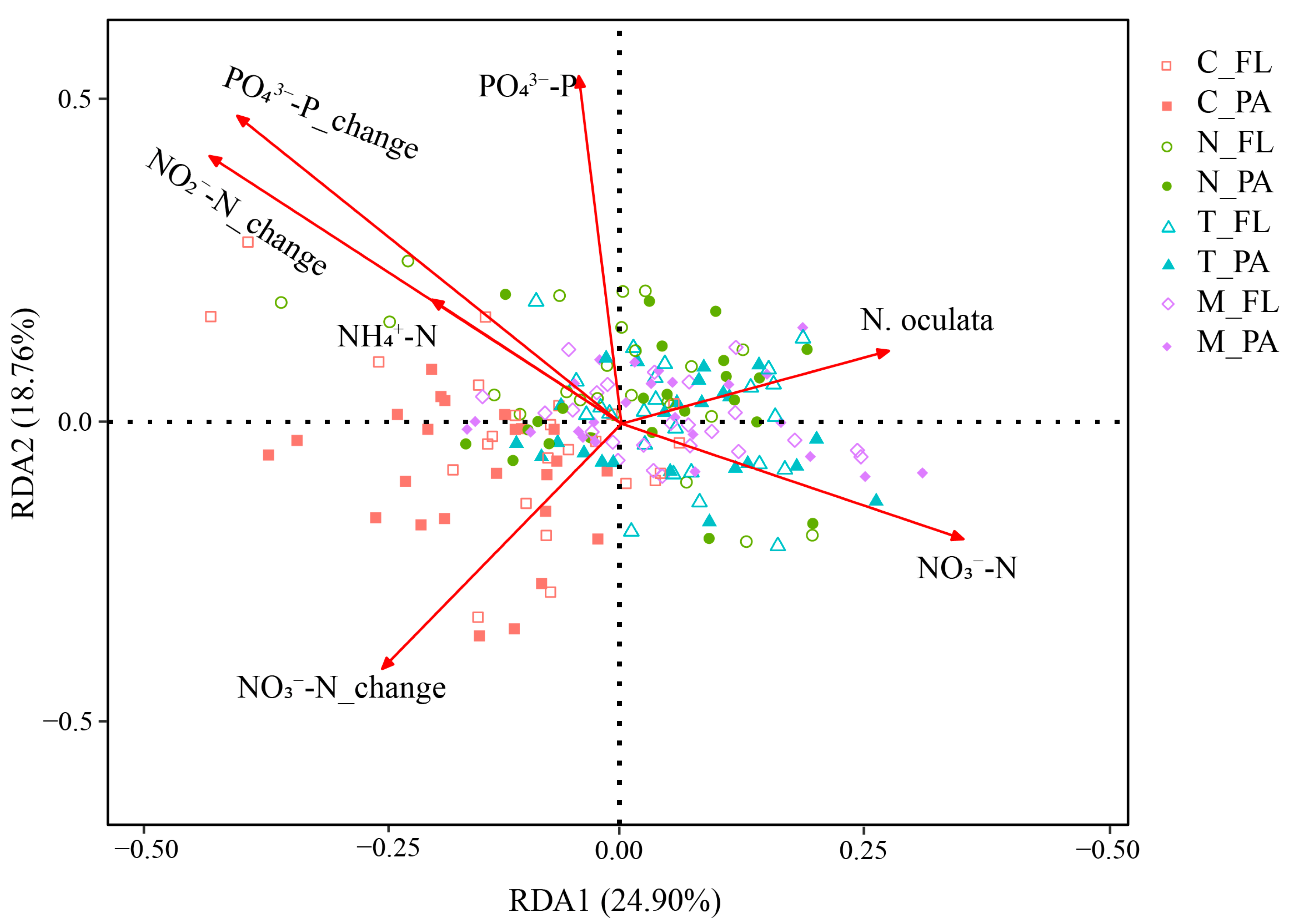
3.4. Relationship between Bacterial Community Structure and Environmental Factors
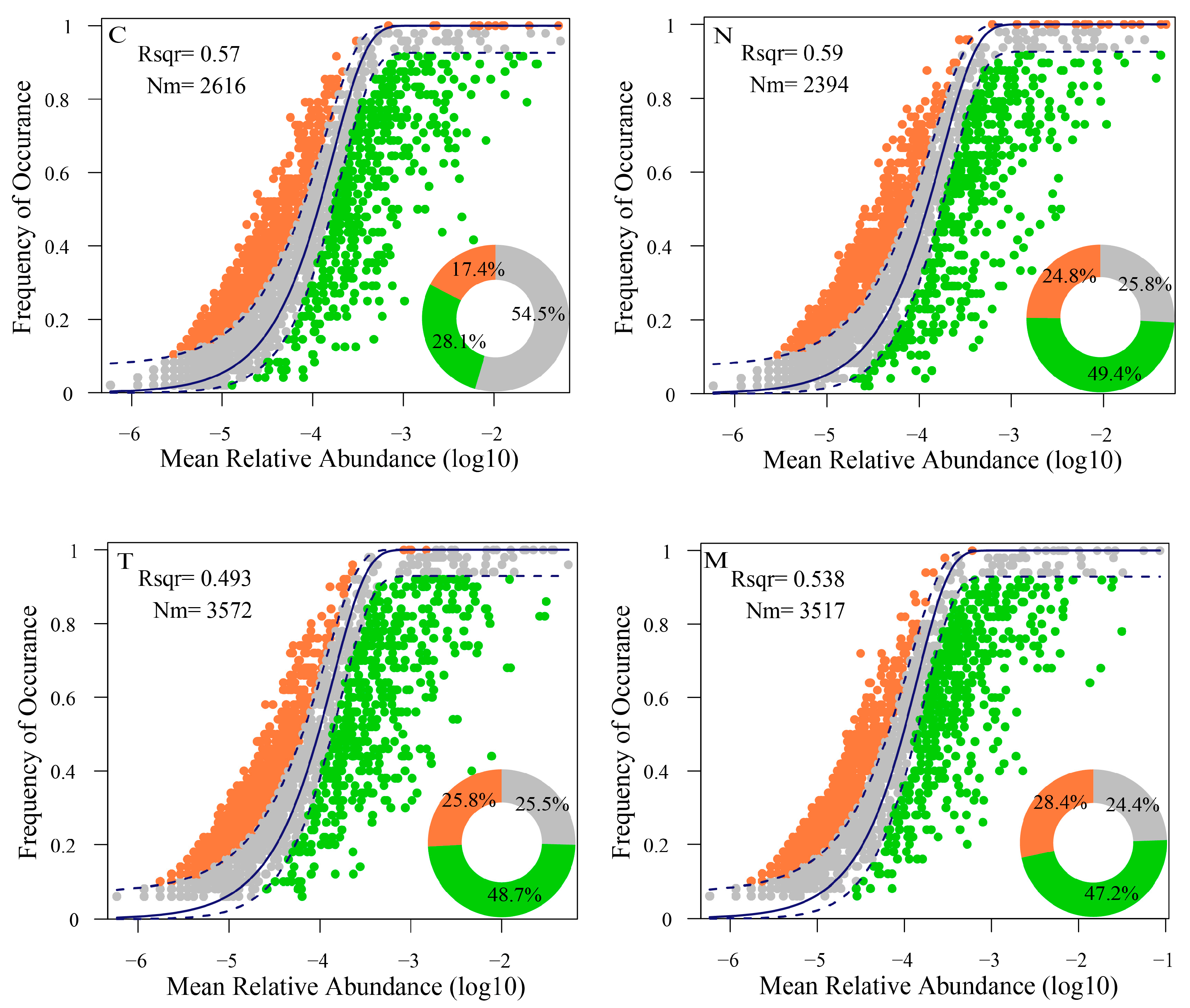
3.5. Assembly Process and Stability of Microbial Communities
4. Discussion
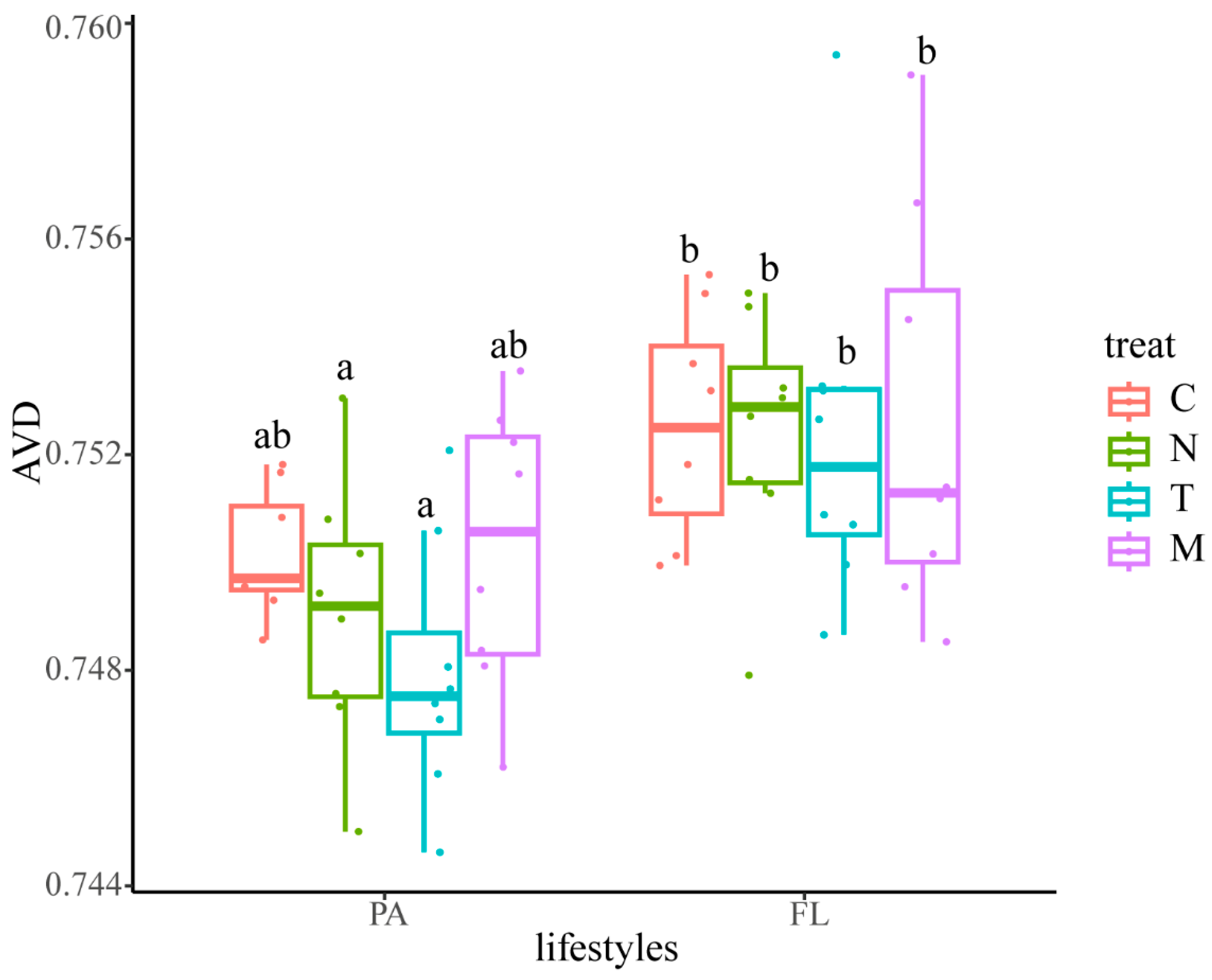
4.1. Microalgae Inoculation and Nutrient Changes Distinctly Influence the Response Patterns of PA and FL Bacterial Communities
4.2. Microalgae Inoculation Induces Alterations in the Assembly Processes of the Bacterial Community and Enhances Community Stability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Edwards, P.; Zhang, W.; Belton, B.; Little, D.C. Misunderstandings, Myths and Mantras in Aquaculture: Its Contribution to World Food Supplies Has Been Systematically over Reported. Mar. Policy 2019, 106, 103547. [Google Scholar] [CrossRef]
- Yang, W.; Zheng, C.; Zheng, Z.; Wei, Y.; Lu, K.; Zhu, J. Nutrient Enrichment during Shrimp Cultivation Alters Bacterioplankton Assemblies and Destroys Community Stability. Ecotoxicol. Environ. Saf. 2018, 156, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Anh, P.T.; Kroeze, C.; Bush, S.R.; Mol, A. Water Pollution by Intensive Brackish Shrimp Farming in South-East Vietnam: Causes and Options for Control. Agric. Water Manag. 2010, 97, 872–882. [Google Scholar] [CrossRef]
- Yang, A.; Zhang, G.; Meng, F.; Zhi, R.; Zhang, P.; Zhu, Y. Nitrogen Metabolism in Photosynthetic Bacteria Wastewater Treatment: A Novel Nitrogen Transformation Pathway. Bioresour. Technol. 2019, 294, 122162. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The Unseen Majority: Soil Microbes as Drivers of Plant Diversity and Productivity in Terrestrial Ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Azam, F.; Malfatti, F. Microbial Structuring of Marine Ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Rajeev, R.; Adithya, K.K.; Kiran, G.S.; Selvin, J. Healthy Microbiome: A Key to Successful and Sustainable Shrimp Aquaculture. Rev. Aquacult. 2021, 13, 238–258. [Google Scholar] [CrossRef]
- Heyse, J.; Props, R.; Kongnuan, P.; De Schryver, P.; Rombaut, G.; Defoirdt, T.; Boon, N. Rearing Water Microbiomes in White Leg Shrimp (Litopenaeus vannamei) Larviculture Assemble Stochastically and Are Influenced by the Microbiomes of Live Feed Products. Environ. Microbiol. 2021, 23, 281–298. [Google Scholar] [CrossRef]
- Cho, D.-H.; Ramanan, R.; Heo, J.; Lee, J.; Kim, B.-H.; Oh, H.-M.; Kim, H.-S. Enhancing Microalgal Biomass Productivity by Engineering a Microalgal–Bacterial Community. Bioresour. Technol. 2015, 175, 578–585. [Google Scholar] [CrossRef]
- Ghulam, M.; Kisay, L. Advanced Treatment of Wastewater Using Symbiotic Co-Culture of Microalgae and Bacteria. Appl. Chem. Eng. 2016, 27, 1–9. [Google Scholar] [CrossRef]
- Rizoulis, A.; Elliott, D.R.; Rolfe, S.A.; Thornton, S.F.; Banwart, S.A.; Pickup, R.W.; Scholes, J.D. Diversity of Planktonic and Attached Bacterial Communities in a Phenol-Contaminated Sandstone Aquifer. Microb. Ecol. 2013, 66, 84–95. [Google Scholar] [CrossRef]
- Zhang, M.; Pan, L.; Huang, F.; Gao, S.; Su, C.; Zhang, M.; He, Z. Metagenomic Analysis of Composition, Function and Cycling Processes of Microbial Community in Water, Sediment and Effluent of Litopenaeus Vannamei Farming Environments under Different Culture Modes. Aquaculture 2019, 506, 280–293. [Google Scholar] [CrossRef]
- Park, B.S.; Choi, W.-J.; Guo, R.; Kim, H.; Ki, J.-S. Changes in Free-Living and Particle-Associated Bacterial Communities Depending on the Growth Phases of Marine Green Algae, Tetraselmis Suecica. J. Mar. Sci. Eng. 2021, 9, 171. [Google Scholar] [CrossRef]
- Bagatini, I.L.; Eiler, A.; Bertilsson, S.; Klaveness, D.; Tessarolli, L.P.; Vieira, A.A.H. Host-Specificity and Dynamics in Bacterial Communities Associated with Bloom-Forming Freshwater Phytoplankton. PLoS ONE 2014, 9, e85950. [Google Scholar] [CrossRef]
- Yang, C.; Wang, Q.; Simon, P.N.; Liu, J.; Liu, L.; Dai, X.; Zhang, X.; Kuang, J.; Igarashi, Y.; Pan, X.; et al. Distinct Network Interactions in Particle-Associated and Free-Living Bacterial Communities during a Microcystis Aeruginosa Bloom in a Plateau Lake. Front. Microbiol. 2017, 8, 1201. [Google Scholar] [CrossRef]
- Crespo, B.G.; Pommier, T.; Fernández-Gómez, B.; Pedrós-Alió, C. Taxonomic Composition of the Particle-Attached and Free-Living Bacterial Assemblages in the Northwest Mediterranean Sea Analyzed by Pyrosequencing of the 16S rRNA. MicrobiologyOpen 2013, 2, 541–552. [Google Scholar] [CrossRef]
- Jiangtao, L.; Bingbing, W.; Jiani, W.; Ying, L.; Shamik, D.; Li, Z.; Jiasong, F. Variation in abundance and community structure of particle-attached and free-living bacteria in the South China Sea. Deep.-Sea Res. 2015, 122, 64–73. [Google Scholar] [CrossRef]
- Jain, A.; Krishnan, K.P. Differences in Free-Living and Particle-Associated Bacterial Communities and Their Spatial Variation in Kongsfjorden, Arctic. J. Basic Microbiol. 2017, 57, 827–838. [Google Scholar] [CrossRef]
- Cai, X.; Yao, L.; Hu, Y.; Jiang, H.; Shen, M.; Hu, Q.; Wang, Z.; Dahlgren, R.A. Particle-Attached Microorganism Oxidation of Ammonia in a Hypereutrophic Urban River. J. Basic Microbiol. 2019, 59, 511–524. [Google Scholar] [CrossRef]
- Tang, X.; Chao, J.; Gong, Y.; Wang, Y.; Wilhelm, S.W.; Gao, G. Spatiotemporal Dynamics of Bacterial Community Composition in Large Shallow Eutrophic Lake Taihu: High Overlap between Free-Living and Particle-Attached Assemblages. Limnol. Oceanogr. 2017, 62, 1366–1382. [Google Scholar] [CrossRef]
- Xue, M.; He, Y.; Chen, D.; Wang, L.; Liang, H.; Liu, J.; Wen, C.-Q. Temporal Dynamics of Aquatic Microbiota and Their Correlation with Environmental Factors during Larviculture of the Shrimp Litopenaeus Vannamei. Aquaculture 2020, 529, 735605. [Google Scholar] [CrossRef]
- Ebeling, J.M.; Timmons, M.B.; Bisogni, J.J. Engineering Analysis of the Stoichiometry of Photoautotrophic, Autotrophic, and Heterotrophic Removal of Ammonia–Nitrogen in Aquaculture Systems. Aquaculture 2006, 257, 346–358. [Google Scholar] [CrossRef]
- Zheng, Y.; Yu, M.; Liu, J.; Qiao, Y.; Wang, L.; Li, Z.; Zhang, X.-H.; Yu, M. Bacterial Community Associated with Healthy and Diseased Pacific White Shrimp (Litopenaeus vannamei) Larvae and Rearing Water across Different Growth Stages. Front. Microbiol. 2017, 8, 1362. [Google Scholar] [CrossRef]
- Ren, Z.; Qu, X.; Peng, W.; Yu, Y.; Zhang, M. Nutrients Drive the Structures of Bacterial Communities in Sediments and Surface Waters in the River-Lake System of Poyang Lake. Water 2019, 11, 930. [Google Scholar] [CrossRef]
- Sun, Y.; Tang, K.; Ma, Y.; Zhu, X.; Li, H.; Zhang, F.; Chen, S.; Huang, H. Variations in Nutrients and Microbes during the Occurrence and Extinction of Algal Blooms: A Mesocosm Experiment with the Addition of Marine Aquaculture Sediment. Front. Mar. Sci. 2022, 9, 959161. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, X.; Cai, H.; Ke, J.; Zhu, J.; Lu, K.; Zheng, Z.; Yang, W. The Assembly Process of Free-Living and Particle-Attached Bacterial Communities in Shrimp-Rearing Waters: The Overwhelming Influence of Nutrient Factors Relative to Microalgal Inoculation. Animals 2023, 13, 3484. [Google Scholar] [CrossRef]
- Ding, Y.; Chen, N.; Ke, J.; Qi, Z.; Chen, W.; Sun, S.; Zheng, Z.; Xu, J.; Yang, W. Response of the Rearing Water Bacterial Community to the Beneficial Microalga Nannochloropsis oculata Cocultured with Pacific White Shrimp (Litopenaeus vannamei). Aquaculture 2021, 542, 736895. [Google Scholar] [CrossRef]
- Liu, B.; Eltanahy, E.E.; Liu, H.; Chua, E.T.; Thomas-Hall, S.R.; Wass, T.J.; Pan, K.; Schenk, P.M. Growth-Promoting Bacteria Double Eicosapentaenoic Acid Yield in Microalgae. Bioresour. Technol. 2020, 316, 123916. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved Error-Correction for Illumina 16S and ITS Amplicon Sequencing. BioRxiv 2016, 8, 1257. [Google Scholar] [CrossRef]
- Xun, W.; Liu, Y.; Li, W.; Ren, Y.; Xiong, W.; Xu, Z.; Zhang, N.; Miao, Y.; Shen, Q.; Zhang, R. Specialized Metabolic Functions of Keystone Taxa Sustain Soil Microbiome Stability. Microbiome 2021, 9, 35. [Google Scholar] [CrossRef]
- Kimbrel, J.A. Host Selection and Stochastic Effects Influence Bacterial Community Assembly on the Microalgal Phycosphere. Algal Res. 2019, 40, 101489. [Google Scholar] [CrossRef]
- Alsufyani, T.; Weiss, A.; Wichard, T. Time Course Exo-Metabolomic Profiling in the Green Marine Macroalga Ulva (Chlorophyta) for Identification of Growth Phase-Dependent Biomarkers. Mar. Drugs 2017, 15, 14. [Google Scholar] [CrossRef]
- Yang, W.; Zheng, Z.; Lu, K.; Zheng, C.; Du, Y.; Wang, J.; Zhu, J. Manipulating the Phytoplankton Community Has the Potential to Create a Stable Bacterioplankton Community in a Shrimp Rearing Environment. Aquaculture 2020, 520, 734789. [Google Scholar] [CrossRef]
- Sun, F.; Wang, C.; Wang, Y.; Tu, K.; Zheng, Z.; Lin, X. Diatom Red Tide Significantly Drive the Changes of Microbiome in Mariculture Ecosystem. Aquaculture 2020, 520, 734742. [Google Scholar] [CrossRef]
- Koops, H.-P.; Pommerening-Röser, A. Distribution and Ecophysiology of the Nitrifying Bacteria Emphasizing Cultured Species. FEMS Microbiol. Ecol. 2001, 37, 1–9. [Google Scholar] [CrossRef]
- Liu, X.; Tang, P.; Liu, Y.; Xie, W.; Chen, C.; Li, T.; He, Q.; Bao, J.; Tiraferri, A.; Liu, B. Efficient Removal of Organic Compounds from Shale Gas Wastewater by Coupled Ozonation and Moving-Bed-Biofilm Submerged Membrane Bioreactor. Bioresour. Technol. 2022, 344, 126191. [Google Scholar] [CrossRef]
- Blaszczyk, M. Effect of Medium Composition on the Denitrification of Nitrate by Paracoccus Denitrificans. Appl. Environ. Microbiol. 1993, 59, 3951–3953. [Google Scholar] [CrossRef]
- Fallahi, A.; Rezvani, F.; Asgharnejad, H.; Khorshidi Nazloo, E.; Hajinajaf, N.; Higgins, B. Interactions of Microalgae-Bacteria Consortia for Nutrient Removal from Wastewater: A Review. Chemosphere 2021, 272, 129878. [Google Scholar] [CrossRef]
- Collao, J.; del Mar Morales-Amaral, M.; Acién-Fernández, F.G.; Bolado-Rodríguez, S.; Fernandez-Gonzalez, N. Effect of Operational Parameters, Environmental Conditions, and Biotic Interactions on Bacterial Communities Present in Urban Wastewater Treatment Photobioreactors. Chemosphere 2021, 284, 131271. [Google Scholar] [CrossRef]
- Tisserand, L.; Dadaglio, L.; Intertaglia, L.; Catala, P.; Panagiotopoulos, C.; Obernosterer, I.; Joux, F. Use of Organic Exudates from Two Polar Diatoms by Bacterial Isolates from the Arctic Ocean. Phil. Trans. R. Soc. A 2020, 378, 20190356. [Google Scholar] [CrossRef]
- Hu, Y.; Xie, G.; Jiang, X.; Shao, K.; Tang, X.; Gao, G. The Relationships between the Free-Living and Particle-Attached Bacterial Communities in Response to Elevated Eutrophication. Front. Microbiol. 2020, 11, 423. [Google Scholar] [CrossRef]
- Bachmann, J.; Heimbach, T.; Hassenrück, C.; Kopprio, G.A.; Iversen, M.H.; Grossart, H.P.; Gärdes, A. Environmental Drivers of Free-Living vs. Particle-Attached Bacterial Community Composition in the Mauritania Upwelling System. Front. Microbiol. 2018, 9, 2836. [Google Scholar] [CrossRef]
- Liu, M.; Liu, L.; Chen, H.; Yu, Z.; Yang, J.R.; Xue, Y.; Huang, B.; Yang, J. Community Dynamics of Free-Living and Particle-Attached Bacteria Following a Reservoir Microcystis Bloom. Sci. Total Environ. 2019, 660, 501–511. [Google Scholar] [CrossRef]
- Mestre, M.; Borrull, E.; Sala, M.M.; Gasol, J.M. Patterns of Bacterial Diversity in the Marine Planktonic Particulate Matter Continuum. ISME J 2017, 11, 999–1010. [Google Scholar] [CrossRef]
- Shen, Z.; Xie, G.; Zhang, Y.; Yu, B.; Shao, K.; Gao, G.; Tang, X. Similar Assembly Mechanisms but Distinct Co-Occurrence Patterns of Free-Living vs. Particle-Attached Bacterial Communities across Different Habitats and Seasons in Shallow, Eutrophic Lake Taihu. Environ. Pollut. 2022, 314, 120305. [Google Scholar] [CrossRef]
- Cydzik-Kwiatkowska, A.; Zielińska, M. Bacterial Communities in Full-Scale Wastewater Treatment Systems. World J. Microbiol. Biotechnol. 2016, 32, 66. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Ren, K.; Isabwe, A.; Chen, H.; Liu, M.; Yang, J. Stochastic Processes Shape Microeukaryotic Community Assembly in a Subtropical River across Wet and Dry Seasons. Microbiome 2019, 7, 138. [Google Scholar] [CrossRef]
- Liu, L.; Yang, J.; Yu, Z.; Wilkinson, D.M. The Biogeography of Abundant and Rare Bacterioplankton in the Lakes and Reservoirs of China. ISME J. 2015, 9, 2068–2077. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Xu, H.; Zeng, J.; Cao, X.; Huang, R.; Shen, F.; Yu, Z. Community Composition and Assembly Processes of the Free-Living and Particle-Attached Bacteria in Taihu Lake. FEMS Microbiol. Ecol. 2017, 93, fix062. [Google Scholar] [CrossRef]
- Yan, Q.; Stegen, J.C.; Yu, Y.; Deng, Y.; Li, X.; Wu, S.; Dai, L.; Zhang, X.; Li, J.; Wang, C.; et al. Nearly a Decade-Long Repeatable Seasonal Diversity Patterns of Bacterioplankton Communities in the Eutrophic Lake Donghu (Wuhan, China). Mol. Ecol. 2017, 26, 3839–3850. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shen, J.; Wu, Y.; Tu, C.; Soininen, J.; Stegen, J.C.; He, J.; Liu, X.; Zhang, L.; Zhang, E. Phylogenetic Beta Diversity in Bacterial Assemblages across Ecosystems: Deterministic versus Stochastic Processes. ISME J. 2013, 7, 1310–1321. [Google Scholar] [CrossRef] [PubMed]
- Shibl, A.A.; Isaac, A.; Ochsenkühn, M.A.; Cárdenas, A.; Fei, C.; Behringer, G.; Arnoux, M.; Drou, N.; Santos, M.P.; Gunsalus, K.C.; et al. Diatom Modulation of Select Bacteria through Use of Two Unique Secondary Metabolites. Proc. Natl. Acad. Sci. USA 2020, 117, 27445–27455. [Google Scholar] [CrossRef]
- Sterling, A.R.; Holland, L.Z.; Bundy, R.M.; Burns, S.M.; Buck, K.N.; Chappell, P.D.; Jenkins, B.D. Potential Interactions between Diatoms and Bacteria Are Shaped by Trace Element Gradients in the Southern Ocean. Front. Mar. Sci. 2023, 9, 876830. [Google Scholar] [CrossRef]
- Sison-Mangus, M.P.; Jiang, S.; Kudela, R.M.; Mehic, S. Phytoplankton-Associated Bacterial Community Composition and Succession during Toxic Diatom Bloom and Non-Bloom Events. Front. Microbiol. 2016, 7, 1433. [Google Scholar] [CrossRef]
| Factors (mg/L) | C | N | T | M |
|---|---|---|---|---|
| N. oculata | - | 24.367 ± 28.849 | - | 20.146 ± 18.320 |
| T. weissflogii | - | - | 128.679 ± 106.771 b | 5.2 ± 3.487 a |
| NH4+-N | 0.344 ± 0.361 | 0.248 ± 0.230 | 0.237 ± 0.229 | 0.254 ± 0.249 |
| NO2−-N | 0.148 ± 0.246 | 0.216 ± 0.401 | 0.165 ± 0.262 | 0.161 ± 0.302 |
| NO3−-N | 1.729 ± 1.978 a | 3.806 ± 4.946 ab | 5.431 ± 4.980 b | 3.945 ± 4.702 ab |
| PO43−-P | 0.889 ± 1.142 a | 1.836 ± 1.239 b | 2.125 ± 1.108 b | 1.903 ± 0.962 b |
| NH4+-N_change | 0.311 ± 0.361 | 0.227 ± 0.229 | 0.212 ± 0.231 | 0.182 ± 0.253 |
| NO2−-N_change | 0.166 ± 0.247 | 0.190 ± 0.400 | 0.142 ± 0.260 | 0.132 ± 0.303 |
| NO3−-N_change | −3.104 ± 2.128 c | −9.727 ± 5.233 a | −6.448 ± 4.971 a | −7.211 ± 5.411 b |
| PO43−-P_change | 0.856 ± 1.145 ab | 1.491 ± 1.234 b | 0.531 ± 1.122 a | 0.676 ± 0.992 a |
| Factors | R2 |
|---|---|
| treat | 0.04394 *** |
| time | 0.13181 *** |
| lifestyles | 0.07824 *** |
| treat: time | 0.03771 *** |
| treat: lifestyles | 0.01214 |
| time: lifestyles | 0.02561 *** |
| treat: time: lifestyles | 0.00933 |
| ρ | F | p | ρ (PA) | ρ (FL) | |
|---|---|---|---|---|---|
| N. oculata | 0.0418 | 1.7796 | 0.003 | 0.1156 | 0.0273 |
| NO3−-N | 0.0761 | 1.7734 | 0.003 | 0.2147 | 0.0446 |
| PO43−-P | 0.0505 | 2.7295 | 0.001 | 0.3125 | 0.2125 |
| NH4+-N | 0.0378 | 1.5658 | 0.010 | 0.0619 | 0.0263 |
| PO43−-P_change | 0.0368 | 2.4080 | 0.001 | 0.2952 | 0.1774 |
| NO2−-N_change | 0.0274 | 1.3424 | 0.037 | 0.2693 | 0.1826 |
| NO3−-N_change | 0.0269 | 1.7725 | 0.001 | 0.1122 | 0.0281 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, C.; Xiang, J.; Cai, H.; Ke, J.; Ni, H.; Zhu, J.; Zheng, Z.; Lu, K.; Yang, W. Microalgae Inoculation Significantly Shapes the Structure, Alters the Assembly Process, and Enhances the Stability of Bacterial Communities in Shrimp-Rearing Water. Biology 2024, 13, 54. https://doi.org/10.3390/biology13010054
Lian C, Xiang J, Cai H, Ke J, Ni H, Zhu J, Zheng Z, Lu K, Yang W. Microalgae Inoculation Significantly Shapes the Structure, Alters the Assembly Process, and Enhances the Stability of Bacterial Communities in Shrimp-Rearing Water. Biology. 2024; 13(1):54. https://doi.org/10.3390/biology13010054
Chicago/Turabian StyleLian, Chen, Jie Xiang, Huifeng Cai, Jiangdong Ke, Heng Ni, Jinyong Zhu, Zhongming Zheng, Kaihong Lu, and Wen Yang. 2024. "Microalgae Inoculation Significantly Shapes the Structure, Alters the Assembly Process, and Enhances the Stability of Bacterial Communities in Shrimp-Rearing Water" Biology 13, no. 1: 54. https://doi.org/10.3390/biology13010054
APA StyleLian, C., Xiang, J., Cai, H., Ke, J., Ni, H., Zhu, J., Zheng, Z., Lu, K., & Yang, W. (2024). Microalgae Inoculation Significantly Shapes the Structure, Alters the Assembly Process, and Enhances the Stability of Bacterial Communities in Shrimp-Rearing Water. Biology, 13(1), 54. https://doi.org/10.3390/biology13010054






