Physiologically Based Pharmacokinetic Modeling of Extracellular Vesicles
Abstract
Simple Summary
Abstract
1. Introduction
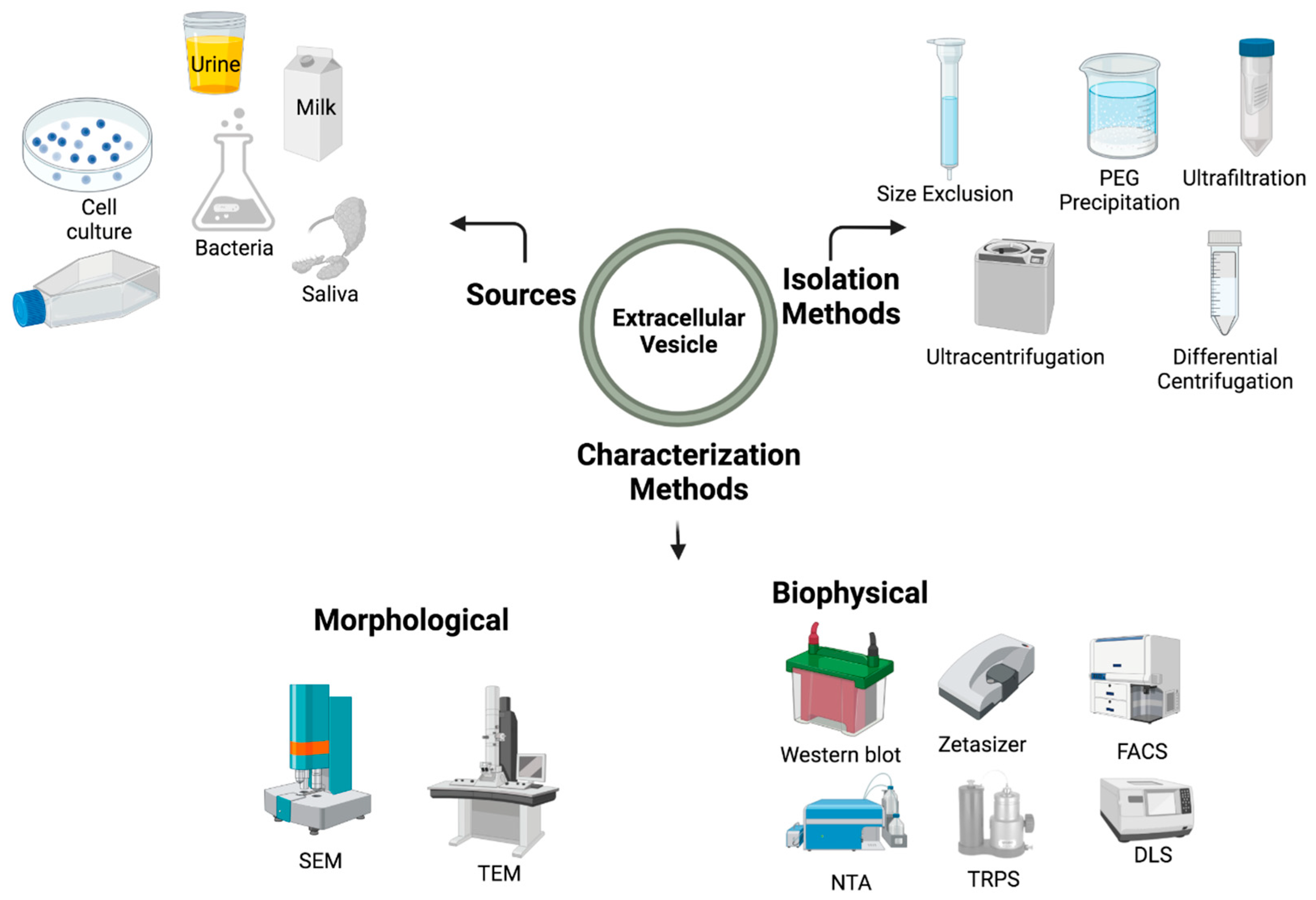
2. Extracellular Vesicles: A Brief Sketch
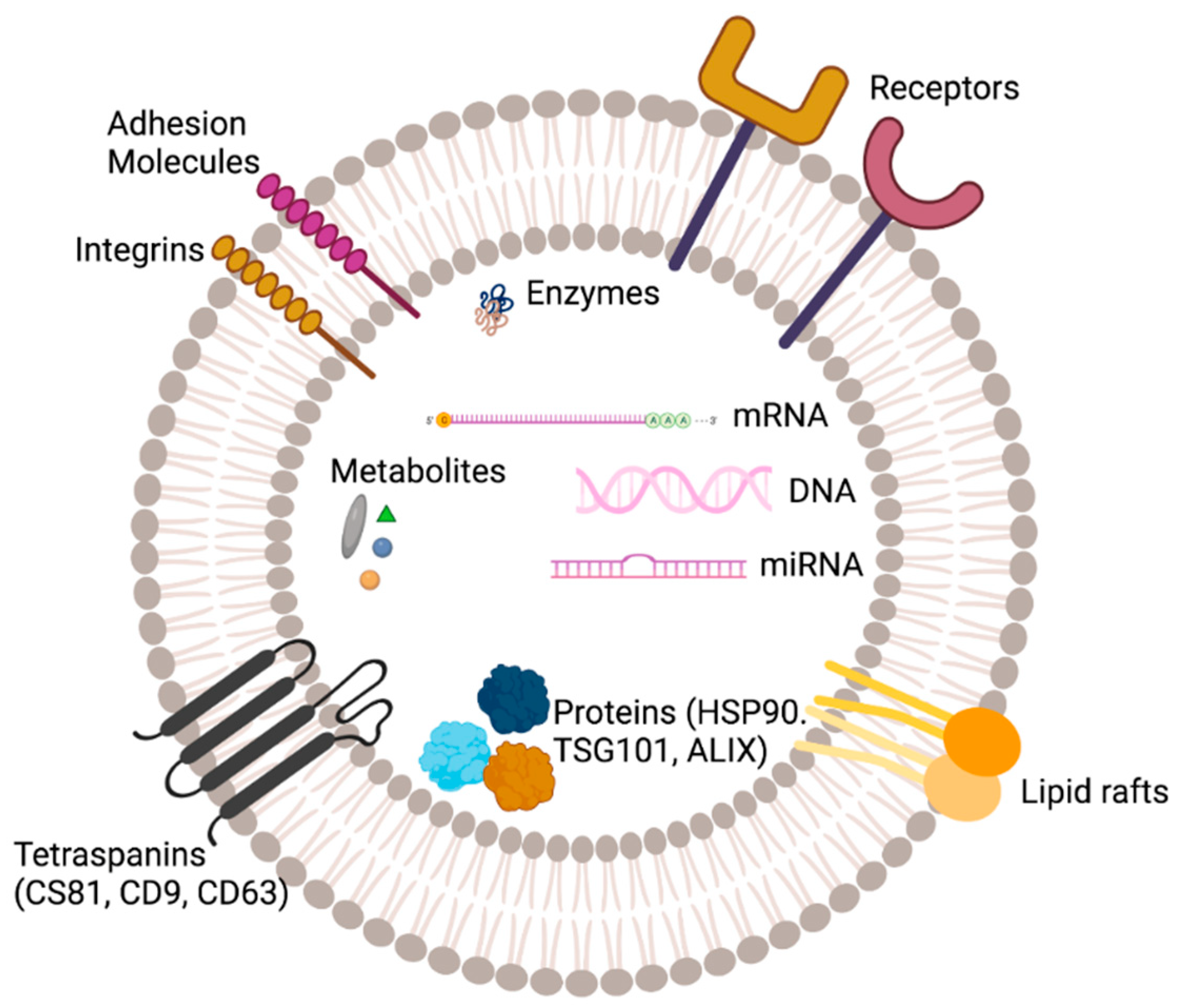
3. Structure and Composition of EVs
4. Biogenesis of EV
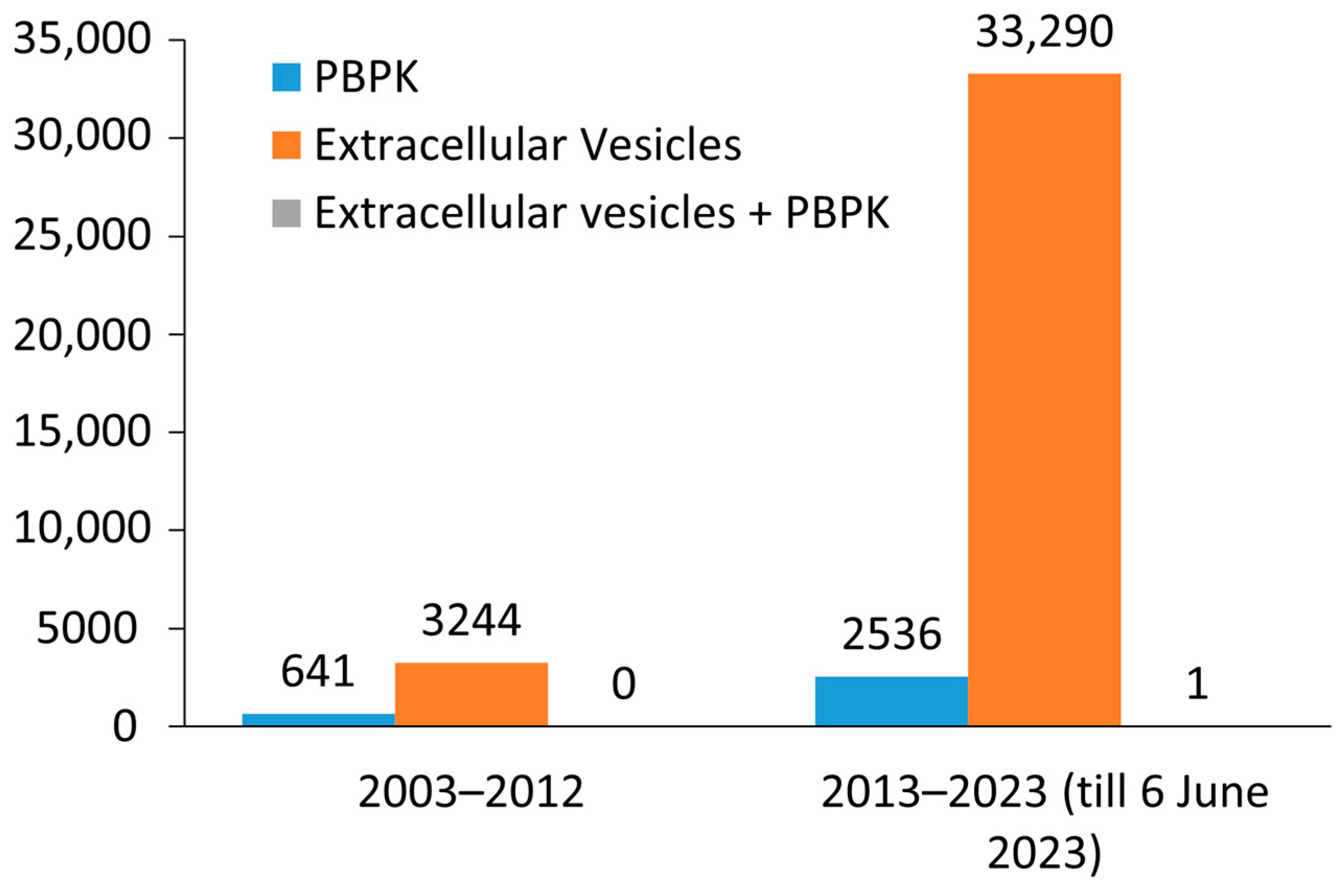
5. Technology Advancement and EV Research
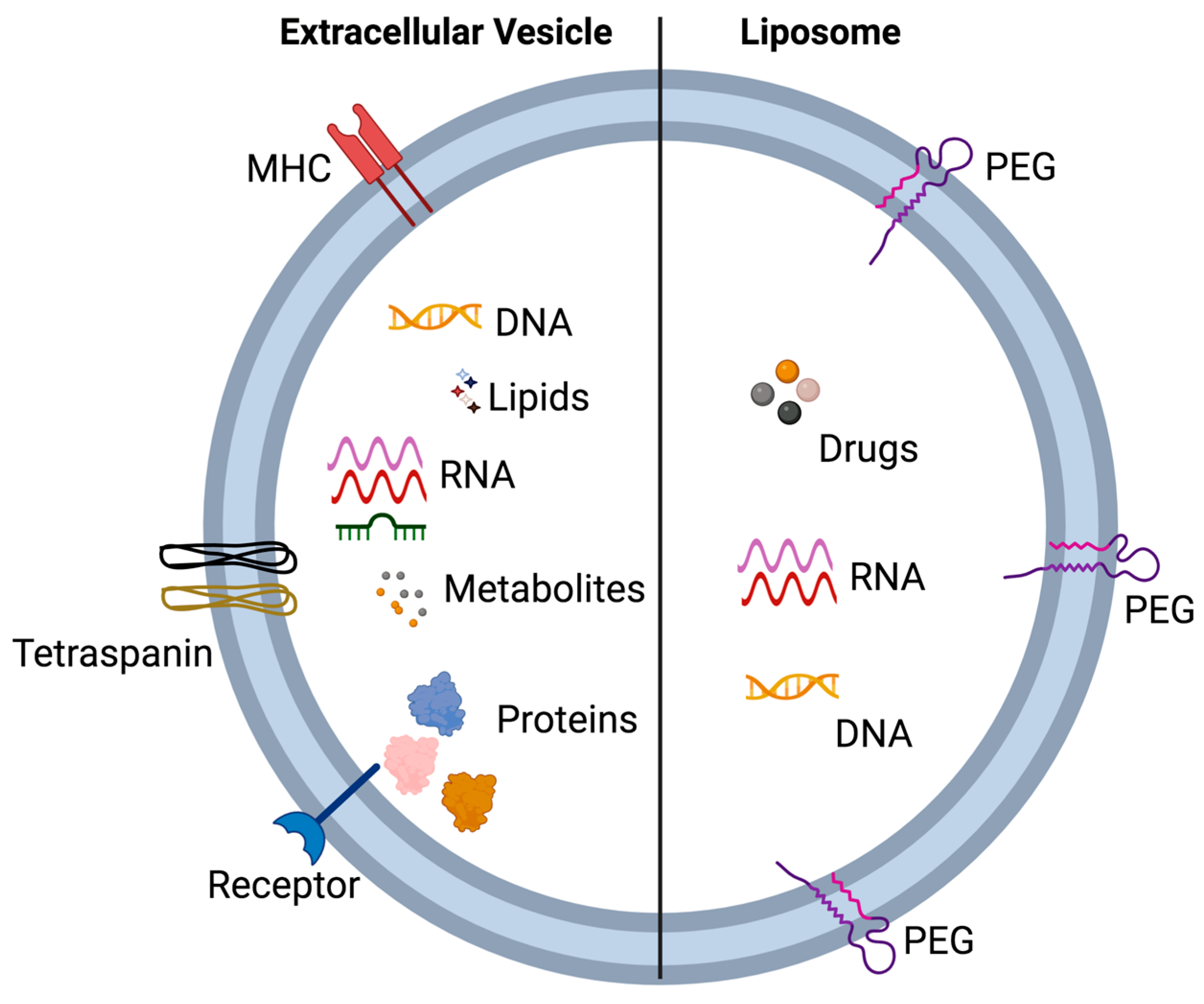
5.1. Extracellular Vesicles and Liposomes—Similarities and Differences
5.2. Extracellular Vesicles and Nanoparticles: PBPK Modeling
6. EVs in Clinical Research
7. ADME of EVs
7.1. Absorption
7.1.1. EV Absorption by Fusion
7.1.2. Phagocytosis
7.1.3. Clathrin-Mediated Endocytosis
7.1.4. Caveolin-Mediated Endocytosis
7.1.5. Lipid-Raft Mediated
7.1.6. Macropinocytosis
7.1.7. Oral Exposure
7.1.8. Intravenous Injections
7.1.9. Other Routes
7.2. Distribution
7.3. Clearance
8. PBPK Modeling in EV Research
8.1. Type of Data Needed to Make a Successful PBPK Model
8.2. PBPK Model Validation Criteria
9. PBPK Modeling and Simulation of Extracellular Vesicles Mediated Drug Delivery
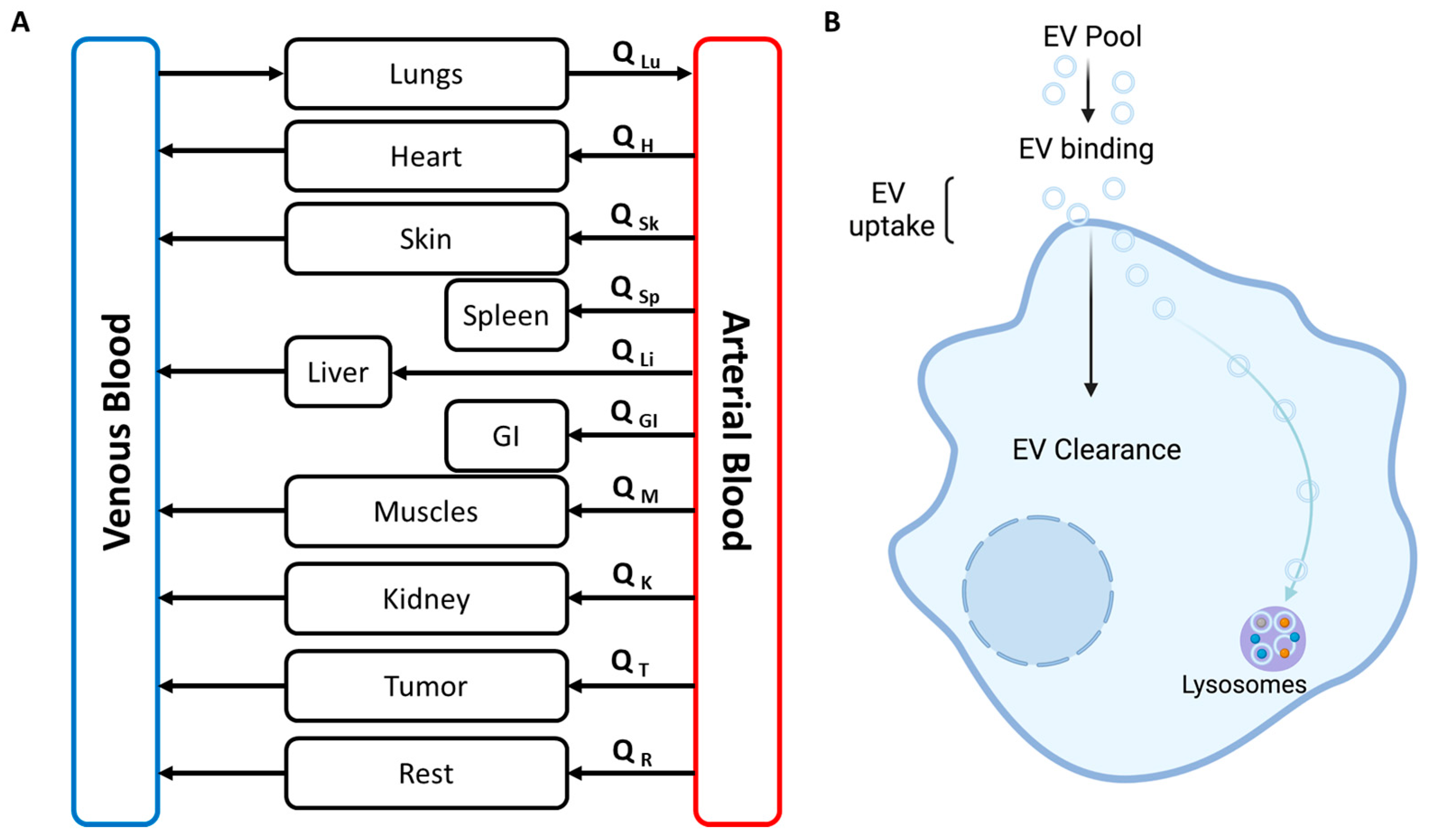
9.1. Whole Body PBPK Model
9.2. Simplified PBPK Model
10. PBPK Modeling Software
10.1. GastroPlus
10.2. Simcyp
10.3. PKSIM
10.4. Berkeley Madonna
11. ADME Mathematical Equations
12. PBPK Modeling Application for EV Therapeutics
12.1. End and Complicated Life Stage Prediction
12.2. IVIVE
12.3. Cancer Model
12.4. Route to Route and Species to Species Extrapolation
13. Perspective and Future Direction
14. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 23, 1535750. [Google Scholar] [CrossRef]
- Li, M.; Liao, L.; Tian, W. Extracellular Vesicles Derived From Apoptotic Cells: An Essential Link Between Death and Regeneration. Front. Cell Dev. Biol. 2020, 8, 573511. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, C.; Migliorino, R.; Leone, A.; Budillon, A. Large extracellular vesicles: Size matters in tumor progression. Cytokine Growth Factor Rev. 2020, 51, 69–74. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Sanwlani, R.; Gangoda, L. Role of Extracellular Vesicles in Cell Death and Inflammation. Cells 2021, 10, 2663. [Google Scholar] [CrossRef] [PubMed]
- Candelario, K.M.; Steindler, D.A. The role of extracellular vesicles in the progression of neurodegenerative disease and cancer. Trends Mol. Med. 2014, 20, 368–374. [Google Scholar] [CrossRef]
- Kodam, S.P.; Ullah, M. Diagnostic and Therapeutic Potential of Extracellular Vesicles. Technol. Cancer Res. Treat. 2021, 20, 15330338211041203. [Google Scholar] [CrossRef]
- Kogure, A.; Yoshioka, Y.; Ochiya, T. Extracellular Vesicles in Cancer Metastasis: Potential as Therapeutic Targets and Materials. Int. J. Mol. Sci. 2020, 21, 4463. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Ciferri, M.C.; Quarto, R.; Tasso, R. Extracellular Vesicles as Biomarkers and Therapeutic Tools: From Pre-Clinical to Clinical Applications. Biology 2021, 10, 359. [Google Scholar] [CrossRef]
- Rashed, M.H.; Bayraktar, E.; KHelal, G.; Abd-Ellah, M.F.; Amero, P.; Chavez-Reyes, A.; Rodriguez-Aguayo, C. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 538. [Google Scholar] [CrossRef] [PubMed]
- Modh, H.; Fang, D.J.; Ou, Y.H.; Yau, J.N.N.; Kovshova, T.; Nagpal, S.; Knoll, J.; Wallenwein, C.M.; Maiti, K.; Bhowmick, S.; et al. Injectable drug delivery systems of doxorubicin revisited: In vitro-in vivo relationships using human clinical data. Int. J. Pharm. 2021, 608, 121073. [Google Scholar] [CrossRef] [PubMed]
- Klyachko, N.L.; Arzt, C.J.; Li, S.M.; Gololobova, O.A.; Batrakova, E.V. Extracellular Vesicle-Based Therapeutics: Preclinical and Clinical Investigations. Pharmaceutics 2020, 12, 1171. [Google Scholar] [CrossRef]
- Yoo, M.H.; Lee, A.R.; Moon, K.S. Characteristics of Extracellular Vesicles and Preclinical Testing Considerations Prior to Clinical Applications. Biomedicines 2022, 10, 869. [Google Scholar] [CrossRef]
- Claridge, B.; Lozano, J.; Poh, Q.H.; Greening, D.W. Development of Extracellular Vesicle Therapeutics: Challenges, Considerations, and Opportunities. Front. Cell Dev. Biol. 2021, 9, 734720. [Google Scholar] [CrossRef] [PubMed]
- Sager, J.E.; Yu, J.; Ragueneau-Majlessi, I.; Isoherranen, N. Physiologically Based Pharmacokinetic (PBPK) Modeling and Simulation Approaches: A Systematic Review of Published Models, Applications, and Model Verification. Drug Metab. Dispos. 2015, 43, 1823–1837. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef]
- Zadjali, F.; Kumar, P.; Yao, Y.; Johnson, D.; Astrinidis, A.; Vogel, P.; Gross, K.W.; Bissler, J.J. Tuberous Sclerosis Complex Axis Controls Renal Extracellular Vesicle Production and Protein Content. Int. J. Mol. Sci. 2020, 21, 1729. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Brennan, K.; Martin, K.; Fitzgerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef]
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Juan, T.; Furthauer, M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin. Cell Dev. Biol. 2018, 74, 66–77. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126 Pt 24, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wijerathne, H.; Godwin, A.K.; Soper, S.A. Isolation and analysis methods of extracellular vesicles (EVs). Extracell. Vesicles Circ. Nucl. Acids 2021, 2, 80–103. [Google Scholar]
- Akbar, A.; Malekian, F.; Baghban, N.; Kodam, S.P.; Ullah, M. Methodologies to Isolate and Purify Clinical Grade Extracellular Vesicles for Medical Applications. Cells 2022, 11, 186. [Google Scholar] [CrossRef]
- Tian, Y.; Gong, M.; Hu, Y.; Liu, H.; Zhang, W.; Zhang, M.; Hu, X.; Aubert, D.; Zhu, S.; Wu, L.; et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles 2020, 9, 1697028. [Google Scholar] [CrossRef]
- Kumar, P.; Zadjali, F.; Yao, Y.; Johnson, D.; Siroky, B.; Astrinidis, A.; Vogel, P.; Gross, K.W.; Bissler, J.J. Tsc2 mutation induces renal tubular cell nonautonomous disease. Genes Dis. 2022, 9, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Zadjali, F.; Yao, Y.; Köttgen, M.; Hofherr, A.; Gross, K.W.; Mehta, D.; Bissler, J.J. Single Gene Mutations in Pkd1 or Tsc2 Alter Extracellular Vesicle Production and Trafficking. Biology 2022, 11, 709. [Google Scholar] [CrossRef]
- Nakai, W.; Yoshida, T.; Diez, D.; Miyatake, Y.; Nishibu, T.; Imawaka, N.; Naruse, K.; Sadamura, Y.; Hanayama, R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016, 6, 33935. [Google Scholar] [CrossRef]
- Stam, J.; Bartel, S.; Bischoff, R.; Wolters, J.C. Isolation of extracellular vesicles with combined enrichment methods. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2021, 1169, 122604. [Google Scholar] [CrossRef]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 2021, 9, 811971. [Google Scholar] [CrossRef]
- Chuo, S.T.; Chien, J.C.; Lai, C.P. Imaging extracellular vesicles: Current and emerging methods. J. Biomed. Sci. 2018, 25, 91. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Zadjali, F.; Yao, Y.; Siroky, B.; Astrinidis, A.; Gross, K.W.; Bissler, J.J. Tsc Gene Locus Disruption and Differences in Renal Epithelial Extracellular Vesicles. Front. Physiol. 2021, 12, 630933. [Google Scholar] [CrossRef]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef]
- Selvaraj, C.; Chandra, I.; Singh, S.K. Artificial intelligence and machine learning approaches for drug design: Challenges and opportunities for the pharmaceutical industries. Mol. Divers. 2022, 26, 1893–1913. [Google Scholar] [CrossRef]
- Sahu, A.; Mishra, J.; Kushwaha, N. Artificial Intelligence (AI) in Drugs and Pharmaceuticals. Comb. Chem. High Throughput Screen. 2022, 25, 1818–1837. [Google Scholar] [CrossRef] [PubMed]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.Y.; Sood, A.K.; Hua, S. Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Chen, G.; Zhang, J. A Review of Liposomes as a Drug Delivery System: Current Status of Approved Products, Regulatory Environments, and Future Perspectives. Molecules 2022, 27, 1372. [Google Scholar] [CrossRef]
- van der Koog, L.; Gandek, T.B.; Nagelkerke, A. Liposomes and Extracellular Vesicles as Drug Delivery Systems: A Comparison of Composition, Pharmacokinetics, and Functionalization. Adv. Healthc. Mater. 2022, 11, e2100639. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Quarto, R.; Tasso, R. Extracellular Vesicles as Natural, Safe and Efficient Drug Delivery Systems. Pharmaceutics 2019, 11, 557. [Google Scholar] [CrossRef]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.-C.; Cheng, Y.-H.; Riviere, J.E.; Monteiro-Riviere, N.A.; Kreyling, W.G.; Lin, Z. Development of a multi-route physiologically based pharmacokinetic (PBPK) model for nanomaterials: A comparison between a traditional versus a new route-specific approach using gold nanoparticles in rats. Part. Fibre Toxicol. 2022, 19, 47. [Google Scholar] [CrossRef]
- Grangier, A.; Branchu, J.; Volatron, J.; Piffoux, M.; Gazeau, F.; Wilhelm, C.; Silva, A.K. Technological advances towards extracellular vesicles mass production. Adv. Drug Deliv. Rev. 2021, 176, 113843. [Google Scholar] [CrossRef]
- Nikfarjam, S.; Rezaie, J.; Zolbanin, N.M.; Jafari, R. Mesenchymal stem cell derived-exosomes: A modern approach in translational medicine. J. Transl. Med. 2020, 18, 449. [Google Scholar] [CrossRef]
- Rezaie, J.; Feghhi, M.; Etemadi, T. A review on exosomes application in clinical trials: Perspective, questions, and challenges. Cell Commun. Signal. 2022, 20, 145. [Google Scholar] [CrossRef]
- Alagga, A.A.; Gupta, V. Drug Absorption; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Morishita, M.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Pharmacokinetics of Exosomes-An Important Factor for Elucidating the Biological Roles of Exosomes and for the Development of Exosome-Based Therapeutics. J. Pharm. Sci. 2017, 106, 2265–2269. [Google Scholar] [CrossRef] [PubMed]
- Vader, P.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Emerging targets for cancer therapy. Trends Mol. Med. 2014, 20, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef]
- Ginini, L.; Billan, S.; Fridman, E.; Gil, Z. Insight into Extracellular Vesicle-Cell Communication: From Cell Recognition to Intracellular Fate. Cells 2022, 11, 1375. [Google Scholar] [CrossRef]
- Prada, I.; Meldolesi, J. Binding and Fusion of Extracellular Vesicles to the Plasma Membrane of Their Cell Targets. Int. J. Mol. Sci. 2016, 17, 1296. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Whitlock, J.M.; Chernomordik, L.V. Flagging fusion: Phosphatidylserine signaling in cell-cell fusion. J. Biol. Chem. 2021, 296, 100411. [Google Scholar] [CrossRef] [PubMed]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Mörgelin, M.; Belting, M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 2013, 288, 17713–17724. [Google Scholar] [CrossRef]
- McKelvey, K.J.; Powell, K.L.; Ashton, A.W.; Morris, J.M.; McCracken, S.A. Exosomes: Mechanisms of Uptake. J. Circ. Biomark. 2015, 4, 7. [Google Scholar] [CrossRef]
- Miyado, K.; Yamada, G.; Yamada, S.; Hasuwa, H.; Nakamura, Y.; Ryu, F.; Suzuki, K.; Kosai, K.; Inoue, K.; Ogura, A.; et al. Requirement of CD9 on the egg plasma membrane for fertilization. Science 2000, 287, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Zhao, W.-L.; Ye, Y.-Y.; Bai, X.-C.; Liu, R.-Q.; Chang, L.-F.; Zhou, Q.; Sui, S.-F. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef]
- Angeloni, N.L.; McMahon, K.M.; Swaminathan, S.; Plebanek, M.P.; Osman, I.; Volpert, O.V.; Thaxton, C.S. Pathways for Modulating Exosome Lipids Identified by High-Density Lipoprotein-Like Nanoparticle Binding to Scavenger Receptor Type B-1. Sci. Rep. 2016, 6, 22915. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Underhill, D.M.; Goodridge, H.S. Information processing during phagocytosis. Nat. Rev. Immunol. 2012, 12, 492–502. [Google Scholar] [CrossRef]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef]
- Eguchi, S.; Takefuji, M.; Sakaguchi, T.; Ishihama, S.; Mori, Y.; Tsuda, T.; Takikawa, T.; Yoshida, T.; Ohashi, K.; Shimizu, Y.; et al. Cardiomyocytes capture stem cell-derived, anti-apoptotic microRNA-214 via clathrin-mediated endocytosis in acute myocardial infarction. J. Biol. Chem. 2019, 294, 11665–11674. [Google Scholar] [CrossRef]
- Kiss, A.L.; Botos, E. Endocytosis via caveolae: Alternative pathway with distinct cellular compartments to avoid lysosomal degradation? J. Cell Mol. Med. 2009, 13, 1228–1237. [Google Scholar] [CrossRef]
- Majkova, Z.; Toborek, M.; Hennig, B. The role of caveolae in endothelial cell dysfunction with a focus on nutrition and environmental toxicants. J. Cell Mol. Med. 2010, 14, 2359–2370. [Google Scholar] [CrossRef] [PubMed]
- Hardin, C.D.; Vallejo, J. Caveolins in vascular smooth muscle: Form organizing function. Cardiovasc. Res. 2006, 69, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-I.; Maeng, Y.-S.; Kim, T.-I.; Lee, Y.; Kim, Y.-S.; Kim, E.K. Lysosomal trafficking of TGFBIp via caveolae-mediated endocytosis. PLoS ONE 2015, 10, e0119561. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nanbo, A.; Kawanishi, E.; Yoshida, R.; Yoshiyama, H. Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J. Virol. 2013, 87, 10334–10347. [Google Scholar] [CrossRef]
- Lajoie, P.; Nabi, I.R. Lipid rafts, caveolae, and their endocytosis. Int. Rev. Cell Mol. Biol. 2010, 282, 135–163. [Google Scholar]
- de Gassart, A.; Geminard, C.; Fevrier, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef]
- Kay, R.R. Macropinocytosis: Biology and mechanisms. Cells Dev. 2021, 168, 203713. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Wang, L.; Chang, C.-C.; Rothenberg, K.E.; Huang, J.; Wang, Y.; Hoffman, B.D.; Liton, P.B.; Yuan, F. Involvement of a Rac1-Dependent Macropinocytosis Pathway in Plasmid DNA Delivery by Electrotransfection. Mol. Ther. 2017, 25, 803–815. [Google Scholar] [CrossRef]
- Fitzner, D.; Schnaars, M.; van Rossum, D.; Krishnamoorthy, G.; Dibaj, P.; Bakhti, M.; Regen, T.; Hanisch, U.-K.; Simons, M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J. Cell Sci. 2011, 124 Pt 3, 447–458. [Google Scholar] [CrossRef]
- Alqahtani, M.S.; Kazi, M.; Alsenaidy, M.A.; Ahmad, M.Z. Advances in Oral Drug Delivery. Front. Pharmacol. 2021, 12, 618411. [Google Scholar] [CrossRef] [PubMed]
- Rautamo, M.; Kvarnström, K.; Sivén, M.; Airaksinen, M.; Lahdenne, P.; Sandler, N. A Focus Group Study about Oral Drug Administration Practices at Hospital Wards-Aspects to Consider in Drug Development of Age-Appropriate Formulations for Children. Pharmaceutics 2020, 12, 109. [Google Scholar] [CrossRef]
- Niu, M.; Tan, Y.; Guan, P.; Hovgaard, L.; Lu, Y.; Qi, J.; Lian, R.; Li, X.; Wu, W. Enhanced oral absorption of insulin-loaded liposomes containing bile salts: A mechanistic study. Int. J. Pharm. 2014, 460, 119–130. [Google Scholar] [CrossRef]
- Wu, W.; Niu, M.; Lu, Y.; Hovgaard, L. Liposomes containing glycocholate as potential oral insulin delivery systems: Preparation, in vitro characterization, and improved protection against enzymatic degradation. Int. J. Nanomed. 2011, 6, 1155–1166. [Google Scholar] [CrossRef]
- Hua, S.; Marks, E.; Schneider, J.J.; Keely, S. Advances in oral nano-delivery systems for colon targeted drug delivery in inflammatory bowel disease: Selective targeting to diseased versus healthy tissue. Nanomedicine 2015, 11, 1117–1132. [Google Scholar] [CrossRef]
- Samuel, M.; Fonseka, P.; Sanwlani, R.; Gangoda, L.; Chee, S.H.; Keerthikumar, S.; Spurling, A.; Chitti, S.V.; Zanker, D.; Ang, C.-S.; et al. Oral administration of bovine milk-derived extracellular vesicles induces senescence in the primary tumor but accelerates cancer metastasis. Nat. Commun. 2021, 12, 3950. [Google Scholar] [CrossRef]
- Zhou, Q.; Jin, J.-F.; Zhu, L.-L.; Chen, M.; Xu, H.-M.; Wang, H.-F.; Feng, X.-Q.; Zhu, X.-P. The optimal choice of medication administration route regarding intravenous, intramuscular, and subcutaneous injection. Patient Prefer. Adherence 2015, 9, 923–942. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Zhao, Y.; Banks, W.A.; Bullock, K.M.; Haney, M.; Batrakova, E.; Kabanov, A.V. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials 2017, 142, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.W.; Wei, P.; Zhang, G.J.; Yan, J.X.; Zhang, S.; Liang, J.; Wang, X.L. Intravenous infusion of the exosomes derived from human umbilical cord mesenchymal stem cells enhance neurological recovery after traumatic brain injury via suppressing the NF-kappaB pathway. Open Life Sci. 2022, 17, 189–201. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nishikawa, M.; Shinotsuka, H.; Matsui, Y.; Ohara, S.; Imai, T.; Takakura, Y. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J. Biotechnol. 2013, 165, 77–84. [Google Scholar] [CrossRef]
- Dinh, P.-U.C.; Paudel, D.; Brochu, H.; Popowski, K.D.; Gracieux, M.C.; Cores, J.; Huang, K.; Hensley, M.T.; Harrell, E.; Vandergriff, A.C.; et al. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J. Control. Release 2015, 199, 145–155. [Google Scholar]
- Zhu, Y.-G.; Shi, M.-M.; Monsel, A.; Dai, C.-X.; Dong, X.; Shen, H.; Li, S.-K.; Chang, J.; Xu, C.-L.; Li, P.; et al. Inhalation of lung spheroid cell secretome and exosomes promotes lung repair in pulmonary fibrosis. Nat. Commun. 2020, 11, 1064. [Google Scholar]
- Hwang, D.W.; Jo, M.J.; Lee, J.H.; Kang, H.; Bao, K.; Hu, S.; Baek, Y.; Moon, H.G.; Lee, D.S.; Kashiwagi, S.; et al. Nebulized exosomes derived from allogenic adipose tissue mesenchymal stromal cells in patients with severe COVID-19: A pilot study. Stem Cell Res. Ther. 2022, 13, 220. [Google Scholar]
- Hwang, D.W.; Jo, M.J.; Lee, J.H.; Kang, H.; Bao, K.; Hu, S.; Baek, Y.; Moon, H.G.; Lee, D.S.; Kashiwagi, S.; et al. Chemical Modulation of Bioengineered Exosomes for Tissue-Specific Biodistribution. Adv. Ther. 2019, 2, 1900111. [Google Scholar] [CrossRef]
- Mathiesen, A.; Hamilton, T.; Carter, N.; Brown, M.; McPheat, W.; Dobrian, A. Endothelial Extracellular Vesicles: From Keepers of Health to Messengers of Disease. Int. J. Mol. Sci. 2021, 22, 4640. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, V.; Yang, X.; Ma, Y.; Wu, M.H.; Yuan, S.Y. Extracellular vesicles: New players in regulating vascular barrier function. Am. J. Physiol. Heart Circ. Physiol. 2020, 319, H1181–H1196. [Google Scholar] [CrossRef]
- Ståhl, A.-L.; Johansson, K.; Mossberg, M.; Kahn, R.; Karpman, D. Exosomes and microvesicles in normal physiology, pathophysiology, and renal diseases. Pediatr. Nephrol. 2019, 34, 11–30. [Google Scholar] [CrossRef]
- Bennit, H.R.F.; Gonda, A.; McMullen, J.R.W.; Kabagwira, J.; Wall, N.R. Peripheral Blood Cell Interactions of Cancer-Derived Exosomes Affect Immune Function. Cancer Microenviron. 2019, 12, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kothandan, V.K.; Kim, H.W.; Kim, K.S.; Kim, J.Y.; Cho, H.J.; Lee, Y.-K.; Lee, D.-E.; Hwang, S.R. Noninvasive Assessment of Exosome Pharmacokinetics In Vivo: A Review. Pharmaceutics 2019, 11, 649. [Google Scholar] [CrossRef]
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Takahashi, Y.; Nishikawa, M.; Kato, K.; Morishita, M.; Yamashita, T.; Matsumoto, A.; Charoenviriyakul, C.; Takakura, Y. Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J. Extracell. Vesicles 2015, 4, 26238. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Zhang, L.; A Grillo, J.; Liu, Q.; Bullock, J.M.; Moon, Y.J.; Song, P.; Brar, S.S.; Madabushi, R.; Wu, T.C.; et al. Applications of physiologically based pharmacokinetic (PBPK) modeling and simulation during regulatory review. Clin. Pharmacol. Ther. 2011, 89, 259–267. [Google Scholar] [CrossRef]
- Zhuang, X.; Lu, C. PBPK modeling and simulation in drug research and development. Acta Pharm. Sin. B 2016, 6, 430–440. [Google Scholar] [CrossRef]
- Jones, H.; Rowland-Yeo, K. Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, e63. [Google Scholar] [CrossRef]
- Hartmanshenn, C.; Scherholz, M.; Androulakis, I.P. Physiologically-based pharmacokinetic models: Approaches for enabling personalized medicine. J. Pharmacokinet. Pharmacodyn. 2016, 43, 481–504. [Google Scholar] [CrossRef]
- Eke, A.C.; Gebreyohannes, R.D. Physiologically based pharmacokinetic modeling (PBPK’s) prediction potential in clinical pharmacology decision making during pregnancy. Int. J. Gynaecol. Obstet. 2020, 150, 414–416. [Google Scholar] [CrossRef]
- Wang, J.; Yeung, B.Z.; Wientjes, M.G.; Cui, M.; Peer, C.J.; Lu, Z.; Figg, W.D.; Woo, S.; Au, J.L.-S. A Quantitative Pharmacology Model of Exosome-Mediated Drug Efflux and Perturbation-Induced Synergy. Pharmaceutics 2021, 13, 997. [Google Scholar] [CrossRef]
- Ruiz, P.; Ray, M.; Fisher, J.; Mumtaz, M. Development of a human Physiologically Based Pharmacokinetic (PBPK) Toolkit for environmental pollutants. Int. J. Mol. Sci. 2011, 12, 7469–7480. [Google Scholar] [CrossRef]
- He, H.; Yuan, D.; Wu, Y.; Cao, Y. Pharmacokinetics and Pharmacodynamics Modeling and Simulation Systems to Support the Development and Regulation of Liposomal Drugs. Pharmaceutics 2019, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Tylutki, Z.; Polak, S.; Wisniowska, B. Top-down, Bottom-up and Middle-out Strategies for Drug Cardiac Safety Assessment via Modeling and Simulations. Curr. Pharmacol. Rep. 2016, 2, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Jamei, M.; Dickinson, G.L.; Rostami-Hodjegan, A. A framework for assessing inter-individual variability in pharmacokinetics using virtual human populations and integrating general knowledge of physical chemistry, biology, anatomy, physiology and genetics: A tale of ‘bottom-up’ vs ‘top-down’ recognition of covariates. Drug Metab. Pharmacokinet. 2009, 24, 53–75. [Google Scholar] [PubMed]
- Tsamandouras, N.; Rostami-Hodjegan, A.; Aarons, L. Combining the ‘bottom up’ and ‘top down’ approaches in pharmacokinetic modelling: Fitting PBPK models to observed clinical data. Br. J. Clin. Pharmacol. 2015, 79, 48–55. [Google Scholar] [CrossRef]
- Utembe, W.; Clewell, H.; Sanabria, N.; Doganis, P.; Gulumian, M. Current Approaches and Techniques in Physiologically Based Pharmacokinetic (PBPK) Modelling of Nanomaterials. Nanomaterials 2020, 10, 1267. [Google Scholar] [CrossRef]
- Wang, Q.; Spenkelink, B.; Boonpawa, R.; Rietjens, I.M. Use of Physiologically Based Pharmacokinetic Modeling to Predict Human Gut Microbial Conversion of Daidzein to S-Equol. J. Agric. Food Chem. 2022, 70, 343–352. [Google Scholar] [CrossRef]
- Kuepfer, L.; Niederalt, C.; Wendl, T.; Schlender, J.; Willmann, S.; Lippert, J.; Block, M.; Eissing, T.; Teutonico, D. Applied Concepts in PBPK Modeling: How to Build a PBPK/PD Model. CPT Pharmacomet. Syst. Pharmacol. 2016, 5, 516–531. [Google Scholar] [CrossRef]
- Stader, F.; Penny, M.A.; Siccardi, M.; Marzolini, C. A Comprehensive Framework for Physiologically-Based Pharmacokinetic Modeling in Matlab. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 444–459. [Google Scholar] [CrossRef]
- Cao, Y.; Jusko, W.J. Applications of minimal physiologically-based pharmacokinetic models. J. Pharmacokinet. Pharmacodyn. 2012, 39, 711–723. [Google Scholar] [CrossRef]
- Xia, B.; Heimbach, T.; Gollen, R.; Nanavati, C.; He, H. A simplified PBPK modeling approach for prediction of pharmacokinetics of four primarily renally excreted and CYP3A metabolized compounds during pregnancy. AAPS J. 2013, 15, 1012–1024. [Google Scholar] [CrossRef]
- Chen, J.; Ruan, Z.; Lou, H.; Yang, D.; Shao, R.; Xu, Y.; Hu, X.; Jiang, B. First-in-human study to investigate the safety and pharmacokinetics of salvianolic acid A and pharmacokinetic simulation using a physiologically based pharmacokinetic model. Front. Pharmacol. 2022, 13, 907208. [Google Scholar] [CrossRef] [PubMed]
- Deepika, D.; Kumar, V. The Role of “Physiologically Based Pharmacokinetic Model (PBPK)” New Approach Methodology (NAM) in Pharmaceuticals and Environmental Chemical Risk Assessment. Int. J. Environ. Res. Public Health 2023, 20, 3473. [Google Scholar] [CrossRef] [PubMed]
- Kesisoglou, F.; Mitra, A. Application of Absorption Modeling in Rational Design of Drug Product Under Quality-by-Design Paradigm. AAPS J. 2015, 17, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lionberger, R.A.; Davit, B.M.; Yu, L.X. Utility of physiologically based absorption modeling in implementing Quality by Design in drug development. AAPS J. 2011, 13, 59–71. [Google Scholar] [CrossRef]
- Huang, W.; Lee, S.L.; Yu, L.X. Mechanistic approaches to predicting oral drug absorption. AAPS J. 2009, 11, 217–224. [Google Scholar] [CrossRef]
- Hussain, A.; Altamimi, M.A.; Alshehri, S.; Imam, S.S.; Singh, S.K. Vesicular elastic liposomes for transdermal delivery of rifampicin: In-Vitro, in-vivo and in silico GastroPlus prediction studies. Eur. J. Pharm. Sci. 2020, 151, 105411. [Google Scholar] [CrossRef] [PubMed]
- Jamei, M.; Marciniak, S.; Edwards, D.; Wragg, K.; Feng, K.; Barnett, A.; Rostami-Hodjegan, A. The simcyp population based simulator: Architecture, implementation, and quality assurance. In Silico Pharmacol. 2013, 1, 9. [Google Scholar] [CrossRef]
- Maharaj, A.R.; Edginton, A.N. Physiologically based pharmacokinetic modeling and simulation in pediatric drug development. CPT Pharmacomet. Syst. Pharmacol. 2014, 3, e150. [Google Scholar] [CrossRef]
- Achour, B.; Al-Majdoub, Z.M.; Grybos-Gajniak, A.; Lea, K.; Kilford, P.; Zhang, M.; Knight, D.; Barber, J.; Schageman, J.; Rostami-Hodjegan, A. Liquid Biopsy Enables Quantification of the Abundance and Interindividual Variability of Hepatic Enzymes and Transporters. Clin. Pharmacol. Ther. 2021, 109, 222–232. [Google Scholar] [CrossRef]
- Ince, I.; Dallmann, A.; Frechen, S.; Coboeken, K.; Niederalt, C.; Wendl, T.; Block, M.; Meyer, M.; Eissing, T.; Burghaus, R.; et al. Predictive Performance of Physiology-Based Pharmacokinetic Dose Estimates for Pediatric Trials: Evaluation With 10 Bayer Small-Molecule Compounds in Children. J. Clin. Pharmacol. 2021, 61 (Suppl. 1), S70–S82. [Google Scholar] [CrossRef]
- Basu, S.; Lien, Y.T.; Vozmediano, V.; Schlender, J.-F.; Eissing, T.; Schmidt, S.; Niederalt, C. Physiologically Based Pharmacokinetic Modeling of Monoclonal Antibodies in Pediatric Populations Using PK-Sim. Front. Pharmacol. 2020, 11, 868. [Google Scholar] [CrossRef]
- Frechen, S.; Solodenko, J.; Wendl, T.; Dallmann, A.; Ince, I.; Lehr, T.; Lippert, J.; Burghaus, R. A generic framework for the physiologically-based pharmacokinetic platform qualification of PK-Sim and its application to predicting cytochrome P450 3A4-mediated drug-drug interactions. CPT Pharmacomet. Syst. Pharmacol. 2021, 10, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, M.; Fisher, J.; Blount, B.; Ruiz, P. Application of physiologically based pharmacokinetic models in chemical risk assessment. J. Toxicol. 2012, 2012, 904603. [Google Scholar] [CrossRef] [PubMed]
- Gufford, B.; Barr, J.; González-Pérez, V.; Layton, M.; White, J.; Oberlies, N.; Paine, M. Quantitative prediction and clinical evaluation of an unexplored herb-drug interaction mechanism in healthy volunteers. CPT Pharmacomet. Syst. Pharmacol. 2015, 4, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Sia, J.E.V.; Tu, S.; Li, X.; Dong, Z.; Yu, Z.; Yao, X.; Hatley, O.; Li, H.; Liu, D. Development of a physiologically based pharmacokinetic (PBPK) population model for Chinese elderly subjects. Br. J. Clin. Pharmacol. 2021, 87, 2711–2722. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Jiang, K.; Wei, X.; Li, Y.; Wang, T.; Song, Y. Physiologically Based Pharmacokinetic Models Are Effective Support for Pediatric Drug Development. AAPS PharmSciTech 2021, 22, 208. [Google Scholar] [CrossRef]
- Coppola, P.; Kerwash, E.; Cole, S. Physiologically Based Pharmacokinetics Model in Pregnancy: A Regulatory Perspective on Model Evaluation. Front. Pediatr. 2021, 9, 687978. [Google Scholar] [CrossRef]
- Sun, L.; Barter, Z.; von Moltke, L.; Yeo, K.R. Using physiologically-based pharmacokinetic modeling for predicting the effects of hepatic impairment on the pharmacokinetics of olanzapine and samidorphan given as a combination tablet. CPT Pharmacomet. Syst. Pharmacol. 2021, 10, 1071–1080. [Google Scholar] [CrossRef]
- Hsueh, C.-H.; Hsu, V.; Zhao, P.; Zhang, L.; Giacomini, K.; Huang, S.-M. PBPK Modeling of the Effect of Reduced Kidney Function on the Pharmacokinetics of Drugs Excreted Renally by Organic Anion Transporters. Clin. Pharmacol. Ther. 2018, 103, 485–492. [Google Scholar] [CrossRef]
- Khalil, F.; Laer, S. Physiologically based pharmacokinetic modeling: Methodology, applications, and limitations with a focus on its role in pediatric drug development. J. Biomed. Biotechnol. 2011, 2011, 907461. [Google Scholar] [CrossRef] [PubMed]
- Hariparsad, N.; Ramsden, D.; Taskar, K.; Badée, J.; Venkatakrishnan, K.; Reddy, M.B.; Cabalu, T.; Mukherjee, D.; Rehmel, J.; Bolleddula, J.; et al. Current Practices, Gap Analysis, and Proposed Workflows for PBPK Modeling of Cytochrome P450 Induction: An Industry Perspective. Clin. Pharmacol. Ther. 2022, 112, 770–781. [Google Scholar] [CrossRef]
- Scotcher, D.; Melillo, N.; Tadimalla, S.; Darwich, A.S.; Ziemian, S.; Ogungbenro, K.; Schütz, G.; Sourbron, S.; Galetin, A. Physiologically Based Pharmacokinetic Modeling of Transporter-Mediated Hepatic Disposition of Imaging Biomarker Gadoxetate in Rats. Mol. Pharm. 2021, 18, 2997–3009. [Google Scholar] [CrossRef] [PubMed]
- Bartelink, I.H.; van de Stadt, E.A.; Leeuwerik, A.F.; Thijssen, V.L.J.L.; Hupsel, J.R.I.; Nieuwendijk, J.F.v.D.; Bahce, I.; Yaqub, M.; Hendrikse, N.H. Physiologically Based Pharmacokinetic (PBPK) Modeling to Predict PET Image Quality of Three Generations EGFR TKI in Advanced-Stage NSCLC Patients. Pharmaceuticals 2022, 15, 796. [Google Scholar] [CrossRef] [PubMed]
- Perry, C.; Davis, G.; Conner, T.M.; Zhang, T. Utilization of Physiologically Based Pharmacokinetic Modeling in Clinical Pharmacology and Therapeutics: An Overview. Curr. Pharmacol. Rep. 2020, 6, 71–84. [Google Scholar] [CrossRef]
- Fairman, K.; Choi, M.-K.; Gonnabathula, P.; Lumen, A.; Worth, A.; Paini, A.; Li, M. An Overview of Physiologically-Based Pharmacokinetic Models for Forensic Science. Toxics 2023, 11, 126. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, J.Y.; Mukadam, S.; Malhi, V.; Kenny, J.R. Application of IVIVE and PBPK modeling in prospective prediction of clinical pharmacokinetics: Strategy and approach during the drug discovery phase with four case studies. Biopharm. Drug Dispos. 2012, 33, 85–98. [Google Scholar] [CrossRef]
- Sharma, A.; Li, M.; Thavathiru, E.; Ibrahim, M.; Garcia-Contreras, L.; Benbrook, D.M.; Woo, S. Physiologically Based Pharmacokinetic Modeling and Tissue Distribution Characteristics of SHetA2 in Tumor-Bearing Mice. AAPS J. 2020, 22, 51. [Google Scholar] [CrossRef]
- Liu, H.; Yu, Y.; Guo, N.; Wang, X.; Han, B.; Xiang, X. Application of Physiologically Based Pharmacokinetic Modeling to Evaluate the Drug-Drug and Drug-Disease Interactions of Apatinib. Front. Pharmacol. 2021, 12, 780937. [Google Scholar] [CrossRef]
- Fan, Y.; Mansoor, N.; Ahmad, T.; Alam Khan, R.; Czejka, M.; Sharib, S.; Yang, D.-H.; Ahmed, M. Physiologically based pharmacokinetic modeling for predicting irinotecan exposure in human body. Oncotarget 2017, 8, 48178–48185. [Google Scholar] [CrossRef]
- He, H.; Liu, C.; Wu, Y.; Zhang, X.; Fan, J.; Cao, Y. A Multiscale Physiologically-Based Pharmacokinetic Model for Doxorubicin to Explore its Mechanisms of Cytotoxicity and Cardiotoxicity in Human Physiological Contexts. Pharm. Res. 2018, 35, 174. [Google Scholar] [CrossRef] [PubMed]
- Chiu, W.A.; White, P. Steady-state solutions to PBPK models and their applications to risk assessment I: Route-to-route extrapolation of volatile chemicals. Risk Anal. 2006, 26, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, E.M. Interspecies extrapolation. Methods Mol. Biol. 2012, 929, 501–520. [Google Scholar] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, P.; Mehta, D.; Bissler, J.J. Physiologically Based Pharmacokinetic Modeling of Extracellular Vesicles. Biology 2023, 12, 1178. https://doi.org/10.3390/biology12091178
Kumar P, Mehta D, Bissler JJ. Physiologically Based Pharmacokinetic Modeling of Extracellular Vesicles. Biology. 2023; 12(9):1178. https://doi.org/10.3390/biology12091178
Chicago/Turabian StyleKumar, Prashant, Darshan Mehta, and John J. Bissler. 2023. "Physiologically Based Pharmacokinetic Modeling of Extracellular Vesicles" Biology 12, no. 9: 1178. https://doi.org/10.3390/biology12091178
APA StyleKumar, P., Mehta, D., & Bissler, J. J. (2023). Physiologically Based Pharmacokinetic Modeling of Extracellular Vesicles. Biology, 12(9), 1178. https://doi.org/10.3390/biology12091178





