eDNA Metabarcoding- and Microscopic Analysis for Diet Determination in Waterfowl, a Comparative Study in Vejlerne, Denmark
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
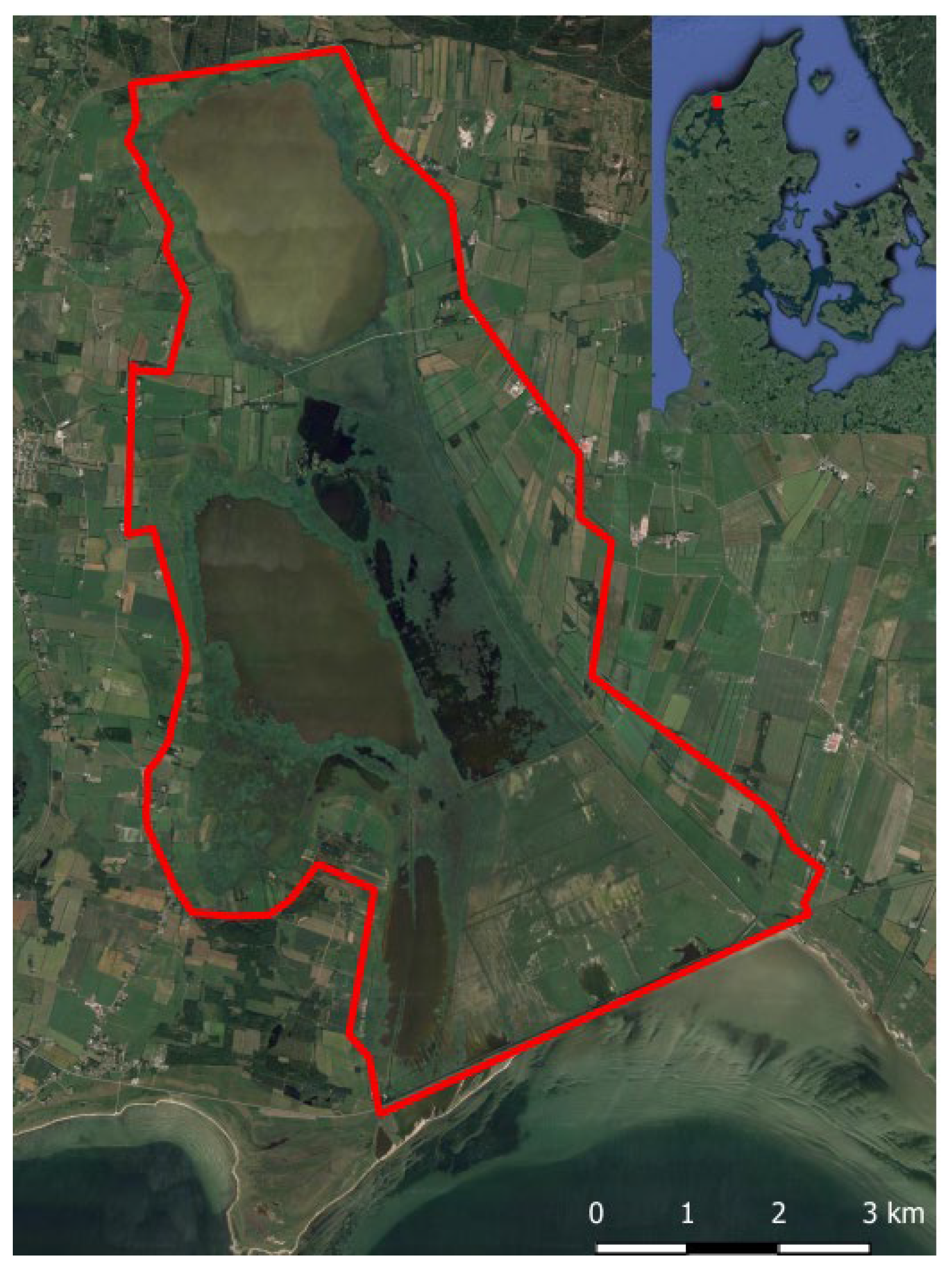
2.1. Study Location
2.2. Samples and Preparations
2.3. Microscopic Analysis
2.4. DNA Extraction
2.5. PCR
2.6. Sequence Analysis and Filtering
2.7. Cumulative Curves
3. Results
3.1. Results of Microscopic Analysis
3.2. Results of eDNA Metabarcoding
3.3. Presence/Absence of Plant Species by Each Method
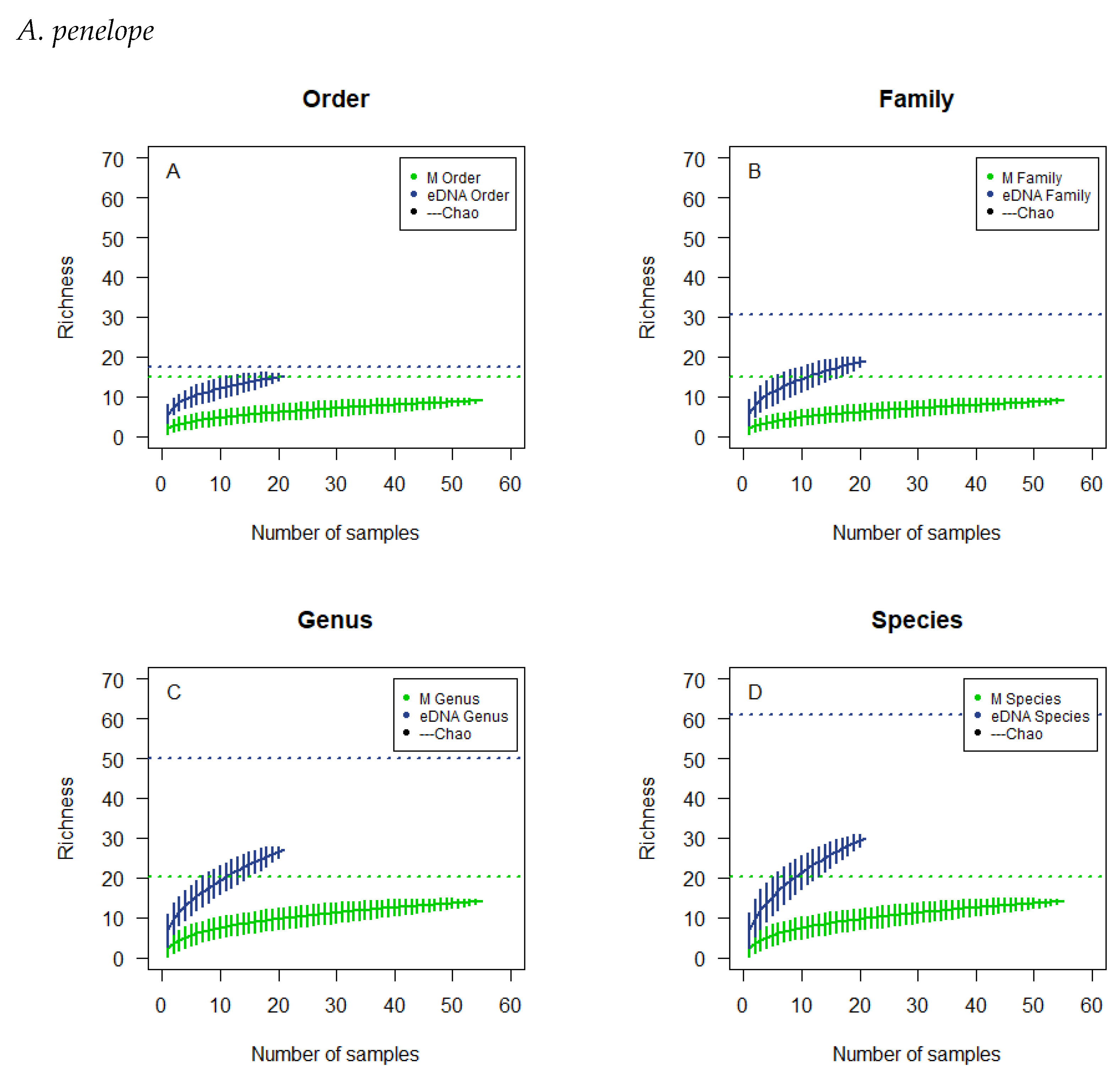
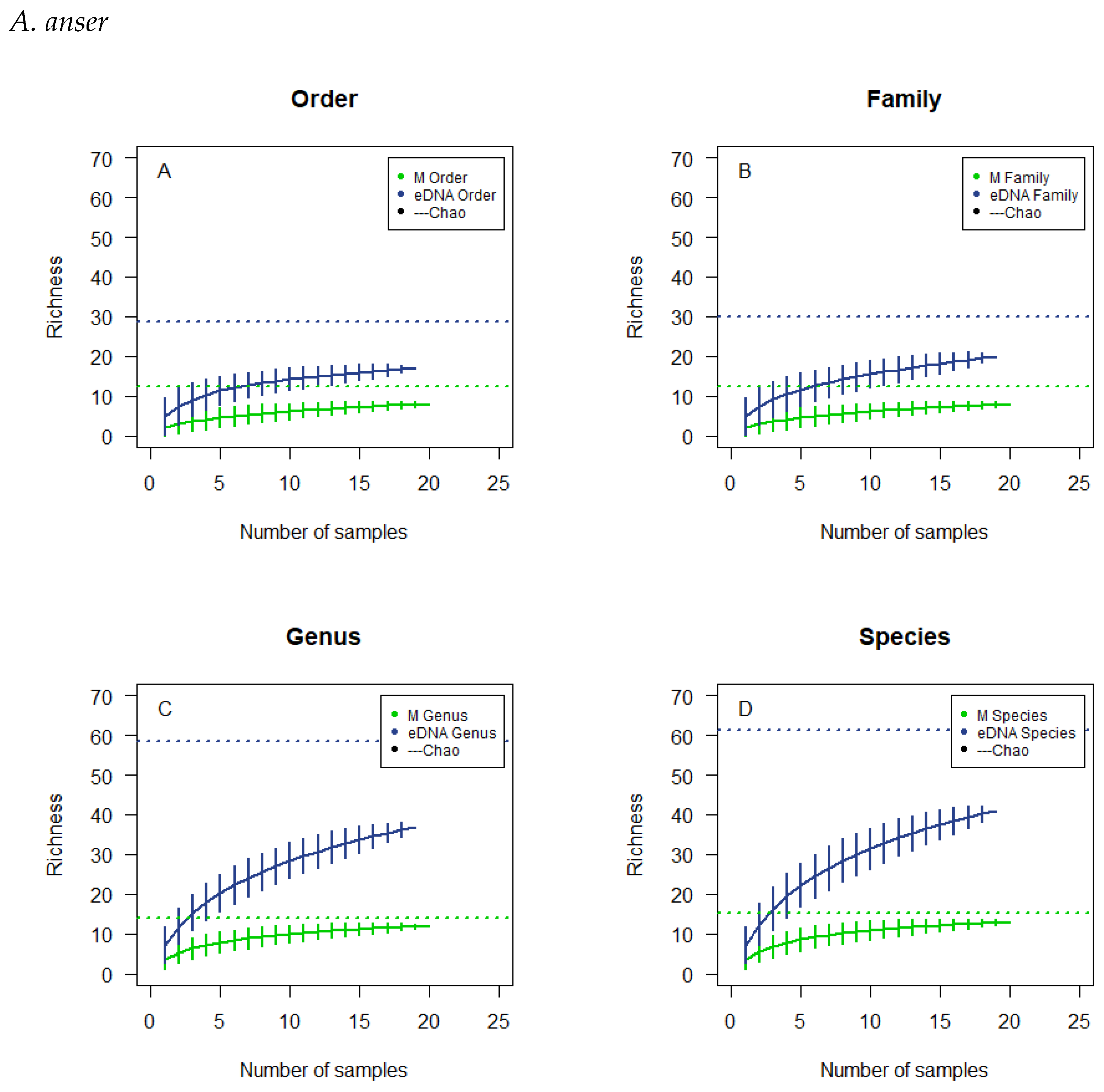
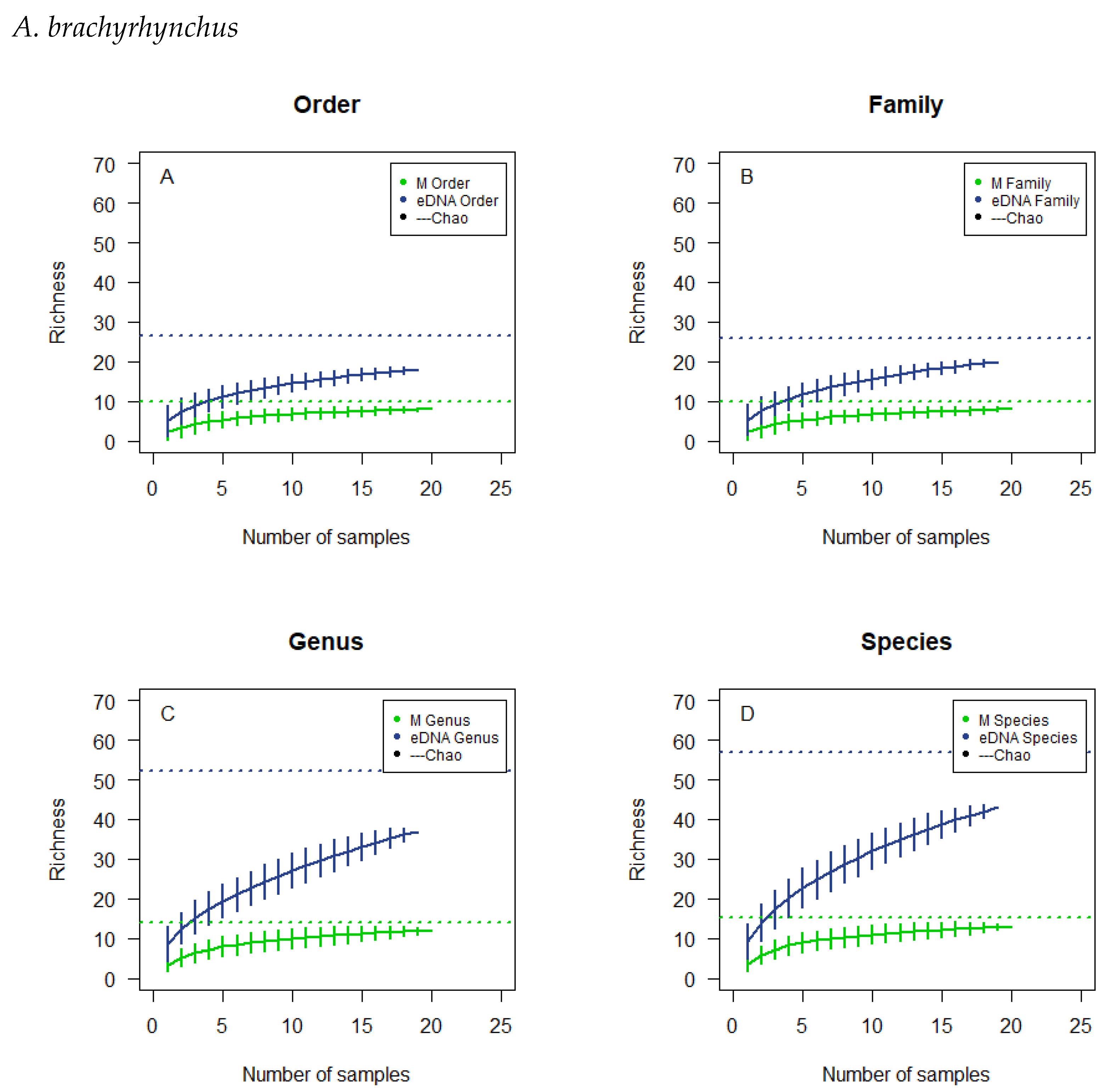
3.4. Observed Richness and Chao Estimates
3.5. Cumulative Curves Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Order | Family | Species |
|---|---|---|
| Asterales | Asteraceae | Aster tripolium |
| Asterales | Asteraceae | Leontodon autumnalis |
| Caryophyllales | Caryophyllaceae | Spergularia marina |
| Cyperales | Cyperaceae | Carex nigra |
| Fabales | Fabaceae | Trifolium pratense |
| Juncales | Juncaceae | Juncus articulatus |
| Plumbaginales | Plumbaginaceae | Armeria maritima |
| Poales | Poaceae | Agrostis capillaris |
| Poales | Poaceae | Agrostis stolonifera |
| Poales | Poaceae | Cynosurus cristatus |
| Poales | Poaceae | Festuca rubra |
| Poales | Poaceae | Holcus lanatus |
| Poales | Poaceae | Hordeum vulgare |
| Poales | Poaceae | Phragmites australis |
| Poales | Poaceae | Puccinellia maritima |
| Poales | Poaceae | Triticum aestivum |
| Poales | Poaceae | Zea mays |
| Primulales | Myrsinaceae | Glaux maritima |
| Rosales | Rosaceae | Potentillaanserina |
| Scrophulariales | Plantaginaceae | Plantago maritima |
| Zosterales | Juncaginaceae | Triglochin maritima |
| Zosterales | Zosteraceae | Zostera Marina |
| A. penelope Sample nr. | Number of Reads | A. anser Sample nr. | Number of Reads | A. brachyrhynchus Sample nr. | Number of Reads | B. leucopsis Sample nr. | Number of Reads |
|---|---|---|---|---|---|---|---|
| 1 | 5938 | 1 | 5436 | 1 | 7552 | 1 | 8312 |
| 2 | 7747 | 2 | 6110 | 2 | 6321 | 2 | 8207 |
| 3 | 7102 | 3 | 6734 | 3 | 5558 | 3 | 5533 |
| 4 | 8978 | 4 | 8339 | 4 | 5904 | 4 | 3593 |
| 5 | 4195 | 5 | 8518 | 5 | 6795 | 5 | 9772 |
| 6 | 8819 | 6 | 7614 | 6 | 7058 | 6 | 4377 |
| 7 | 10,363 | 7 | 7278 | 7 | 6740 | 7 | 5175 |
| 8 | 5426 | 8 | 3693 | 8 | 6380 | 8 | 4060 |
| 9 | 8027 | 9 | 7585 | 9 | 6986 | 9 | 4782 |
| 10 | 4881 | 10 | 7339 | 10 | 7726 | 10 | 6401 |
| 11 | 7122 | 11 | 12,271 | 11 | 11,804 | 11 | 7402 |
| 12 | 8304 | 12 | 7980 | 12 | 5545 | 12 | 5937 |
| 13 | 5446 | 13 | 8029 | 13 | 7806 | 13 | 9759 |
| 14 | 446 | 14 | 5607 | 14 | 14,973 | 14 | 5455 |
| 15 | 9189 | 15 | 11,932 | 15 | 6511 | 15 | 8557 |
| 16 | 11,112 | 16 | 9127 | 16 | 7053 | 16 | 7477 |
| 17 | 8069 | 17 | 20,479 | 17 | 8304 | 17 | 6523 |
| 18 | 7265 | 18 | 13,648 | 18 | 5180 | 18 | 8194 |
| 19 | 10,229 | 19 | 13,220 | 19 | 9967 | 19 | 10,154 |
| 20 | 9959 | 20 | 7524 | ||||
| 21 | 11,144 | ||||||
| Total number of reads | 159,761 | 170,939 | 144,163 | 137,194 | |||
| Grand total | 612,057 |
References
- Kennedy, C.M.; Oakleaf, J.R.; Theobald, D.M.; Baruch-Mordo, S.; Kiesecker, J. Managing the middle: A shift in conservation priorities based on the global human modification gradient. Glob. Change Biol. 2019, 25, 811–826. [Google Scholar] [CrossRef] [PubMed]
- Primack, R.B. Essentials of Conservation Biology, 5th ed.; Sinauer Associates: Sunderland, MA, USA, 2010; p. 142. [Google Scholar]
- Casper, R.M.; Jarman, S.N.; Deagle, B.E.; Gales, N.J.; Hindell, M.A. Detecting prey from DNA in predator scats: A comparison with morphological analysis, using Arctocephalus seals fed a known diet. J. Exp. Mar. Biol. Ecol. 2007, 347, 144–154. [Google Scholar] [CrossRef]
- Klare, U.; Kamler, J.F.; Macdonald, D.W. A comparison and critique of different scat-analysis methods for determining carnivore diet. Mammal Rev. 2011, 41, 294–312. [Google Scholar] [CrossRef]
- Barba, M.D.; Boyer, C.M.F.; Mercier, C.; Rioux, D.; Coissac, E.; Taberlet, P. DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: Application to omnivorous diet. Mol. Ecol. Resour. 2014, 14, 306–323. [Google Scholar] [CrossRef]
- Thalinger, B.; Oehm, J.; Mayr, H.; Obwexer, A.; Zeisler, C.; Traugott, M. Molecular prey identification in Central European piscivores. Mol. Ecol. Resour. 2016, 16, 123–137. [Google Scholar] [CrossRef]
- Nielsen, J.M.; Clare, E.L.; Hayden, B.; Brett, M.T.; Kratina, P. Diet tracing in ecology: Method comparison and selection. Methods Ecol. Evol. 2017, 9, 278–291. [Google Scholar] [CrossRef]
- Hartvig, I.; Howe, A.G.; Schmidt, E.N.B.; Pertoldi, C.; Nielsen, J.L.; Buttenschøn, R.M. Diet of the European bison (Bison bonasus) in a forest habitat estimated by DNA barcoding. Mammal Res. 2020, 66, 123–136. [Google Scholar] [CrossRef]
- Nørgaard, L.; Olesen, C.R.; Trøjelsgaard, K.; Pertoldi, C.; Nielsen, J.L.; Taberlet, P.; Ruiz-González, A.; De Barba, M.; Iacolina, L. Author Correction: eDNA metabarcoding for biodiversity assessment, generalist predators as sampling assistants. Sci. Rep. 2021, 11, 6820. [Google Scholar] [CrossRef]
- Pertoldi, C.; Schmidt, J.B.; Thomsen, P.; Nielsen, L.B.; Jonge, N.D.; Iacolina, L.; Muro, F.; Nielsen, K.T.; Pagh, S.; Andersen, L.H.; et al. Comparing DNA metabarcoding with faecal analysis for diet determination of the Eurasian otter (Lutralutra) in Vejlerne, Denmark. Mammal Res. 2021, 66, 115–122. [Google Scholar] [CrossRef]
- Davis, J.B.; Guillemain, M.; Karminski, R.M.; Arzel, C.; Eadie, J.M.; Rees, E.C. Habitat and resource use by waterfowl in the northern hemisphere in autumn and winter. Wildfowl 2014, 17–69. Available online: https://wildfowl.wwt.org.uk/index.php/wildfowl/article/view/2602/1720 (accessed on 29 May 2023).
- Batt, B.D.J. Ecology and Management of Breeding Waterfowl; University of Minnesota Press: Minneapolis, MN, USA, 1992; pp. 1–664. ISBN 10: 0816668329. [Google Scholar]
- Owen, M.; Krebes, R.H. On the autumn food of Barnacle Geese at Caerlaverock National Nature Reserve. Windfowl 1971, 22, 114–119. [Google Scholar] [CrossRef]
- Bartolomé, J.; Franch, J.; Gutman, M.; Seligman, N.G. Technical Note: Physical Factors that Influence Fecal Analysis Estimates of Herbivore Diets. Soc. Manag. 1995, 48, 267–270. [Google Scholar] [CrossRef]
- Galimberti, A.; Spinelli, S.; Bruno, A.; Mezzasalma, V.; De Mattia, F.; Cortis, P.; Labra, M. Evaluating the efficacy of restoration plantings through DNA barcoding of frugivorous bird diets. Conserv. Biol. 2016, 30, 763–773. [Google Scholar] [CrossRef]
- Rodway, M.S.; Cooke, F. Use of fecal analysis to determine seasonal changes in the diet of wintering Harlequin Ducks at a herring spawning site. J. Field Ornithol. 2002, 73, 363–371. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Pansu, J.; Bonin, A.E.; Coissac, E.; Giguet-Covex, C.; Barba, M.D.; Gielly, L.; Lopes, C.M.; Boyer, F.; Raye, F.P.G.; et al. Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Resour. 2015, 15, 543–556. [Google Scholar] [CrossRef] [PubMed]
- BirdLife International. Marecapenelope 2021. The IUCN Red List of Threatened Species 2021: e.T22680157A166199138; BirdLife International: Cambridge, UK, 2021. [Google Scholar] [CrossRef]
- BirdLife International. Anserbrachyrhynchus 2021. The IUCN Red List of Threatened Species 2021: e.T22679872A166191820; BirdLife International: Cambridge, UK, 2021. [Google Scholar] [CrossRef]
- BirdLife International. Brantaleucopsis2021. The IUCN Red List of Threatened Species 2021: e.T22679943A166195703; BirdLife International: Cambridge, UK, 2021. [Google Scholar] [CrossRef]
- BirdLife International. Anseranser, 2018. The IUCN Red List of Threatened Species 2018: e.T22679889A131907747; BirdLife International: Cambridge, UK, 2018. [Google Scholar] [CrossRef]
- Fox, A.D.; Dalby, L.; Christensen, T.K.; Nagy, S.; Balsby, T.J.S.; Crowe, O.; Clausen, P.; Deceuninck, B.; Devos, K.; Holt, C.A.; et al. Seeking explanations for recent changes in abundance of wintering Eurasian Wigeon (Anas penelope) in northwest Europe. Ornis Fenn. 2015, 93, 1–14. Available online: https://www.researchgate.net/publication/285176027_Seeking_explanations_for_recent_changes_in_abundance_of_wintering_Eurasian_Wigeon_Anas_penelope_in_northwest_Europe (accessed on 29 May 2023).
- Nielsen, H.H.; Clausen, P. Ynglende og Rastende Fugle i Vejlerne 2020; Teknisk Rapport nr. 232; Aarhus Universitet, DCE—Nationalt Center for Miljø og Energi: Roskilde, Denmark, 2022; p. 60. Available online: http://dce2.au.dk/pub/TR232.pdf (accessed on 30 May 2023).
- Andersen, L.H.; Nummi, P.; Bahrndorff, S.; Pertoldi, C.; Trøjelsgaard, K.; Lauridsen, T.L.; Rafn, J.; Frederiksen, C.M.S.; Kristjansen, M.P.; Bruhn, D. Reed bed vegetation structure and plant species diversity depend on management type and the time period since last management. Appl. Veg. Sci. 2020, 24, e12531. [Google Scholar] [CrossRef]
- Andersen, L.H.; Skærbæk, A.S.K.; Sørensen, T.B.; Knudsen, J.S.; Pertoldi, C.; Bahrndorff, S.; Bruhn, D. Turnover and change in plant species composition in a shielded salt marsh following variation in precipitation and temperature. J. Veg. Sci. 2020, 31, 465–475. [Google Scholar] [CrossRef]
- Andersen, L.H.; Knudsen, J.S.; Sørensen, T.B.; Skærbæk, A.S.K.; Bahrndorff, S.; Pertoldi, C.; Trøjelsgaard, K.; Bruhn, D. Coastal Meadow Vegetation following a Century of Shielding behind a Dike. Estuaries Coasts 2021, 44, 2087–2099. [Google Scholar] [CrossRef]
- Aage, V.; Jensen Naturfond. Vejlerne. Available online: http://www.avjf.dk/avjnf/naturomrader/vejlerne/ (accessed on 30 May 2023).
- Clausen, P.; Holm, T.E.; Kjeldsen, J.P. Naturgenopretning af Søerne i Vejlerne—En Vurdering af Effekterne på Yngle- og Trækfugle, Faglig Rapport fra DMU nr. 583. 2006, pp. 1–122. Available online: http://faglige-rapporter.dmu.dk (accessed on 28 May 2023).
- Nielsen, H.H.; Clausen, P. Ynglende og Rastende Fugle i Vejlerne 2015–2017; Teknisk Rapport nr. 136; Aarhus Universitet, DCE—Nationalt Center for Miljø og Energi: Roskilde, Denmark, 2019; p. 68. Available online: http://dce2.au.dk/pub/TR136.pdf (accessed on 29 May 2023).
- DOFbasen. Danmarks Fugle. Available online: https://dofbasen.dk/danmarksfugle/ (accessed on 1 July 2023).
- Ralph, C.P.; Nagata, S.E.; Ralph, C.J. Analysis of Droppings to Describe Diets of Small Birds. J. Field Ornithol. 1985, 56, 165–174. [Google Scholar]
- Haus, M.J.; Kelsch, R.D.; Jacobs, T.W. Application of Optical Topometry to Analysis of the Plant Epidermis. Plant Physiol. 2015, 169, 946–959. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.D.; Kahlert, J.; Ettrup, H. Diet and habitat use of moultingGreylag Geese Anseranser on the Danish Island of Saltholm. IBIS 1998, 140, 676–683. [Google Scholar] [CrossRef]
- Billerman, S.M.; Keeney, B.K.; Rodewald, P.G.; Schulenberg, T.S. (Eds.) Birds of the World; Cornell Laboratory of Ornithology: Ithaca, NY, USA, 2022; Available online: https://birdsoftheworld.org/bow/home (accessed on 7 September 2023).
- Nierychlo, M.; Andersen, K.S.; Xu, Y.; Green, N.; Jiang, C.; Albertsen, M.; Dueholm, M.S.; Nielsen, P.H. MiDAS 3: An ecosystem-specific reference database, taxonomy and knowledge platform for activated sludge and anaerobic digesters reveals species-level microbiome composition of activated sludge. Water Res. 2020, 182, 115955. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miguel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007, 35, e14. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: MolecularEvolutionary Genetics Analysisversion 7.0. Mol. Biol. Evol. 2015. Available online: http://www.kumarlab.net/publications (accessed on 7 August 2023).
- Oksanen, F.J.; Blanchet, G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, M.P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 7 August 2023).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 7 August 2023).
- Aude, E.; Frederiksen, R.F. Mosserne i Nationalpark Thy. Habitat Vision, Rapport 15. p. 152. 2005. Available online: https://nationalparkthy.dk/media/234151/mosser-i-nationalpark-thy_print.pdf (accessed on 28 May 2023).
- Frederiksen, S.; Rasmussen, F.N.; Seberg, O. Dansk Flora, 3rd ed.; Gyldendal: Copenhagen, Denmark, 2019; pp. 1–701. [Google Scholar]
- Naturbasen. Vandranunkel sp. (Batrachium sp.). Available online: https://www.naturbasen.dk/art/7442/vandranunkel-sp (accessed on 29 May 2023).
- Naturbasen. Gold Hejre (Anisantha sterilis). Available online: https://www.naturbasen.dk/art/4108/gold-hejre (accessed on 29 May 2023).
- Naturbasen. Stiv Seglmos (Drepanocladus sendtneri). Available online: https://www.naturbasen.dk/art/8673/stiv-seglmos (accessed on 29 May 2023).
- Stewart, D.R.M. Analysis of Plant Epidermis in Faeces: A Technique for Studying the Food Preferences of Grazing Herbivores. J. Appl. Ecol. 1967, 4, 83–111. [Google Scholar] [CrossRef]
- Owen, M. The Selection of Winter Food by Whiterfronted Geese. J. Appl. Ecol. 1976, 13, 715–729. [Google Scholar] [CrossRef]
- Andersen, K.; Bird, K.L.; Rasmussen, M.; Haile, J.; Breuning-Madsen, H.; Kjær, K.H.; Orlando, L.; Gilbert, M.T.P.; Willerslev, E. Meta-barcoding of “dirt” DNA from soil reflects vertebrate biodiversity. Mol. Ecol. 2012, 21, 1966–1979. [Google Scholar] [CrossRef]
- Dessborn, L.; Brochet, A.L.; Elmberg, J.; Legagneux, P.; Gauthier-Clerc, M.; Guillemain, M. Geographical and temporal patterns in the diet of pintail Anas acuta, wigeon Anas penelope, mallard Anas platyrhynchos and teal Anas crecca in the Western Palearctic. Eur. J. Wildl. Res. 2011, 57, 1119–1129. [Google Scholar] [CrossRef]
- QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation Project. 2023. Available online: http://qgis.osgeo.org (accessed on 29 May 2023).

| Plant Species | A. penelope | A. anser | A. brachyrhynchus | B. leucopsis | ||||
|---|---|---|---|---|---|---|---|---|
| FO | % | FO | % | FO | % | FO | % | |
| Agrostis capillaris | 7 | 9.86 | 8 | 10.96 | 6 | 7.32 | ||
| Agrostis stolonifera | 21 | 16.41 | 8 | 11.27 | 12 | 16.44 | 12 | 14.63 |
| Armeria maritima | 1 | 1.41 | 2 | 2.74 | ||||
| Carex nigra | 1 | 0.78 | 2 | 2.44 | ||||
| Cynosurus cristatus | 1 | 0.78 | 5 | 7.04 | 2 | 2.74 | 6 | 7.32 |
| Festuca rubra | 19 | 14.84 | 10 | 14.08 | 9 | 12.33 | 15 | 18.29 |
| Glaux maritima | 50 | 39.06 | 11 | 15.49 | 5 | 6.85 | 10 | 12.20 |
| Holcus lanatus | 9 | 7.03 | 16 | 22.54 | 15 | 20.55 | 18 | 21.95 |
| Hordeum vulgare | 2 | 2.82 | ||||||
| Juncus articulatus | 5 | 3.91 | 1 | 1.37 | 2 | 2.44 | ||
| Leontodon autumnalis | 12 | 9.38 | 3 | 4.23 | 5 | 6.85 | 2 | 2.44 |
| Phragmites australis | 2 | 1.56 | ||||||
| Plantago maritima | 1 | 0.78 | 1 | 1.41 | 1 | 1.22 | ||
| Potentilla anserina | 1 | 0.78 | 1 | 1.41 | 7 | 9.59 | ||
| Puccinellia maritima | 7 | 8.54 | ||||||
| Trifolium pratense | 3 | 2.34 | 4 | 5.63 | 5 | 6.85 | 1 | 1.22 |
| Triglochin maritima | 2 | 2.82 | 1 | 1.37 | ||||
| Triticum aestivum | 2 | 1.56 | 1 | 1.37 | ||||
| Zostera marina | 1 | 0.78 | ||||||
| Total FO | 128 | 100 | 71 | 100 | 73 | 100 | 82 | 100 |
| Total number of samples | 55 | 20 | 20 | 25 | ||||
| A. penelope | A. anser | A. brachyrhynchus | B. leucopsis | |||
|---|---|---|---|---|---|---|
| Order | Family | Species | Number of Samples | |||
| Asparagales | 1 | 1 | ||||
| Asterales | Asteraceae | Achillea spp. | 1 | 1 | 1 | |
| Asterales | Asteraceae | Bellis perennis | 1 | |||
| Asterales | Asteraceae | Cirsium spp. | 2 | |||
| Asterales | Asteraceae | Leontodon autumnalis | 7 | 7 | 12 | 15 |
| Asterales | Asteraceae | Leontodon spp. | 1 | |||
| Capparales | Brassicaceae | Brassica napus | 3 | 1 | ||
| Capparales | Brassicaceae | Cardamine hirsuta | 1 | |||
| Capparales | Brassicaceae | Cardamine impatiens * | 1 | 7 | ||
| Caryophyllales | Caryophyllaceae | Cerastium fontanum | 1 | 2 | ||
| Caryophyllales | Caryophyllaceae | Lychnis flos-cuculi | 1 | |||
| Caryophyllales | Caryophyllaceae | Sagina spp. | 1 | 1 | 3 | |
| Caryophyllales | Caryophyllaceae | Spergularia media | 1 | 1 | ||
| Cyperales | Cyperaceae | Carex lasiocarpa | 1 | 2 | 10 | |
| Cyperales | Cyperaceae | Eleocharis uniglumis | 3 | 1 | 1 | |
| Cyperales | Cyperaceae | Schoenoplectus tabernaemontani | 3 | |||
| Cyperales | Cyperaceae | Schoenus nigricans * | 1 | 3 | 1 | |
| Fagales | Betulaceae | Betula pendula | 1 | |||
| Fabales | Fabaceae | Trifolium pratense | 2 | |||
| Fabales | Fabaceae | Trifolium repens | 16 | 8 | 13 | 13 |
| Fabales | Fabaceae | Vicia sativa | 1 | |||
| Gentianales | Gentianaceae | 1 | ||||
| Gentianales | Rubiaceae | Galium spp. | 1 | 1 | ||
| Hypnales | Amblystegiaceae | Drepanocladus sendtneri * | 3 | 1 | 1 | 1 |
| Hypnales | Brachytheciaceae | Kindbergia praelonga | 3 | 2 | 2 | 2 |
| Juncales | Juncaceae | Juncus bufonius | 4 | 4 | 5 | 1 |
| Juncales | Juncaceae | Juncus bulbosus | 2 | 2 | 4 | |
| Juncales | Juncaceae | Juncus conglomeratus | 1 | 1 | ||
| Juncales | Juncaceae | Juncus gerardii | 17 | 9 | 12 | 11 |
| Papaverales | Papaveraceae | Meconopsis spp. * | 1 | |||
| Pinales | Pinaceae | Pinus mugo | 1 | 2 | 2 | 2 |
| Poales | Poaceae | Agrostis capillaris | 2 | 2 | 7 | 10 |
| Poales | Poaceae | Alopecurus geniculatus | 2 | 7 | 15 | 18 |
| Poales | Poaceae | Anthoxanthum spp. | 1 | 1 | ||
| Poales | Poaceae | Briza spp. | 5 | 2 | 7 | |
| Poales | Poaceae | Anisantha sterilis * | 1 | |||
| Poales | Poaceae | Calamagrostis arundinacea * | 1 | |||
| Poales | Poaceae | Catabrosa aquatica | 1 | |||
| Poales | Poaceae | Cynosurus cristatus | 2 | 7 | 5 | |
| Poales | Poaceae | Festuca arundinacea | 4 | 9 | 19 | 19 |
| Poales | Poaceae | Festuca spp. | 11 | 13 | 17 | 20 |
| Poales | Poaceae | Glyceria declinata * | 1 | 4 | ||
| Poales | Poaceae | Holcus lanatus | 3 | 1 | ||
| Poales | Poaceae | Hordeum vulgare | 6 | 2 | ||
| Poales | Poaceae | Phragmites australis | 3 | 4 | 1 | |
| Poales | Poaceae | Poa pratensis | 1 | 3 | 3 | 9 |
| Poales | Poaceae | Poa supina * | 1 | 5 | 3 | |
| Poales | Poaceae | Poa trivialis | 18 | 13 | 18 | 18 |
| Poales | Poaceae | Triticum aestivum | 1 | 1 | 1 | |
| Polygonales | Polygonaceae | Rumex crispus | 1 | |||
| Primulales | Myrsinaceae | Glaux maritima | 15 | 4 | 2 | 1 |
| Ranunculales | Ranunculaceae | Batrachium spp. | 1 | |||
| Ranunculales | Ranunculaceae | Myosurus minimus * | 1 | |||
| Ranunculales | Ranunculaceae | Ranunculus repens | 1 | 6 | 4 | |
| Rosales | Rosaceae | Potentilla anserina | 17 | 10 | 9 | 13 |
| Rosales | Rosaceae | Potentilla spp. | 1 | |||
| Scrophulariales | Lentibulariaceae | Utricularia australis * | 1 | |||
| Scrophulariales | Orobanchaceae | 3 | 1 | 2 | ||
| Scrophulariales | Plantaginaceae | Plantago major | 2 | 1 | ||
| Scrophulariales | Plantaginaceae | Plantago maritima | 12 | 5 | 4 | 2 |
| Solanales | Boraginaceae | Myosotis arvensis | 1 | |||
| Solanales | Convolvulaceae | Cuscuta spp. * | 1 | |||
| Solanales | Solanaceae | Solanum spp. | 1 | |||
| Zosterales | Juncaginaceae | Triglochin maritima | 1 | |||
| Zosterales | Potamogetonaceae | Potamogeton perfoliatus * | 2 | 2 | ||
| Total number of samples | 21 | 19 | 19 | 20 | ||
| Species | A. penelope | A. anser | A. brachyrhynchus | B. leucopsis |
|---|---|---|---|---|
| Achillea spp. | E | E | E | |
| Agrostis capillaris | E | M/E | M/E | M/E |
| Agrostis stolonifera | M | M | M | M |
| Alopecurus geniculatus | E | E | E | E |
| Anisanthasterilis | E | |||
| Anthoxanthum spp. | E | E | ||
| Armeria maritima | M | M | ||
| Batrachium spp. | E | |||
| Bellis perennis | E | |||
| Betula pendula | E | |||
| Brassica napus | E | E | ||
| Briza spp. | E | E | E | |
| Calamagrostis arundinacea | E | |||
| Cardamine hirsuta | E | |||
| Cardamine impatiens | E | E | ||
| Carex lasiocarpa | E | E | E | |
| Carex nigra | M | M | ||
| Catabrosa aquatica | E | |||
| Cerastium fontanum | E | E | ||
| Cirsium spp. | E | |||
| Cuscuta spp. | E | |||
| Cynosurus cristatus | M | M/E | M/E | M/E |
| Drepanocladus sendtneri | E | E | E | E |
| Eleocharis uniglumis | E | E | E | |
| Festuca arundinacea | E | E | E | E |
| Festuca rubra | M | M | M | M |
| Festuca spp. | E | E | E | E |
| Galium spp. | E | E | ||
| Glaux maritima | M/E | M/E | M/E | M/E |
| Glyceria declinata | E | E | ||
| Holcus lanatus | M | M/E | M | M/E |
| Hordeum vulgare | M/E | E | ||
| Juncus articulatus | M | M | M | |
| Juncus bufonius | E | E | E | E |
| Juncus bulbosus | E | E | E | |
| Juncus conglomeratus | E | E | ||
| Juncus gerardii | E | E | E | E |
| Kindbergia praelonga | E | E | E | E |
| Leontodon autumnalis | M/E | M/E | M/E | M/E |
| Leontodon spp. | E | |||
| Lychnis flos-cuculi | E | |||
| Meconopsis spp. | E | |||
| Myosotis arvensis | E | |||
| Myosurus minimus | E | |||
| Phragmites australis | M/E | E | E | |
| Pinus mugo | E | E | E | E |
| Plantago major | E | E | ||
| Plantago maritima | M/E | M/E | E | M/E |
| Poa pratensis | E | E | E | E |
| Poa supina | E | E | E | |
| Poa trivialis | E | E | E | E |
| Potamogeton perfoliatus | E | E | ||
| Potentilla anserina | M/E | M/E | M/E | E |
| Potentilla spp. | E | |||
| Puccinellia maritima | M | |||
| Ranunculus repens | E | E | E | |
| Rumex crispus | E | |||
| Sagina spp. | E | E | E | |
| Schoenoplectus tabernaemontani | E | |||
| Schoenus nigricans | E | E | E | |
| Solanum spp. | E | |||
| Spergularia media | E | E | ||
| Trifolium pratense | M | M | M | M/E |
| Trifolium repens | E | E | E | E |
| Triglochin maritima | M/E | M | ||
| Triticum aestivum | M | E | M/E | E |
| Utricularia australis | E | |||
| Vicia sativa | E | |||
| Zostera marina | M |
| A. penelope | A. anser | A. brachyrhynchus | B. leucopsis | |||||
|---|---|---|---|---|---|---|---|---|
| Observed | Estimated | Observed | Estimated | Observed | Estimated | Observed | Estimated | |
| Microscopic | ||||||||
| Order | 9 | 14.89 | 8 | 12.28 | 8 | 9.90 | 7 | 7.64 |
| Family | 9 | 14.89 | 8 | 12.28 | 8 | 9.90 | 7 | 7.64 |
| Genus | 14 | 20.14 | 12 | 14.14 | 12 | 14.14 | 11 | 11.64 |
| Species | 14 | 20.14 | 13 | 15.14 | 13 | 15.14 | 12 | 12.64 |
| eDNA | ||||||||
| Order | 15 | 17.54 | 17 | 28.84 | 18 | 26.53 | 13 | 13.24 |
| Family | 19 | 30.67 | 20 | 30.11 | 20 | 25.80 | 14 | 14.95 |
| Genus | 27 | 49.86 | 37 | 58.32 | 37 | 52.21 | 26 | 41.20 |
| Species | 30 | 61.11 | 41 | 61.21 | 43 | 56.95 | 33 | 42.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Svendsen, A.-S.L.; Nielsen, L.B.; Schmidt, J.B.; Bruhn, D.; Andersen, L.H.; Pertoldi, C. eDNA Metabarcoding- and Microscopic Analysis for Diet Determination in Waterfowl, a Comparative Study in Vejlerne, Denmark. Biology 2023, 12, 1272. https://doi.org/10.3390/biology12091272
Svendsen A-SL, Nielsen LB, Schmidt JB, Bruhn D, Andersen LH, Pertoldi C. eDNA Metabarcoding- and Microscopic Analysis for Diet Determination in Waterfowl, a Comparative Study in Vejlerne, Denmark. Biology. 2023; 12(9):1272. https://doi.org/10.3390/biology12091272
Chicago/Turabian StyleSvendsen, Anna-Sofie Lützhøft, Louise Bach Nielsen, Jakob Braüner Schmidt, Dan Bruhn, Line Holm Andersen, and Cino Pertoldi. 2023. "eDNA Metabarcoding- and Microscopic Analysis for Diet Determination in Waterfowl, a Comparative Study in Vejlerne, Denmark" Biology 12, no. 9: 1272. https://doi.org/10.3390/biology12091272
APA StyleSvendsen, A.-S. L., Nielsen, L. B., Schmidt, J. B., Bruhn, D., Andersen, L. H., & Pertoldi, C. (2023). eDNA Metabarcoding- and Microscopic Analysis for Diet Determination in Waterfowl, a Comparative Study in Vejlerne, Denmark. Biology, 12(9), 1272. https://doi.org/10.3390/biology12091272







