A Greater Increase in Complement C5a Receptor 1 Level at Onset and a Smaller Decrease in Immunoglobulin G Level after Recovery in Severer Coronavirus Disease 2019 Patients: A New Analysis of Existing Data with a New Two-Tailed t-Test
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Data Sources
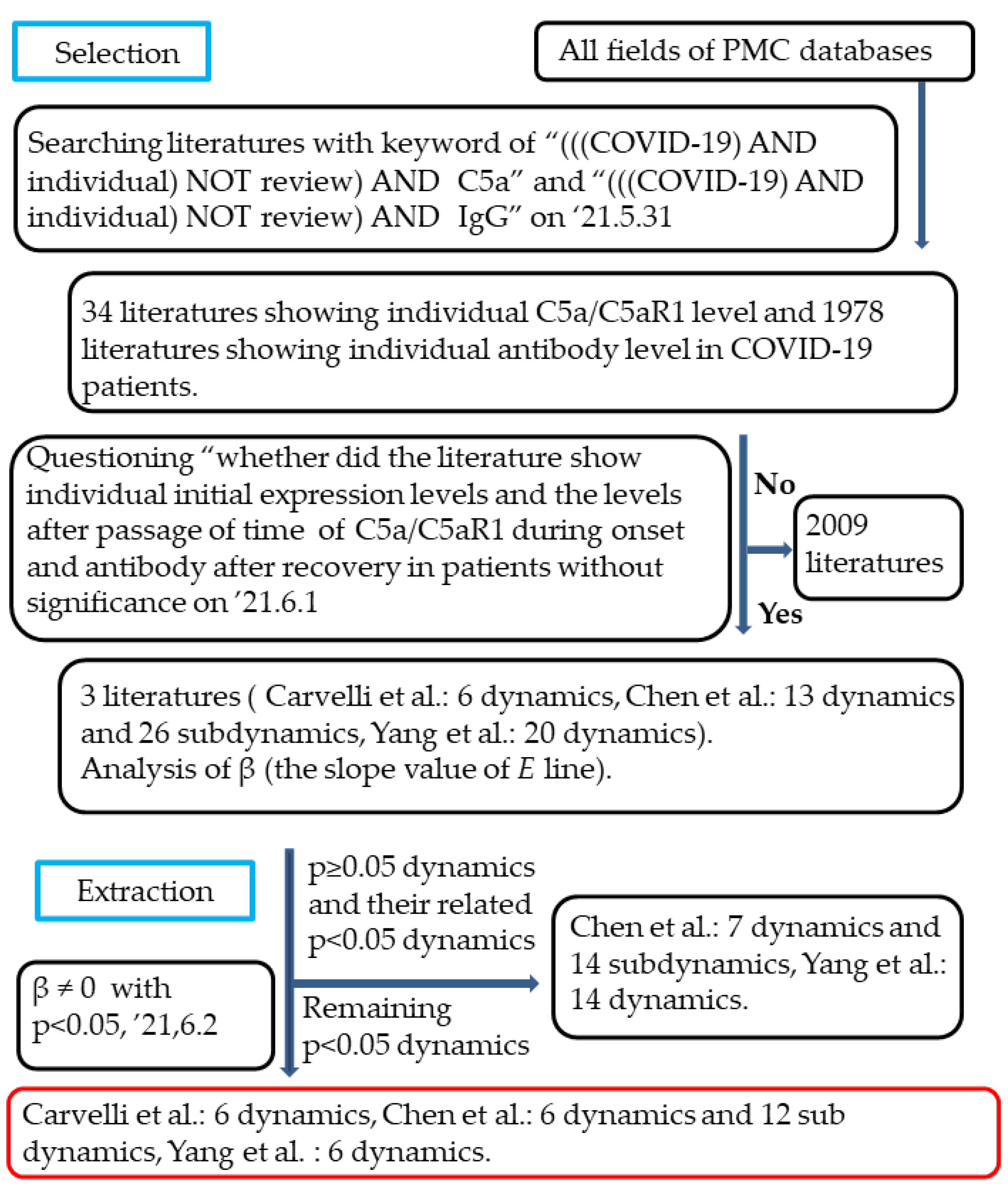
2.2. Selection of the Studies
2.3. Data Extraction and Synthesis
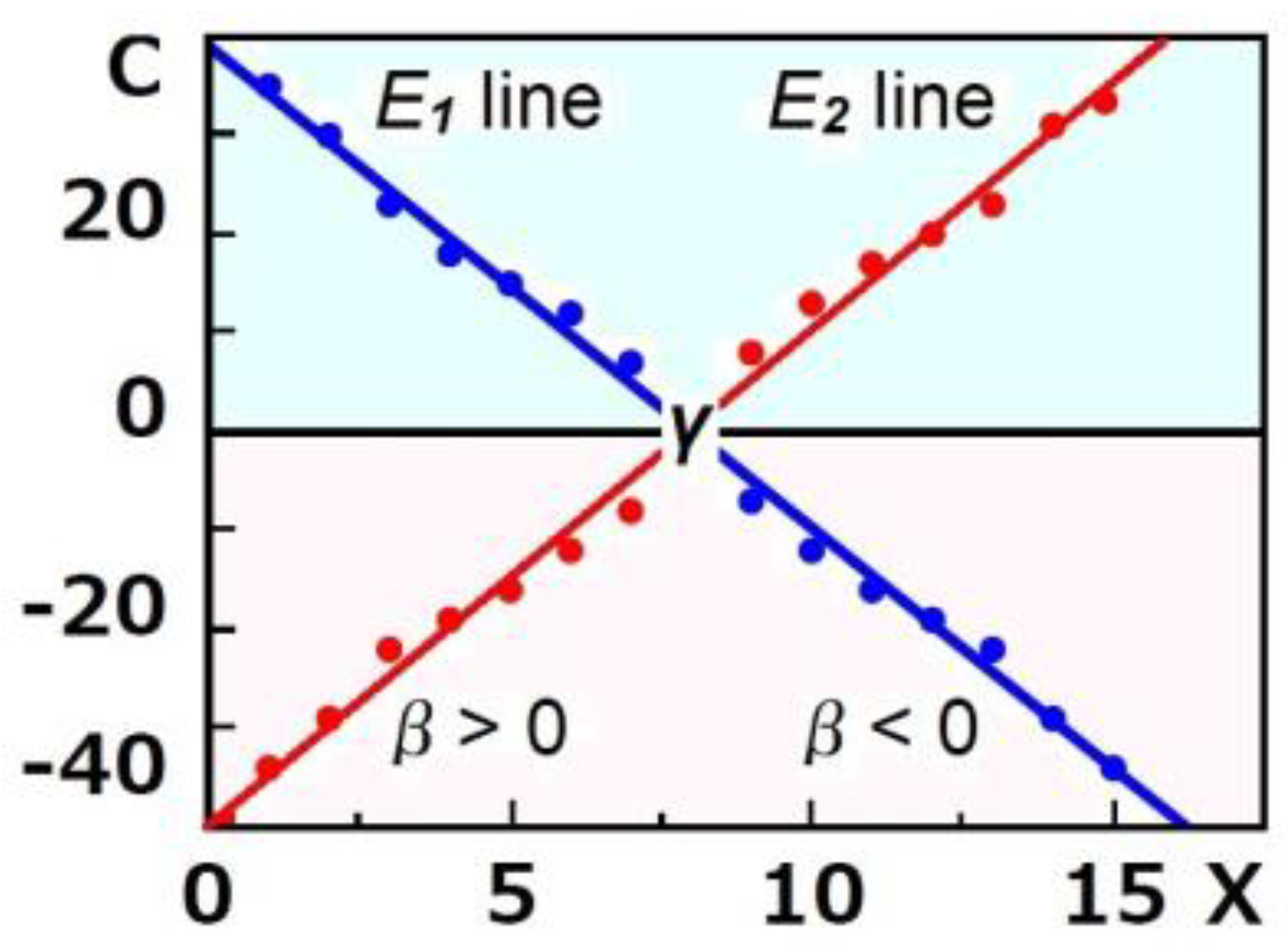
2.4. Ishida’s t-Test1
2.5. Ishida’s t-Test2
2.6. Validation
2.7. Risk of Bias in the Methods
3. Results
3.1. Selection of Studies
3.2. Extraction of Dynamics Dependent on X
3.3. Analysis of the Extracted Dynamics with Ishida’s t-Test1
3.4. Analysis of the Extracted Dynamics with Ishida’s t-Test2
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.S.C.; Zumla, A. Severe acute respiratory syndrome: Historical, epidemiologic, and clinical features. Infect. Dis. Clin. N. Am. 2019, 33, 869–889. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xiao, H.; Guo, R.; Li, Y.; Shen, B. The role of C5a in acute lung injury induced by highly pathogenic viral infections. Emerg. Microbes Infect. 2015, 4, 1–7. [Google Scholar] [CrossRef]
- Carvelli, J.; Demaria, O.; Vély, F.; Batista, L.; Benmansour, N.C.; Fares, J.; Carpentier, S.; Thibult, M.-L.; Morel, A.; Remark, R.; et al. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature 2020, 588, 146–150. [Google Scholar] [CrossRef]
- Apostolidis, S.A.; Sarkar, A.; Giannini, H.M.; Goel, R.R.; Mathew, D.; Suzuki, A.; Baxter, A.E.; Greenplate, A.R.; Alanio, C.; Abdel-Hakeem, M.; et al. Signaling through FcγRIIA and the C5a-C5aR pathway mediate platelet hyperactivation in COVID-19. Front. Immunol. 2022, 13, 834988. [Google Scholar] [CrossRef]
- Vlaar, A.P.J.; Witzenrath, M.; van Paassen, P.; Heunks, L.M.A.; Mourvillier, B.; de Bruin, S.; Lim, E.H.T.; Brouwer, M.C.; Tuinman, P.R.; Saraiva, J.F.K.; et al. Anti-C5a antibody (vilobelimab) therapy for critically ill, invasively mechanically ventilated patients with COVID-19 (PANAMO): A multicentre, double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Respir. Med. 2022, 10, 1137–1146. [Google Scholar] [CrossRef]
- Subramaniam, S.; Kothari, H.; Bosmann, M. Tissue factor in COVID-19-associated coagulopathy. Thromb. Res. 2022, 220, 35–47. [Google Scholar] [CrossRef]
- Plášek, J.; Gumulec, J.; Máca, J.; Škarda, J.; Procházka, V.; Grézl, T.; Václavík, J. COVID-19 associated coagulopathy: Mechanisms and host-directed treatment. Am. J. Med. Sci. 2022, 363, 465–475. [Google Scholar] [CrossRef]
- Li, J.; Liu, B. The roles and potential therapeutic implications of C5a in the pathogenesis of COVID-19-associated coagulopathy. Cytokine Growth Fact. Rev. 2021, 58, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Skendros, P.; Mitsios, A.; Chrysanthopoulou, A.; Mastellos, D.C.; Metallidis, S.; Rafailidis, P.; Ntinopoulou, M.; Sertaridou, E.; Tsironidou, V.; Tsigalou, C.; et al. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Investig. 2020, 130, 6151–6157. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, D.; Kuźniewska, A.; Senent, Y.; Tavira, B.; Inogés, S.; López-Díaz de Cerio, A.; Pio, R.; Okrój, M.; Yuste, J.R. C5a elevation in convalescents from severe COVID-19 is not associated with early complement activation markers C3bBbP or C4d. Front. Immunol. 2022, 13, 946522. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, T.M.; Shukla, A.K. The Complement C5a-C5aR1 GPCR axis in COVID-19 Therapeutics. Trends Immunol. 2020, 41, 965–967. [Google Scholar] [CrossRef] [PubMed]
- Vlaar, A.P.J.; Lim, E.H.T.; de Bruin, S.; Rückinger, S.; Pilz, K.; Brouwer, M.C.; Guo, R.F.; Heunks, L.M.A.; Busch, M.H.; van Paassen, P.; et al. The anti-C5a antibody vilobelimab efficiently inhibits C5a in patients with severe COVID-19. Clin. Transl. Sci. 2022, 15, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Bu, G.; Sun, Y.; Ren, C.; Qu, M.; Gao, Y.; Zhu, Y.; Wang, L.; Sun, L.; Liu, Y.J. Evaluation of serum IgM and IgG antibodies in COVID-19 patients by enzyme linked immunosorbent assay. Med. Virol. 2021, 93, 2857–2866. [Google Scholar] [CrossRef]
- Ali, H.; Alahmad, B.; Al-Shammari, A.A.; Alterki, A.; Hammad, M.; Cherian, P.; Alkhairi, I.; Sindhu, S.; Thanaraj, T.A.; Mohammad, A.; et al. Previous COVID-19 Infection and Antibody Levels After Vaccination. Front. Public Health 2021, 9, 778243. [Google Scholar] [CrossRef]
- Song, K.H.; Kim, D.M.; Lee, H.; Ham, S.Y.; Oh, S.M.; Jeong, H.; Jung, J.; Kang, C.K.; Park, J.Y.; Kang, Y.M.; et al. Dynamics of viral load and anti-SARS-CoV-2 antibodies in patients with positive RT-PCR results after recovery from COVID-19. Korean J. Intern. Med. 2021, 36, 11–14. [Google Scholar] [CrossRef]
- Kong, W.H.; Zhao, R.; Zhou, J.B.; Wang, F.; Kong, D.G.; Sun, J.B.; Ruan, Q.F.; Liu, M.Q. Serologic Response to SARS-CoV-2 in COVID-19 patients with different severity. Virol. Sin. 2020, 35, 752–757. [Google Scholar] [CrossRef]
- Bichara, C.D.A.; da Silva Graça Amoras, E.; Vaz, G.L.; da Silva Torres, M.K.; Queiroz, M.A.F.; do Amaral, I.P.C.; Vallinoto, I.M.V.C.; Bichara, C.N.C.; Vallinoto, A.C.R. Dynamics of anti-SARS-CoV-2 IgG antibodies post-COVID-19 in a Brazilian Amazon population. BMC Infect. Dis. 2021, 21, 443. [Google Scholar] [CrossRef] [PubMed]
- Wilcoxon, F. Individual comparisons by Ranking Methods. Biom. Bull. 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Kenward, M.G.; Roger, J.H. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 1997, 53, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Kenward, M.G.; Roger, J.H. An improved approximation to the precision of fixed effects from restricted maximum likelihood. Comput. Stat. Data Anal. 2009, 53, 2583–2595. [Google Scholar] [CrossRef]
- Chen, Y.; Tong, X.; Li, Y.; Gu, B.; Yan, J.; Liu, Y.; Shen, H.; Huang, R.; Wu, C. A comprehensive, longitudinal analysis of humoral responses specific to four recombinant antigens of SARS-CoV-2 in severe and non-severe COVID-19 patients. PLoS Pathog. 2020, 16, e1008796. [Google Scholar] [CrossRef] [PubMed]
- Student (Gosset, W.S.). The probable error of a mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Yang, C.; Jiang, M.; Wang, X.; Tang, X.; Fang, S.; Li, H.; Zuo, L.; Jiang, Y.; Zhong, Y.; Chen, O.; et al. Viral RNA level, serum antibody responses, and transmission risk in recovered COVID-19 patients with recurrent positive SARS-CoV-2 RNA test results: A population-based observational cohort study. Emerg. Microbes Infect. 2020, 9, 2368–2378. [Google Scholar] [CrossRef]
- Paris, R.M.; Petrovas, C.; Ferrando-Martinez, S.; Eirini Moysi, E.; Boswell, K.L.; Archer, E.; Yamamoto, T.; Ambrozak, D.; Casazza, J.P.; Haubrich, R.; et al. Selective loss of early differentiated, highly functional PD1high CD4 T cells with HIV progression. PLoS ONE 2015, 10, e0144767. [Google Scholar] [CrossRef]
- Gauss, C.F. Theoria combinationis observationum erroribus minimis obnoxiae. (Classic Reprint); Forgotten Books: London, UK.
- Keppel, G. Design and Analysis: A Researcher’s Handbook, 3rd ed.; Prentice-Hall, Inc.: Englewood Cliffs, NJ, USA, 1991. [Google Scholar]
- Fisher, R.A. On the "probable error" of a coefficient of correlation deduced from a small sample. Metron 1921, 1, 3–32. [Google Scholar]
- Dunn, O.J. Multiple comparisons among means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
- Box, G.E.P. A general distribution theory for a class of likelihood criteria. Biometrika 1949, 36, 317–346. [Google Scholar] [CrossRef]
- Breusch, T.S.; Pagan, A.R. A simple test for heteroskedasticity and random coefficient variation. Econometrica 1979, 47, 1287–1294. [Google Scholar] [CrossRef]
- Brunner, E.; Munzel, U. The nonparametric behrens-fisher problem: Asymptotic theory and a small-sample approximation. Biom. J. 2000, 42, 17–25. [Google Scholar] [CrossRef]
- Cochran, W.G. Some methods for strengthening the common chi-squared tests. Biom. Int. Biom. Society 1954, 10, 417–451. [Google Scholar] [CrossRef]
- Armitage, P. Tests for linear trends in proportions and frequencies. Biom. Int. Biom. Society 1955, 11, 375–386. [Google Scholar] [CrossRef]
- Cochran, W.G. The Comparison of percentages in matched samples. Biometrika 1950, 37, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Mantel, N. Chi-square tests with one degree of freedom, extensions of the Mantel-Haenszel procedure. J. Am. Stat. Assoc. 1963, 58, 690–700. [Google Scholar] [CrossRef]
- Mantel, N.; Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959, 22, 719–748. [Google Scholar] [CrossRef]
- Hirotsu, C. Cumulative chi-squared statistic as a tool for testing goodness of fit. Biometrika 1986, 73, 165–173. [Google Scholar] [CrossRef]
- Dixon, W.J. Processing data for outliers. Biometrics 1953, 9, 74–89. [Google Scholar] [CrossRef]
- Dunnett, C.W. A multiple comparison procedure for comparing several treatments with a control. J. Am. Stat. Assoc. 1955, 50, 1096–1121. [Google Scholar] [CrossRef]
- Friedman, M. The use of ranks to avoid the assumption of normality implicit in the analysis of variance. J. Am. Stat. Assoc. 1937, 32, 675–701. [Google Scholar] [CrossRef]
- Friedman, M. A comparison of alternative tests of significance for the problem of m rankings. Ann. Math. Stat. 1940, 11, 86–92. [Google Scholar] [CrossRef]
- Snedecor, G.W.; Cochran, W.G. Statistical Methods, 8th ed.; Iowa State University Press: Ames, IA, USA, 1989; ISBN 0-8138-1561-4. [Google Scholar]
- McDonald, J.H. G–test of goodness-of-fit. In Handbook of Biological Statistics, 3rd ed.; Sparky House Publishing: Baltimore, MA, USA, 2014; pp. 53–58. [Google Scholar]
- Gehan, E.A. A generalized Wilcoxon Test for comparing arbitrarily singly-censored samples. Biometrika 1965, 52, 203–223. [Google Scholar] [CrossRef]
- Jarque, C.M.; Bera, A.K. A test for normality of observations and regression residuals. Int. Stat. Rev. 1987, 55, 163–172. [Google Scholar] [CrossRef]
- Jonckheere, A.R. A distribution-free k-sample test against ordered alternatives. Biometrika. 1954, 41, 133–145. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Levene, H. Robust tests for equality of variances. In Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling; Olkin, I., Ghurye, S.G., Hoeffding, W., Madow, W.G., Mann, H.B., Eds.; Stanford University Press: Stanford, CA, USA, 1960; pp. 278–292. [Google Scholar]
- Lilliefors, H. On the Kolmogorov–Smirnov test for normality with mean and variance unknown. J. Am. Stat. Assoc. 1967, 62, 399–402. [Google Scholar] [CrossRef]
- Lilliefors, H. On the Kolmogorov–Smirnov test for the exponential distribution with mean unknown. J. Am. Stat. Assoc. 1969, 64, 387–389. [Google Scholar] [CrossRef]
- Mantel, N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother. Rep. 1966, 50, 163–170. [Google Scholar] [PubMed]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Hakulinen, T. A Mantel-Haenszel statistic for testing the association between a polychotomous exposure and a rare outcome. Am. J. Epidemiol. 1981, 113, 192–197. [Google Scholar] [CrossRef] [PubMed]
- McNemar, Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Pearson, K. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Philos. Mag. Ser. 1900, 50, 157–175. [Google Scholar] [CrossRef]
- Scheffe, H. The Analysis of Variance; Wiley: New York, NY, USA, 1959; ISBN 978-0-471-34505-3. [Google Scholar]
- Shirley, E. Non-parametric equivalent of Williams test for contrasting increasing dose levels of a treatment. Biometrics 1977, 33, 386–389. [Google Scholar] [CrossRef]
- Williams, D.A. Note on Shirley’s nonparametric test for comparing several dose levels with a zero-dose control. Biometrics 1986, 42, 183–186. [Google Scholar] [CrossRef]
- Grubbs, F.E. Sample criteria for testing outlying observations. Ann. Math. Stat. 1950, 21, 27–58. [Google Scholar] [CrossRef]
- Steel, R.G.D. A multiple comparison rank sum test treatment versus control. Biometrika 1959, 15, 560–572. [Google Scholar] [CrossRef]
- Thompson, W.R. On a criterion for rejection of observations and the distribution of the ratio of deviation to sampling standard deviation. Ann. Math. Statist. 1935, 6, 214–219. [Google Scholar] [CrossRef]
- Tukey, J.W. Comparing individual means in the analysis of variance. Biometrics 1959, 5, 99–114. [Google Scholar] [CrossRef]
- Welch, B.L. On the comparison of several mean values: An alternative approach. Biometrika 1951, 38, 330–336. [Google Scholar] [CrossRef]
- Williams, D.A. A test for differences between treatment means when several dose levels are compared with a zero dose control. Biometrics 1971, 27, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.A. The comparison of several dose levels with a zero dose control. Biometrics 1972, 28, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Yates, B.A. Contingency table involving small numbers and the χ2 test. Suppl. J. R. Stat. Soc. 1934, 1, 217–235. [Google Scholar] [CrossRef]
- Sprinthall, R.C. Basic Statistical Analysis, 9th ed.; Pearson Education: London, UK, 2011; ISBN 978-0-205-05217-2. [Google Scholar]
- Barnett, A.G.; van der Pols, J.C.; Dobson, A.J. Regression to the mean: What it is and how to deal with it. Int. J. Epidemiol. 2005, 34, 215–220. [Google Scholar] [CrossRef]
- Cohen, J. Quantitative methods in psychology: A power primer. Psychol. Bull. 1992, 112, 155–159. [Google Scholar] [CrossRef]

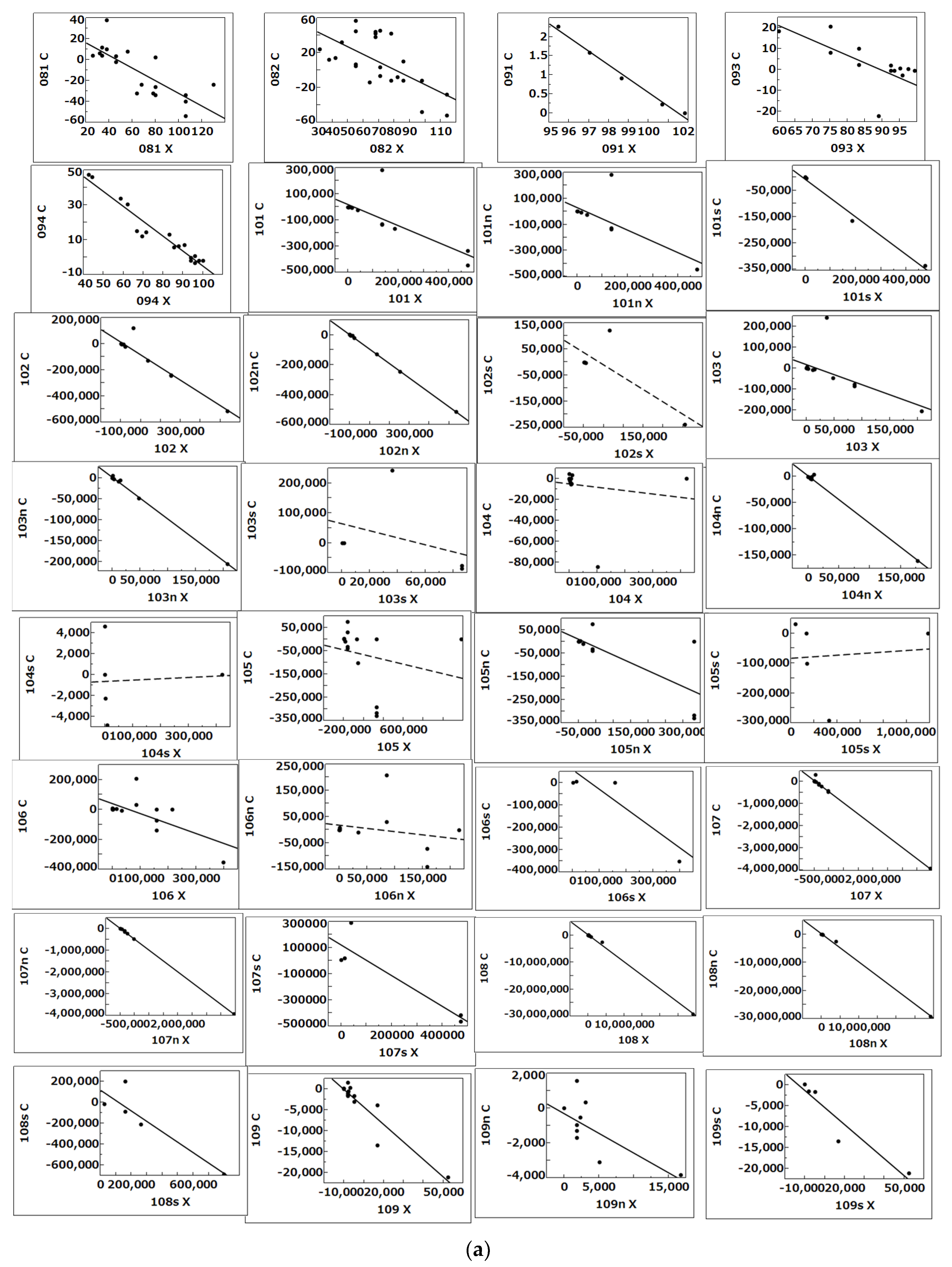

| DYN Name | Contents and Sources of Dynamics (DYN) |
|---|---|
| 081, 082 | Dynamics of concentration (ng/mL) of C5a desArg in plasma from pneumonia (non-COVID-19) (green: 081, n = 19) and ARDS (COVID-19) (red: 082, n = 25) patients between T0 and T1in extended Figure 1 by Carvelli et al. [8] |
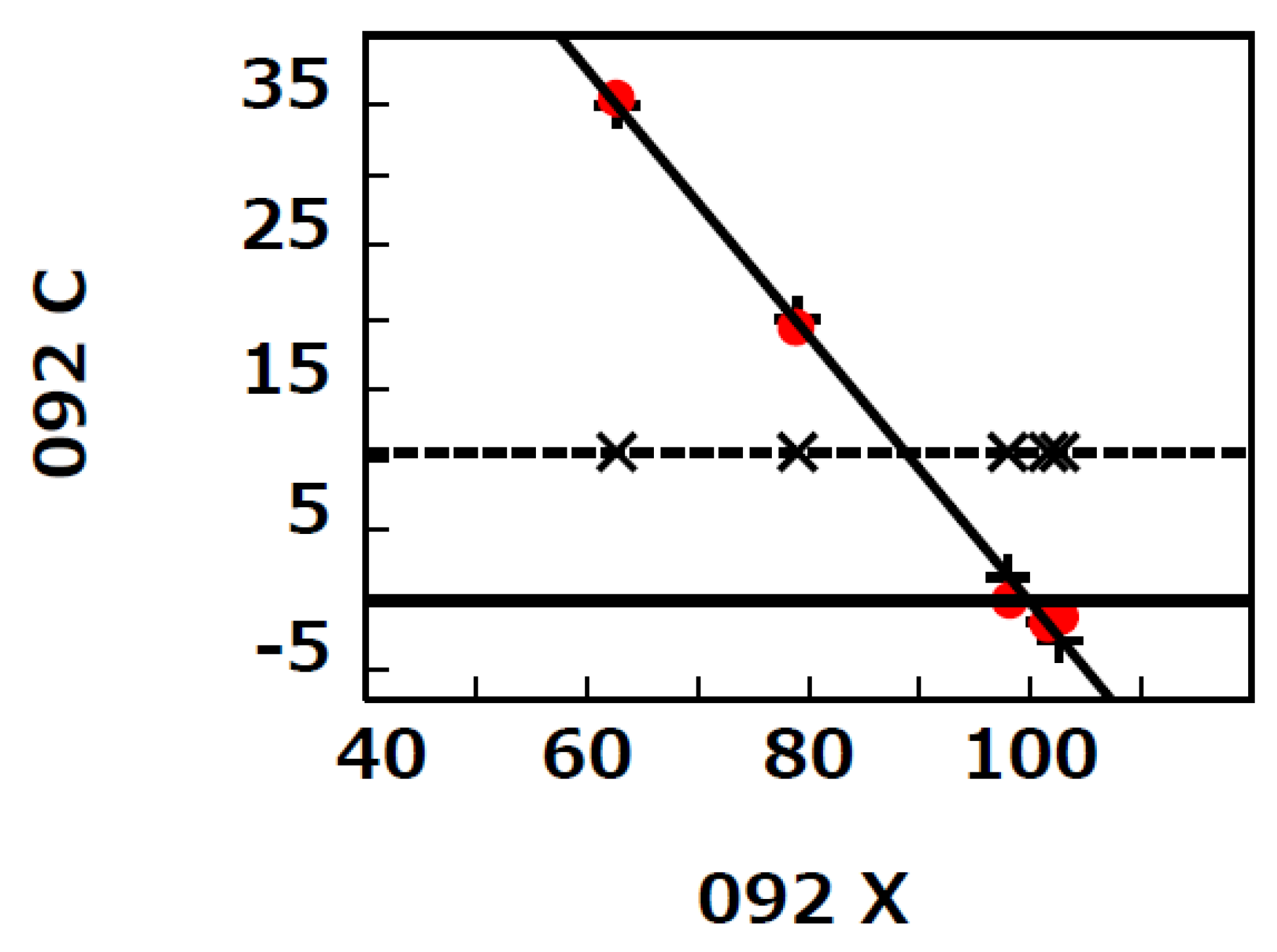
| 091 092 | Dynamics of % C5aR1-expressing neutrophils in pneumonia (non-COVID-19) (green: 091, n = 5) and ARDS (COVID-19) (red: 092, n = 5) patients between T0 and T1 in extended Figure 2 by Carvelli et al. [8] |
| 093 094 | Dynamics of % C5aR1-expressing monocytes in pneumonia (non-COVID-19) (green: 093, n = 15) and ARDS (COVID-19) (red: 094, n = 17) patients between T0 and T1 in extended Figure 2 by Carvelli et al. [8] |
| 101 | Dynamics of the IgM titer specific to RBD from non-severe (101n, n = 15) and severe (101s, n = 5) patients between T3 and T4 in Figure 6 (anti-RBD IgM) by Chen et al. [26] |
| 107 | Dynamics of the IgG titer specific to S1 from non-severe (107n, n = 11) and severe (107s, n = 5) patients between T3 and T4 in Figure 6 (anti-S1 IgG) by Chen et al. [26] |
| 108 | Dynamics of the IgG titer specific to NP from non-severe (108n, n = 11) and severe (108s, n = 5) patients between T3 and T4 in Figure 6 (anti-NP IgG) by Chen et al. [26] |
| 109 | Dynamics of the IgA titer specific to RBD from non-severe (109n, n = 10) and severe (109s, n = 5) patients between T3 and T4 in Figure 6 (anti-RBD IgA) by Chen et al. [26] |
| 10y | Dynamics of the IgA titer specific to S1 from non-severe (10yn, n = 11) and severe (10ys, n = 5) patients between T3 and T4 in Figure 6 (anti-S1 IgA) by Chen et al. [26] |
| 10w | Dynamics of the NAb titer (IC50) from non-severe (10wn, n = 11) and severe (10ws, n = 5) patients between T3 and T4 described in Figure 6 (Nab) by Chen et al. [26] |
| 211 212 | Dynamics of IgM levels specific to RBD (titers) from non-recurrent-positive (211, n = 35) and recurrent-positive (212, n = 6) patients between T5 and T6 described in Figure 4 (IgM levels) by Yang et al. [29] |
| 221 222 | Dynamics of IgM levels specific to RBD (titers) from recurrent-positive patients between T7 and T8 (221, n = 27) and between T8 and T9 (222, n = 23) described in Figure 4 (IgM levels) by Yang et al. [29] |
| 229 22x | Dynamics of NAb levels specific to RBD (titers) from recurrent-positive patients between T7 and T8 (229, n = 21) and between T8 and T9 (22x, n = 18) described in Figure 4 (Nab levels) by Yang et al. [29] |
| Grp | β | γ | SD | p | d | Mp | Mn | Me | SD1 | p1 | d1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 081 | −0.61 | 47.0 | 0.51 | <0.0001 | 1.19 | 2.60 | −13.91 | −11.30 | 14.77 | 0.004 | 0.77 |
| 082 | −0.87 | 81.1 | 1.11 | 0.0007 | 0.79 | 12.64 | −3.89 | 8.75 | 22.88 | 0.068 | 0.38 |
| 091 | −0.36 | 101.5 | 0.05 | 0.0005 | 7.12 | 1.02 | −0.02 | 1 | 0.10 | <0.0001 | 9.78 |
| 092 | −0.94 | 99.9 | 0.09 | 0.0002 | 10.9 | 11.29 | −0.87 | 10.42 | 1.17 | <0.0001 | 8.90 |
| 093 | −0.65 | 94.6 | 0.40 | <0.0001 | 1.61 | 5.73 | −0.41 | 5.32 | 4.11 | 0.0002 | 1.29 |
| 094 | −0.86 | 94.1 | 0.24 | <0.0001 | 3.62 | 13.73 | −0.67 | 13.06 | 4.17 | <0.0001 | 3.13 |
| 101 | −0.81 | 3 × 104 | 0.71 | 0.0007 | 1.15 | 8381 | −8 × 104 | −7 × 104 | 1 × 105 | 0.017 | 0.70 |
| 101n | −0.87 | 4 × 104 | 0.98 | 0.0162 | 0.89 | 1 × 104 | −7 × 104 | −5 × 104 | 1 × 105 | 0.174 | 0.44 |
| 101s | −0.72 | −9583 | 0.11 | 0.0053 | 6.87 | 0 | −1 × 105 | −1 × 105 | 2 × 104 | 0.0006 | 7.68 |
| 107 | −1.01 | 4 × 104 | 0.09 | <0.0001 | 11.3 | 2 × 104 | −4 × 105 | −3 × 105 | 8 × 104 | <0.0001 | 4.25 |
| 107n | −1.00 | 1 × 104 | 0.02 | <0.0001 | 60.9 | 5310 | −5 × 105 | −4 × 105 | 2 × 104 | <0.0001 | 25.77 |
| 107s | −1.18 | 1 × 105 | 0.68 | 0.0306 | 1.73 | 6 × 104 | −2 × 105 | −1 × 105 | 1 × 105 | 0.128 | 0.86 |
| 108 | −1.00 | 2 × 105 | 0.05 | <0.0001 | 19.5 | 7 × 104 | −2 × 106 | −2 × 106 | 4 × 105 | <0.0001 | 6.13 |
| 108n | −1.00 | 2 × 105 | 0.05 | <0.0001 | 18.9 | 1 × 105 | −3 × 106 | −3 × 106 | 4 × 105 | <0.0001 | 7.38 |
| 108s | −1.00 | 1 × 105 | 0.60 | 0.0333 | 1.67 | 2 × 104 | −2 × 105 | −2 × 105 | 1 × 105 | 0.059 | 1.17 |
| 109 | −0.42 | −306 | 0.17 | <0.0001 | 2.47 | 0 | −3260 | −3260 | 2125 | <0.0001 | 1.53 |
| 109n | −0.23 | −1346 | 0.27 | 0.0303 | 0.83 | 0 | −1127 | −1127 | 1167 | 0.014 | 0.97 |
| 109s | −0.41 | −3275 | 0.18 | 0.0139 | 2.32 | 0 | −7526 | −7526 | 2960 | 0.005 | 2.54 |
| 10y | −0.82 | 90.2 | 0.28 | <0.0001 | 2.96 | 8.24 | −2 × 104 | −2 × 104 | 9263 | <0.0001 | 1.90 |
| 10yn | −0.70 | −52.0 | 0.28 | <0.0001 | 2.55 | 0 | −1 × 104 | −1 × 104 | 8674 | 0.0008 | 1.43 |
| 10ys | −1.01 | 1303 | 0.03 | <0.0001 | 39.7 | 228 | −3 × 104 | −3 × 104 | 781 | <0.0001 | 37.35 |
| 10w | −0.94 | 546 | 0.13 | <0.0001 | 7.28 | 120 | −2093 | −1973 | 637 | <0.0001 | 3.10 |
| 10wn | −0.94 | 679 | 0.13 | <0.0001 | 7.27 | 254 | −2380 | −2126 | 748 | <0.0001 | 2.84 |
| 10ws | −0.93 | 255 | 0.04 | <0.0001 | 25.2 | 0 | −1636 | −1636 | 42 | <0.0001 | 38.92 |
| 211 | −0.26 | 0.87 | 0.16 | <0.0001 | 1.66 | 0.04 | −0.56 | −0.53 | 0.57 | <0.0001 | 0.92 |
| 212 | −0.34 | 0.72 | 0.17 | 0.0073 | 2.06 | 0.06 | −0.72 | −0.67 | 0.36 | 0.006 | 1.85 |
| 221 | −0.23 | 0.33 | 0.11 | <0.0001 | 2.14 | 0.01 | −0.12 | −0.11 | 0.11 | <0.0001 | 1.04 |
| 222 | −0.45 | 0.94 | 0.77 | 0.011 | 0.58 | 0.22 | −0.13 | 0.10 | 0.82 | 0.579 | 0.12 |
| 229 | −0.27 | 46.1 | 0.30 | 0.0047 | 0.70 | 5.33 | −18.13 | −12.80 | 57.92 | 0.323 | 0.22 |
| 22x | −0.80 | 69.57 | 0.29 | <0.0001 | 2.78 | 38.65 | −53.97 | −15.32 | 70.12 | 0.367 | 0.22 |
| Gr k | Gr l | dMe | SD2 | p2 | d2 | Gr k | Gr l | dMe | SD2 | p2 | d2 |
| 081 | 082 | −20.1 | 19.8 | 0.002 | 1.01 | 109n | 109s | 6399 | 1908 | <0.0001 | 3.35 |
| 091 | 092 | −9.42 | 0.83 | <0.0001 | 11.4 | 10yn | 10ys | 2 × 104 | 7343 | 0.0008 | 2.72 |
| 093 | 094 | −7.74 | 4.14 | <0.0001 | 1.87 | 10wn | 10ws | −489 | 633 | 0.173 | 0.77 |
| 101n | 101s | 7 × 104 | 1 × 105 | 0.276 | 0.66 | 211 | 212 | 0.14 | 0.55 | 0.559 | 0.26 |
| 107n | 107s | −3 × 105 | 7 × 104 | <0.0001 | 4.65 | 221 | 222 | −0.21 | 0.56 | 0.198 | 0.37 |
| 108n | 108s | −3 × 106 | 4 × 105 | <0.0001 | 8.23 | 229 | 22x | 2.52 | 63.8 | 0.903 | 0.04 |
| DYN Name | Contents and Sources of Dynamics (DYN) |
|---|---|
| 102 | Dynamics of the IgM titer specific to ECD from non-severe (102n, n = 11) and severe (102s, n = 5) patients between T3 and T4 in Figure 6 (anti-ECD IgM) by Chen et al. [26] |
| 103 | Dynamics of the IgM titer specific to S1 from non-severe (103n, n = 11) and severe (103s, n = 5) patients between T3 and T4 in Figure 6 (anti-S1 IgM) by Chen et al. [26] |
| 104 | Dynamics of the IgM titer specific to NP from non-severe (104n, n = 11) and severe (104s, n = 5) patients between T3 and T4 in Figure 6 (anti-NP IgM) by Chen et al. [26] |
| 105 | Dynamics of the IgG titer specific to RBD from non-severe (105n, n = 11) and severe (105s, n = 5) patients between T3 and T4 in Figure 6 (anti-RBD IgG) by Chen et al. [26] |
| 106 | Dynamics of the IgG titer specific to ECD from non-severe (106n, n = 11) and severe (106s, n = 5) patients between T3 and T4 in Figure 6 (anti-ECD IgG) by Chen et al. [26] |
| 10x | Dynamics of the IgA titer specific to ECD from non-severe (10xn, n = 9) and severe (10xs, n = 5) patients between T3 and T4 in Figure 6 (anti-ECD IgA) by Chen et al. [26] |
| 10z | Dynamics of the IgA titer specific to NP from non-severe (10zn, n = 11) and severe (10zs, n = 5) patients between T3 and T4 in Figure 6 (anti-NP IgA) by Chen et al. [26] |
| 213 214 | Dynamics of IgG levels specific to RBD (titers) from non-recurrent-positive (213, n = 32) and recurrent-positive (214, n = 6) patients between T5 and T6 described in Figure 4 (IgG levels) by Yang et al. [29] |
| 215 216 | Dynamics of IgA levels specific to RBD (titers) from non-recurrent-positive (215, n = 32) and recurrent-positive (216, n = 5) patients between T5 and T6 described in Figure 4 (IgA levels) by Yang et al. [29] |
| 217 218 | Dynamics of all Ig levels specific to RBD (titers) from non-recurrent-positive (217, n = 33) and recurrent-positive (218, n = 6) patients between T5 and T6 described in Figure 4 (all Ig levels) by Yang et al. [29] |
| 219 21x | Dynamics of Nab levels specific to RBD (titers) from non-recurrent-positive (217, n = 33) and recurrent-positive (218, n = 6) patients between T5 and T6 described in Figure 4 (Nab levels) by Yang et al. [29] |
| 223 224 | Dynamics of IgG levels specific to RBD (titers) from recurrent-positive patients between T7 and T8 (223, n = 27) and between T8 and T9 (224, n = 23) described in Figure 4 (IgG levels) by Yang et al. [29] |
| 225 226 | Dynamics of IgA levels specific to RBD (titers) from recurrent-positive patients between T7 and T8 (225, n = 21) and between T8 and T9 (226, n = 20) described in Figure 4 (IgA levels) by Yang et al. [29] |
| 227 228 | Dynamics of all Ig levels specific to RBD (titers) from recurrent-positive patients between T7 and T8 (227, n = 27) and between T8 and T9 (228, n = 25) described in Figure 4 (all Ig levels) by Yang et al. [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishida, T.; Takagi, K.; Wang, G.; Tanahashi, N.; Kawanokuchi, J.; Takagi, H.; Guo, Y.; Ma, N. A Greater Increase in Complement C5a Receptor 1 Level at Onset and a Smaller Decrease in Immunoglobulin G Level after Recovery in Severer Coronavirus Disease 2019 Patients: A New Analysis of Existing Data with a New Two-Tailed t-Test. Biology 2023, 12, 1176. https://doi.org/10.3390/biology12091176
Ishida T, Takagi K, Wang G, Tanahashi N, Kawanokuchi J, Takagi H, Guo Y, Ma N. A Greater Increase in Complement C5a Receptor 1 Level at Onset and a Smaller Decrease in Immunoglobulin G Level after Recovery in Severer Coronavirus Disease 2019 Patients: A New Analysis of Existing Data with a New Two-Tailed t-Test. Biology. 2023; 12(9):1176. https://doi.org/10.3390/biology12091176
Chicago/Turabian StyleIshida, Torao, Ken Takagi, Guifeng Wang, Nobuyuki Tanahashi, Jun Kawanokuchi, Hisayo Takagi, Yi Guo, and Ning Ma. 2023. "A Greater Increase in Complement C5a Receptor 1 Level at Onset and a Smaller Decrease in Immunoglobulin G Level after Recovery in Severer Coronavirus Disease 2019 Patients: A New Analysis of Existing Data with a New Two-Tailed t-Test" Biology 12, no. 9: 1176. https://doi.org/10.3390/biology12091176
APA StyleIshida, T., Takagi, K., Wang, G., Tanahashi, N., Kawanokuchi, J., Takagi, H., Guo, Y., & Ma, N. (2023). A Greater Increase in Complement C5a Receptor 1 Level at Onset and a Smaller Decrease in Immunoglobulin G Level after Recovery in Severer Coronavirus Disease 2019 Patients: A New Analysis of Existing Data with a New Two-Tailed t-Test. Biology, 12(9), 1176. https://doi.org/10.3390/biology12091176










