Loss of Function of the Retinoblastoma Gene Affects Gap Junctional Intercellular Communication and Cell Fate in Osteoblasts
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Conditions
2.2. Cell Lines and Treatment
2.3. Quantitative Realtime PCR
2.4. Western Blot Analysis
2.5. Immunofluorescence
2.6. Dye Transfer GJIC Assay
2.7. Statistical Analyses
3. Results
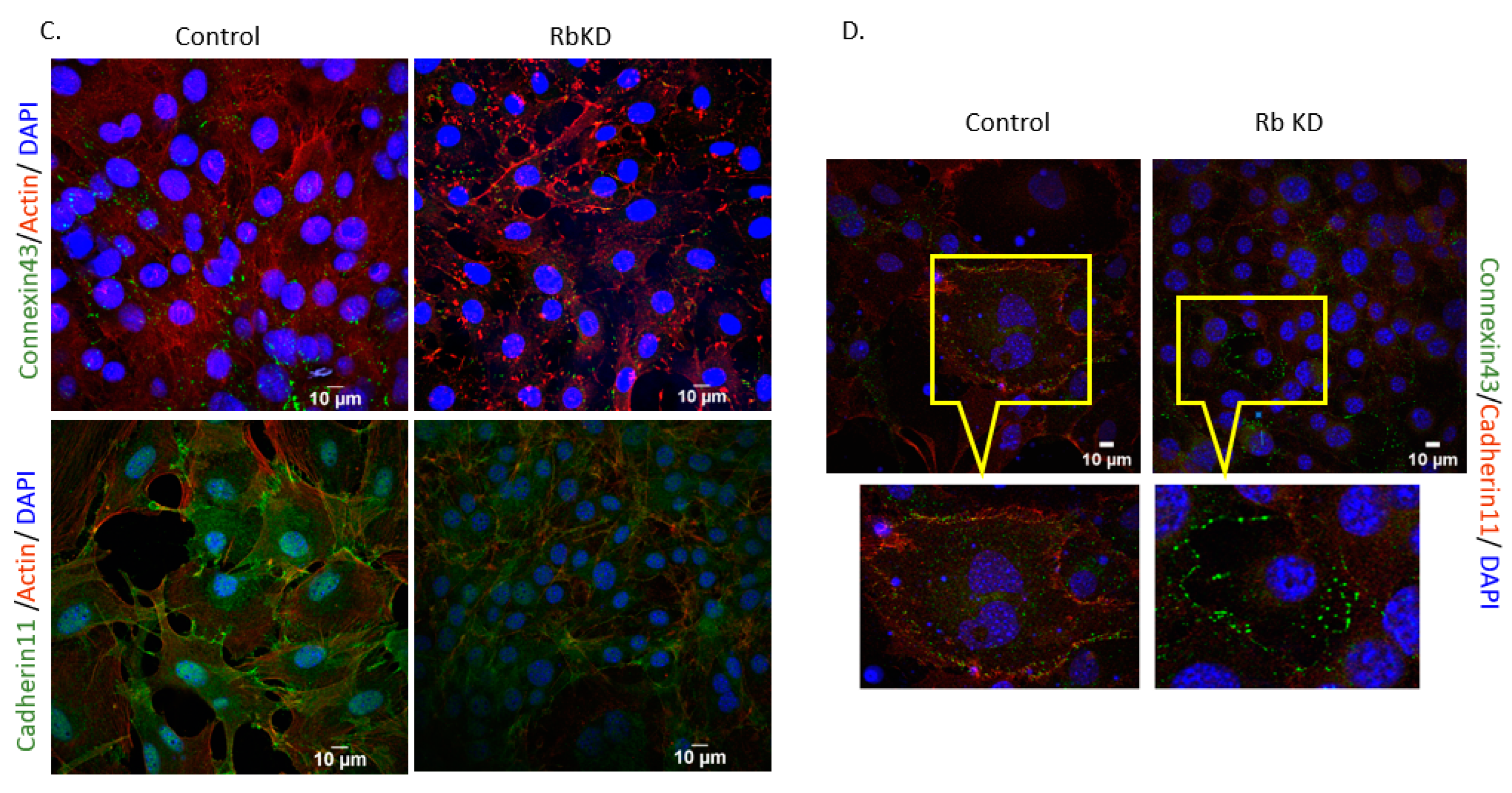
3.1. Reduction in pRb Expression in Osteoblasts Produces Changes to the Cell’s Phenotype
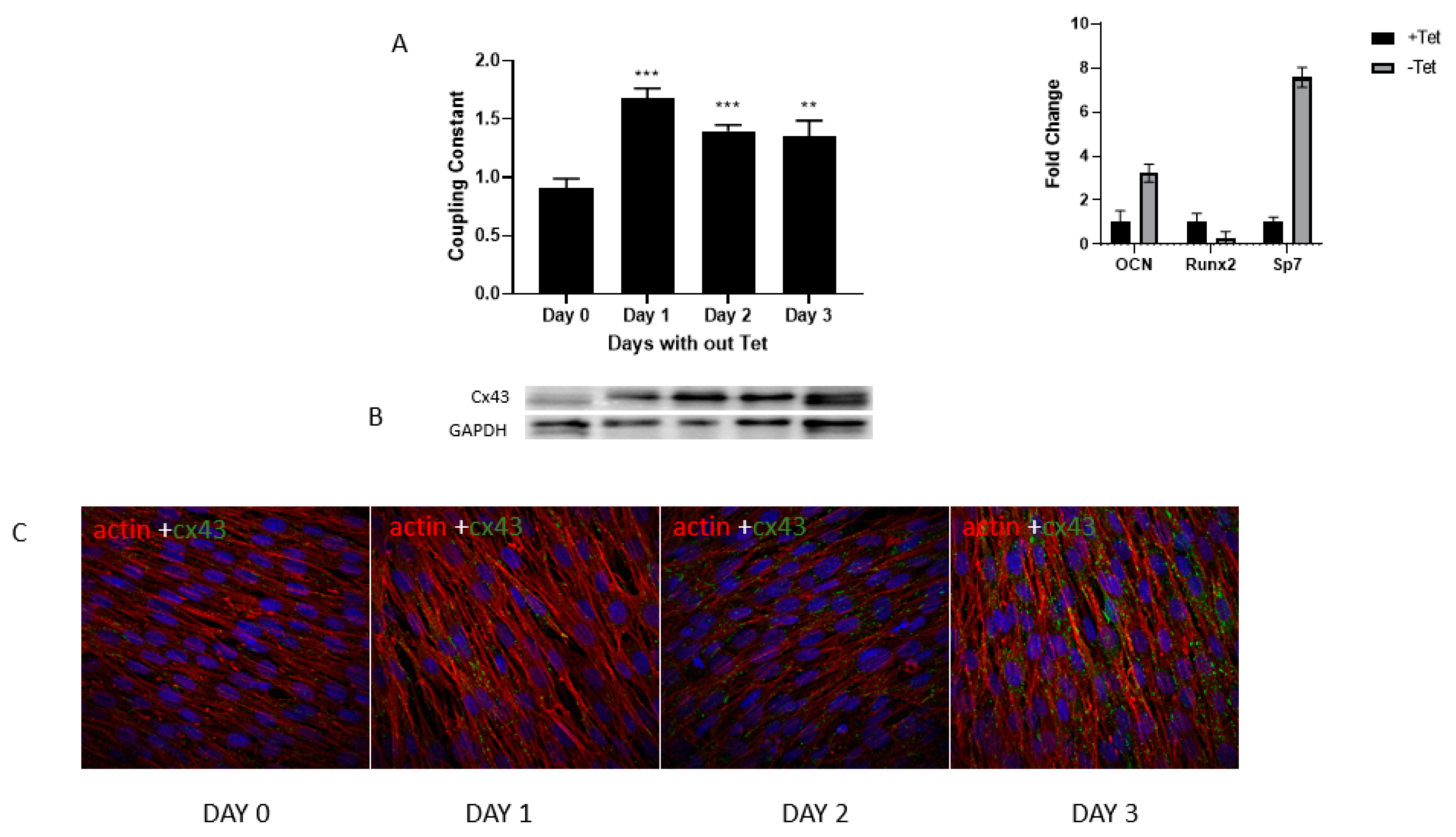
3.2. Gap Junctional Intercellular Communication Increases with Osteoblast Differentiation
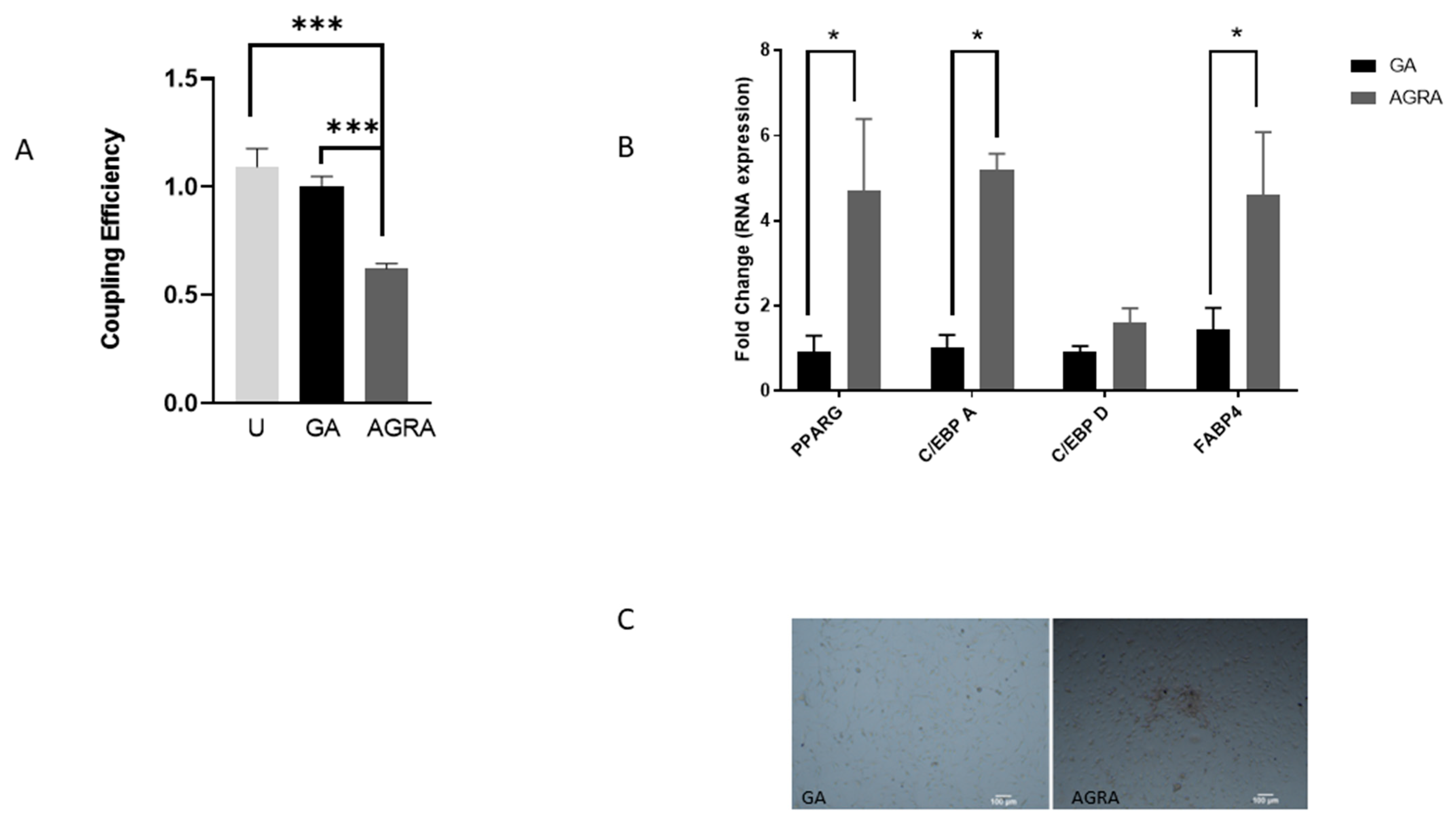
3.3. Inhibition of Gap Junctional Communication Increases Adipocytic Gene Expression in Osteoblasts
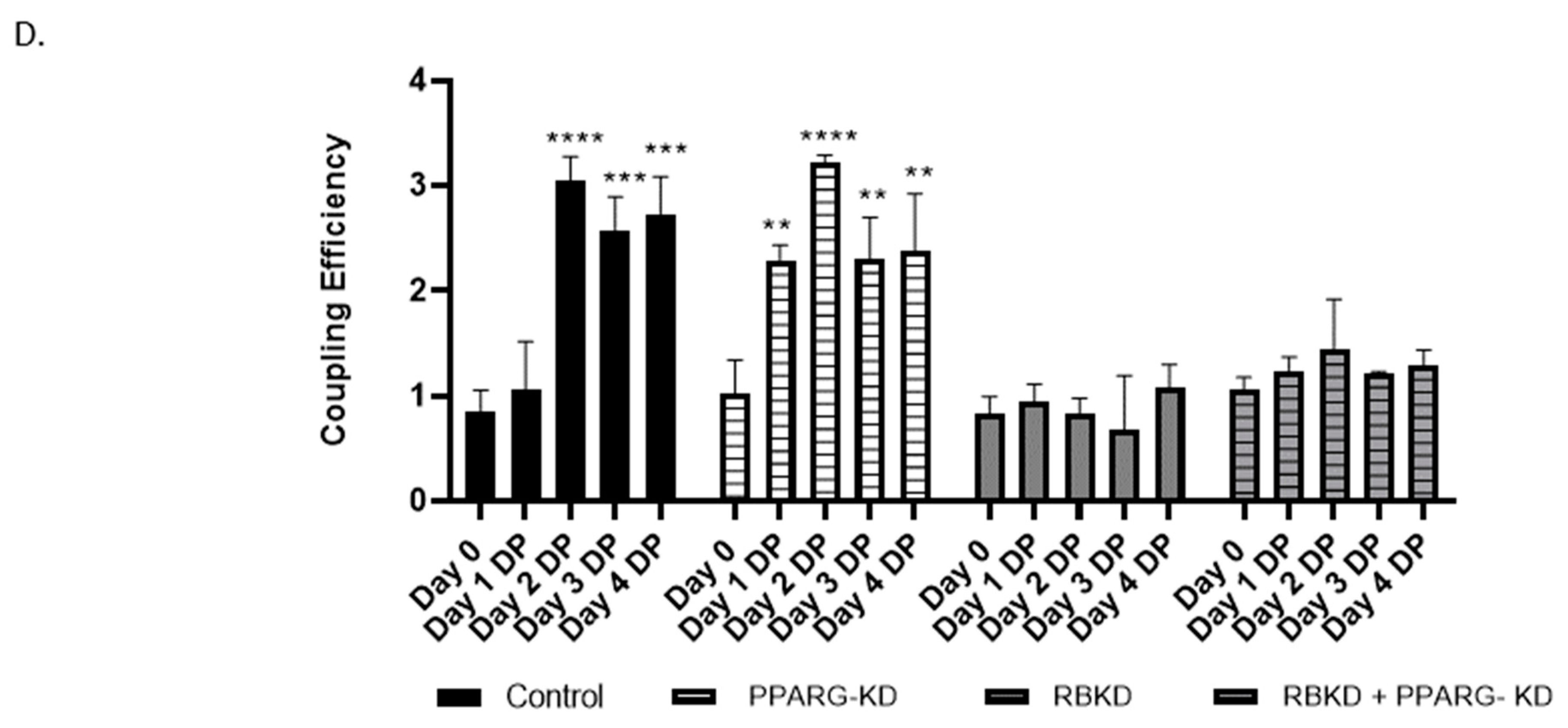
3.4. Increase in GJA1 Gene (Cx43) Expression during Differentiation Is Not Seen in RbKD Cells
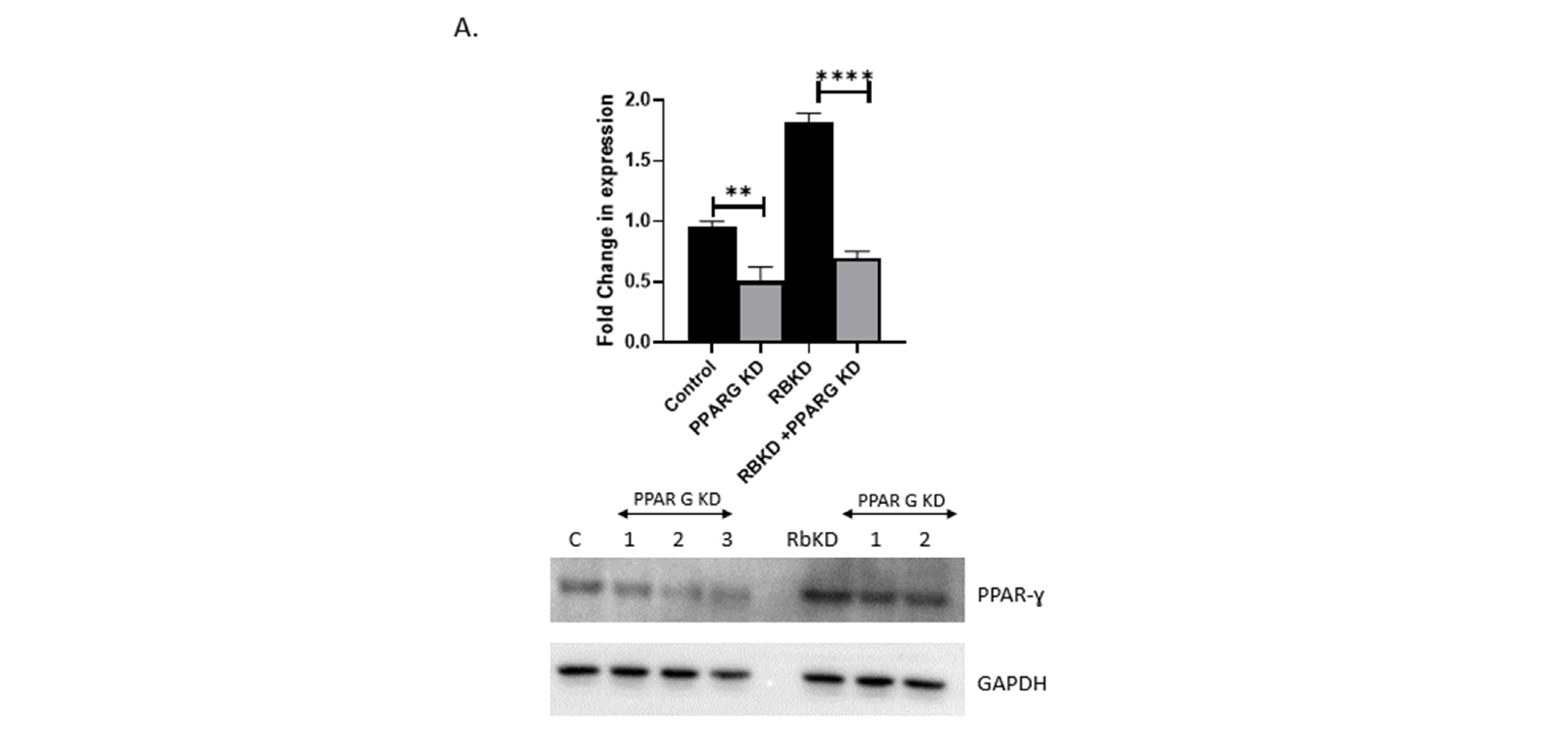
3.5. Reduction in PPAR-ɣ Levels Does Not Completely Reverse the Phenotype Produced with Rb2 Loss
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ottaviani, G.; Jaffe, N. The etiology of osteosarcoma. Cancer Treat. Res. 2009, 152, 15–32. [Google Scholar] [PubMed]
- Chandar, N.; Billig, B.; McMaster, J.; Novak, J. Inactivation of p53 gene in human and murine osteosarcoma cells. Br. J. Cancer 1992, 65, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Chandar, N.; Campbell, P.; Novak, J.; Smith, M. Dependence of induction of osteocalcin gene expression on the presence of wild type p53 in a murine osteosarcoma cell line. Mol. Carcinog. 1993, 8, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Hays, E.; Liboon, J.; Neely, C.; Kolman, K.; Chandar, N. Osteocalcin gene expression is regulated by wild type p53. Calcif. Tissue Int. 2011, 89, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Venkatachalam, S.; Shi, Y.; Jones, S.N.; Vogel, H.; Bradley, A.; Pinkel, D.; Donehower, L.A. Retention of wild type p53 in tumors from p53 heterozygous mice: Reduction of p53 dosage can promote cancer formation. EMBO J. 1998, 17, 4657–4667. [Google Scholar] [CrossRef] [PubMed]
- Chandar, N.; Donehower, L.; Lanciloti, N. Reduction in p53 gene dosage diminishes differentiation capacity of osteoblasts. Anticancer Res. 2000, 20, 2553–2559. [Google Scholar]
- Jacks, T.; Fazeli, A.; Schmitt, E.M.; Bronson, R.T.; Goodell, M.A.; Weinberg, R.A. Effects of an Rb mutation in the mouse. Nature 1992, 359, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Roufayel, R.; Mezher, R.; Storey, K.B. The Role of Retinoblastoma Protein in Cell Cycle Regulation: An Updated Review. Curr. Mol. Med. 2021, 21, 620–629. [Google Scholar] [CrossRef]
- Hansen, M.F.; Cavenee, W.K. Retinoblastoma and osteosarcoma: The prototypic cancer family. Acta Paediatr. Jpn. 1987, 29, 526–533. [Google Scholar] [CrossRef]
- Dyson, N.J. RB1: A prototype tumor suppressor and an enigma. Genes Dev. 2016, 30, 1492–1502. [Google Scholar] [CrossRef]
- Calo, E.; Quintero-Estades, J.A.; Danielian, P.S.; Nedelcu, S.; Berman, S.D.; Lees, J.A. Rb regulates fate choice and lineage commitment in vivo. Nature 2010, 466, 1110–1114. [Google Scholar] [CrossRef] [PubMed]
- Sosa-García, B.; Gunduz, V.; Vázquez-Rivera, V.; Cress, W.D.; Wright, G.; Bian, H.; Hinds, P.W.; Santiago-Cardona, P.G. A role for the retinoblastoma protein as a regulator of mouse osteoblast cell adhesion: Implications for osteogenesis and osteosarcoma formation. PLoS ONE 2010, 5, e13954. [Google Scholar] [CrossRef] [PubMed]
- Laurie, N.; Mohan, A.; McEvoy, J.; Reed, D.; Zhang, J.; Schweers, B.; Ajioka, I.; Valentine, V.; Johnson, D.; Ellison, D.; et al. Changes in retinoblastoma cell adhesion associated with optic nerve invasion. Mol. Cell. Biol. 2009, 29, 6268–6282. [Google Scholar] [CrossRef]
- Luan, Y.; Yu, X.-P.; Xu, K.; Ding, B.; Yu, J.; Huang, Y.; Yang, N.; Lengyel, P.; Di Cesare, P.E.; Liu, C.-J. The retinoblastoma protein is an essential mediator of osteogenesis that links the p204 protein to the Cbfa1 transcription factor thereby increasing its activity. J. Biol. Chem. 2007, 282, 16860–16870. [Google Scholar] [CrossRef] [PubMed]
- Yamanouchi, K.; Yada, E.; Ishiguro, N.; Nishihara, M. 18alpha-glycyrrhetinic acid induces phenotypic changes of skeletal muscle cells to enter adipogenesis. Cell. Physiol. Biochem. 2007, 20, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Schiller, P.C.; D’Ippolito, G.; Brambilla, R.; Roos, B.A.; Howard, G.A. Inhibition of gap-junctional communication induces the trans-differentiation of osteoblasts to an adipocytic phenotype in vitro. J. Biol. Chem. 2001, 276, 14133–14138. [Google Scholar] [CrossRef]
- Stains, J.P.; Watkins, M.P.; Grimston, S.K.; Hebert, C.; Civitelli, R. Molecular mechanisms of osteoblast/osteocyte regulation by connexin43. Calcif. Tissue Int. 2014, 94, 55–67. [Google Scholar] [CrossRef][Green Version]
- Stains, J.P.; Civitelli, R. Gap junctions regulate extracellular signal-regulated kinase signaling to affect gene transcription. Mol. Biol. Cell 2005, 16, 64–72. [Google Scholar] [CrossRef]
- Stains, J.P.; Lecanda, F.; Screen, J.; Towler, D.A.; Civitelli, R. Gap junctional communication modulates gene transcription by altering the recruitment of Sp1 and Sp3 to connexin-response elements in osteoblast promoters. J. Biol. Chem. 2003, 278, 24377–24387. [Google Scholar] [CrossRef]
- Civitelli, R. Cell-cell communication in the osteoblast/osteocyte lineage. Arch. Biochem. Biophys. 2008, 473, 188–192. [Google Scholar] [CrossRef]
- Stains, J.P.; Civitelli, R. Connexins in the skeleton. Semin. Cell Dev. Biol. 2016, 50, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, L.I.; Davis, H.M.; Cisterna, B.A.; Sáez, J.C. Connexins and Pannexins in Bone and Skeletal Muscle. Curr. Osteoporos. Rep. 2017, 15, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Upham, B.L.; Sovadinova, I.; Babica, P. Gap Junctional Intercellular Communication: A Functional Biomarker to Assess Adverse Effects of Toxicants and Toxins, and Health Benefits of Natural Products. J. Vis. Exp. 2016, 118, e54281. [Google Scholar]
- Czyż, J.; Irmer, U.; Schulz, G.; Mindermann, A.; Hülser, D.F. Gap-junctional coupling measured by flow cytometry. Exp. Cell Res. 2000, 255, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, G.S.; Bechberger, J.F.; Naus, C.C. A pre-loading method of evaluating gap junctional communication by fluorescent dye transfer. Biotechniques 1995, 18, 490–497. [Google Scholar]
- Pendleton, E.; Chandar, N. In Vitro Differentiation of Preosteoblast-Like Cells, MC3T3-E1, to Adipocytes Is Enhanced by 1,25(OH)2 Vitamin D3. Front. Endocrinol. 2017, 8, 128. [Google Scholar] [CrossRef]
- Mukherjee, P.; Winter, S.L.; Alexandrow, M.G. Cell cycle arrest by transforming growth factor beta1 near G1/S is mediated by acute abrogation of prereplication complex activation involving an Rb-MCM interaction. Mol. Cell. Biol. 2010, 30, 845–856. [Google Scholar] [CrossRef]
- Zhang, C.; Cho, K.; Huang, Y.; Lyons, J.P.; Zhou, X.; Sinha, K.; McCrea, P.D.; De Crombrugghe, B. Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc. Natl. Acad. Sci. USA 2008, 105, 6936–6941. [Google Scholar] [CrossRef]
- Ducy, P.; Zhang, R.; Geoffroy, V.; Ridall, A.L.; Karsenty, G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 1997, 89, 747–754. [Google Scholar] [CrossRef]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef]
- Lee, J.-E.; Schmidt, H.; Lai, B.; Ge, K. Transcriptional and Epigenomic Regulation of Adipogenesis. Mol. Cell. Biol. 2019, 39, e00601-18. [Google Scholar] [CrossRef] [PubMed]
- Berman, S.D.; Yuan, T.L.; Miller, E.S.; Lee, E.Y.; Caron, A.; Lees, J.A. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol. Cancer Res. 2008, 6, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- Ambele, M.A.; Dhanraj, P.; Giles, R.; Pepper, M.S. Adipogenesis: A Complex Interplay of Multiple Molecular Determinants and Pathways. Int. J. Mol. Sci. 2020, 21, 4283. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.P.L.; Dass, C.R. Transdifferentiation of adipocytes to osteoblasts: Potential for orthopaedic treatment. J. Pharm. Pharmacol. 2018, 70, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Hashida, Y.; Nakahama, K.-I.; Shimizu, K.; Akiyama, M.; Harada, K.; Morita, I. Communication-dependent mineralization of osteoblasts via gap junctions. Bone 2014, 61, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Jeansonne, B.; Feagin, F.; McMinn, R.; Shoemaker, R.; Rehm, W. Cell-to-cell communication of osteoblasts. J. Dent. Res. 1979, 58, 1415–1423. [Google Scholar] [CrossRef]
- Talbot, J.; Brion, R.; Lamora, A.; Mullard, M.; Morice, S.; Heymann, D.; Verrecchia, F. Connexin43 intercellular communication drives the early differentiation of human bone marrow stromal cells into osteoblasts. J. Cell. Physiol. 2018, 233, 946–957. [Google Scholar] [CrossRef]
- Batra, N.; Kar, R.; Jiang, J.X. Gap junctions and hemichannels in signal transmission, function and development of bone. Biochim. Biophys. Acta 2012, 1818, 1909–1918. [Google Scholar] [CrossRef]
- Matsuo, K. Crosstalk among bone cells. Curr. Opin. Nephrol. Hypertens. 2009, 18, 292–297. [Google Scholar] [CrossRef]
- Gupta, A.; Anderson, H.; Buo, A.M.; Moorer, M.C.; Ren, M.; Stains, J.P. Communication of cAMP by connexin43 gap junctions regulates osteoblast signaling and gene expression. Cell. Signal. 2016, 28, 1048–1057. [Google Scholar] [CrossRef]
- Azarnia, R.; Russell, T.R. Cyclic AMP effects on cell-to-cell junctional membrane permeability during adipocyte differentiation of 3T3-L1 fibroblasts. J. Cell Biol. 1985, 100, 265–269. [Google Scholar] [CrossRef]
- Umezawa, A.; Hata, J. Expression of gap-junctional protein (connexin 43 or alpha 1 gap junction) is down-regulated at the transcriptional level during adipocyte differentiation of H-1/A marrow stromal cells. Cell Struct. Funct. 1992, 17, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Yeganeh, A.; Stelmack, G.L.; Fandrich, R.R.; Halayko, A.J.; Kardami, E.; Zahradka, P. Connexin 43 phosphorylation and degradation are required for adipogenesis. Biochim. Biophys. Acta 2012, 1823, 1731–1744. [Google Scholar] [CrossRef] [PubMed]
- Sinha, K.M.; Yasuda, H.; Coombes, M.M.; Dent, S.Y.R.; de Crombrugghe, B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 2010, 29, 68–79. [Google Scholar] [CrossRef]
- Strauss, R.E.; Gourdie, R.G. Cx43 and the Actin Cytoskeleton: Novel Roles and Implications for Cell-Cell Junction-Based Barrier Function Regulation. Biomolecules 2020, 10, 1656. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka, A.M.; Synoradzki, K.; Firlej, W.; Bartnik, E.; Sobczuk, P.; Fiedorowicz, M.; Grieb, P.; Rutkowski, P. Molecular Biology of Osteosarcoma. Cancers 2020, 12, 2130. [Google Scholar] [CrossRef] [PubMed]
- Gokgoz, N.; Wunder, J.S.; Mousses, S.; Eskandarian, S.; Bell, R.S.; Andrulis, I.L. Comparison of p53 mutations in patients with localized osteosarcoma and metastatic osteosarcoma. Cancer 2001, 92, 2181–2189. [Google Scholar] [CrossRef] [PubMed]
- Wunder, J.S.; Gokgoz, N.; Parkes, R.; Bull, S.B.; Eskandarian, S.; Davis, A.M.; Beauchamp, C.P.; Conrad, E.U.; Grimer, R.J.; Healey, J.H.; et al. TP53 mutations and outcome in osteosarcoma: A prospective, multicenter study. J. Clin. Oncol. 2005, 23, 1483–1490. [Google Scholar] [CrossRef]
- Feugeas, O.; Guriec, N.; Babin-Boilletot, A.; Marcellin, L.; Simon, P.; Babin, S.; Thyss, A.; Hofman, P.; Terrier, P.; Kalifa, C.; et al. Loss of heterozygosity of the RB gene is a poor prognostic factor in patients with osteosarcoma. J. Clin. Oncol. 1996, 14, 467–472. [Google Scholar] [CrossRef]
- Toguchida, J.; Ishizaki, K.; Sasaki, M.S.; Ikenaga, M.; Sugimoto, M.; Kotoura, Y.; Yamamuro, T. Chromosomal reorganization for the expression of recessive mutation of retinoblastoma susceptibility gene in the development of osteosarcoma. Cancer Res. 1988, 48, 3939–3943. [Google Scholar]
- Sellers, W.R.; Kaelin, W.G., Jr. Role of the retinoblastoma protein in the pathogenesis of human cancer. J. Clin. Oncol. 1997, 15, 3301–3312. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Gu, X.; Xu, X.; Ge, S.; Jia, R. Novel insights into RB1 mutation. Cancer Lett. 2022, 547, 215870. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, C.; Palazzini, S.; Marotti, G. Morphological study of intercellular junctions during osteocyte differentiation. Bone 1990, 11, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, F. Variable conformation of GAP junctions linking bone cells: A transmission electron microscopic study of linear, stacked linear, curvilinear, oval, and annular junctions. Calcif. Tissue Int. 1997, 61, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Lecanda, F.; Towler, D.A.; Ziambaras, K.; Cheng, S.-L.; Koval, M.; Steinberg, T.H.; Civitelli, R. Gap junctional communication modulates gene expression in osteoblastic cells. Mol. Biol. Cell 1998, 9, 2249–2258. [Google Scholar] [CrossRef]
- Schiller, P.C.; D’ippolito, G.; Balkan, W.; Roos, B.; Howard, G. Gap-junctional communication is required for the maturation process of osteoblastic cells in culture. Bone 2001, 28, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhou, Z.; Saunders, M.M.; Donahue, H.J. Modulation of connexin43 alters expression of osteoblastic differentiation markers. Am. J. Physiol.-Cell Physiol. 2006, 290, C1248–C1255. [Google Scholar] [CrossRef]
- Gündüz, V.; Kong, E.; Bryan, C.D.; Hinds, P.W. Loss of the retinoblastoma tumor suppressor protein in murine calvaria facilitates immortalization of osteoblast-adipocyte bipotent progenitor cells characterized by low expression of N-cadherin. Mol. Cell. Biol. 2012, 32, 2561–2569. [Google Scholar] [CrossRef]
- Engel, B.E.; Cress, W.D.; Santiago-Cardona, P.G. The Retinoblastoma Protein: A Master Tumor Suppressor Acts as a Link between Cell Cycle and Cell Adhesion. Cell Health Cytoskelet. 2015, 7, 1–10. [Google Scholar]
- Smas, C.M.; Sul, H.S. Molecular mechanisms of adipocyte differentiation and inhibitory action of pref-1. Crit. Rev. Eukaryot. Gene Expr. 1997, 7, 281–298. [Google Scholar] [CrossRef]





| Primer | Forward | Reverse |
|---|---|---|
| Sp7 | ACTCATCCCTATGGCTCGTG | GGTAGGGAGCTGGGTTAAGG |
| RUNX2 | AACAAGACCCTGCCCGTG | TGAAACTCTTGCCTCGTCCG |
| FABP4 | CCGCAGACGACAGGAAGGT | AGGGCCCCGCCATCT |
| Cad11 | AGACGTTGGATCCGAGAAAGAG | GGTTGTCCTTCCAGGATACT |
| Cn43 | ACAGCGGTTGAGTCAGCTTG | GAGAGATGGGGAAGGACTTGT |
| ALP | GCTGATCATTCCCACGTTTT | CTGGGCCTGGTAGTTGTTGT |
| COL1A1 | ACGTCCTGGTGAAGTTGGTC | CAGGGAAGCCTCTTTCTCCT |
| C/EBPα | CAAGAACAGCAACGAGTACCG | GTCACTGGTCAACTCCAGCAC |
| C/EBPβ | GGGTTGTTGATGTTTTTGGTTT | GAAACGGAAAAGGTTCTCAAAA |
| C/EBPδ | CGACTTCAGCGCCTACATTGA | GAAGAGGTCGGCGAAGAGTT |
| PPARγ | TGAAAGAAGCGGTGAACCACTG | TGGCATCTCTGTGTCAACCATG |
| β-Actin | TGTCCACCTTCCAGCAGATGT | AGCTCAGTAACAGTCCGCCTAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pendleton, E.; Ketner, A.; Ransick, P.; Ardekani, D.; Bodenstine, T.; Chandar, N. Loss of Function of the Retinoblastoma Gene Affects Gap Junctional Intercellular Communication and Cell Fate in Osteoblasts. Biology 2024, 13, 39. https://doi.org/10.3390/biology13010039
Pendleton E, Ketner A, Ransick P, Ardekani D, Bodenstine T, Chandar N. Loss of Function of the Retinoblastoma Gene Affects Gap Junctional Intercellular Communication and Cell Fate in Osteoblasts. Biology. 2024; 13(1):39. https://doi.org/10.3390/biology13010039
Chicago/Turabian StylePendleton, Elisha, Anthony Ketner, Phil Ransick, Doug Ardekani, Thomas Bodenstine, and Nalini Chandar. 2024. "Loss of Function of the Retinoblastoma Gene Affects Gap Junctional Intercellular Communication and Cell Fate in Osteoblasts" Biology 13, no. 1: 39. https://doi.org/10.3390/biology13010039
APA StylePendleton, E., Ketner, A., Ransick, P., Ardekani, D., Bodenstine, T., & Chandar, N. (2024). Loss of Function of the Retinoblastoma Gene Affects Gap Junctional Intercellular Communication and Cell Fate in Osteoblasts. Biology, 13(1), 39. https://doi.org/10.3390/biology13010039







