Characterising the Gene Expression, Enzymatic Activity and Subcellular Localisation of Arabidopsis thaliana Metacaspase 5 (AtMCA-IIb)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Method
2.1. Plant Growth Condition
2.2. β-Glucuronidase (GUS) Histochemical Staining
2.3. Sytox Green Staining
2.4. DNA Sequencing
2.5. Point Mutation
2.6. Total RNA Extraction and cDNA Synthesis
2.7. Quantitative Reverse Transcription PCR (qRT-PCR)
2.8. Biolistic Particle Bombardment
2.9. Plasmid Construction of AtMC5::RFP
2.10. Purification of His-Tagged Recombinant Protein
2.11. Protein Concentration
2.12. SDS-PAGE
2.13. Western Blot
2.14. Optimisation of AtMC5 Enzymatic Assay
2.15. Fluorescence Imaging
3. Results
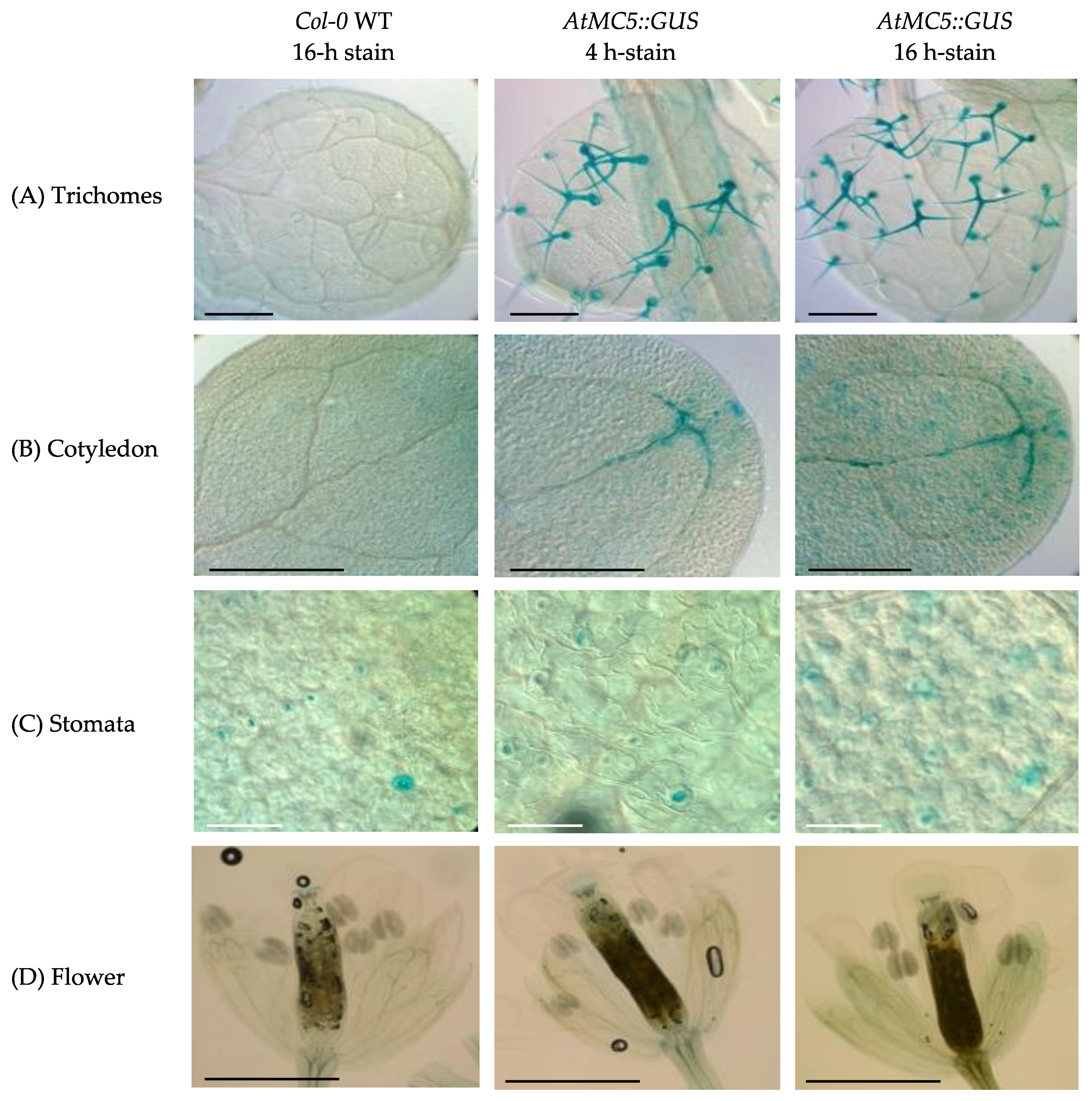
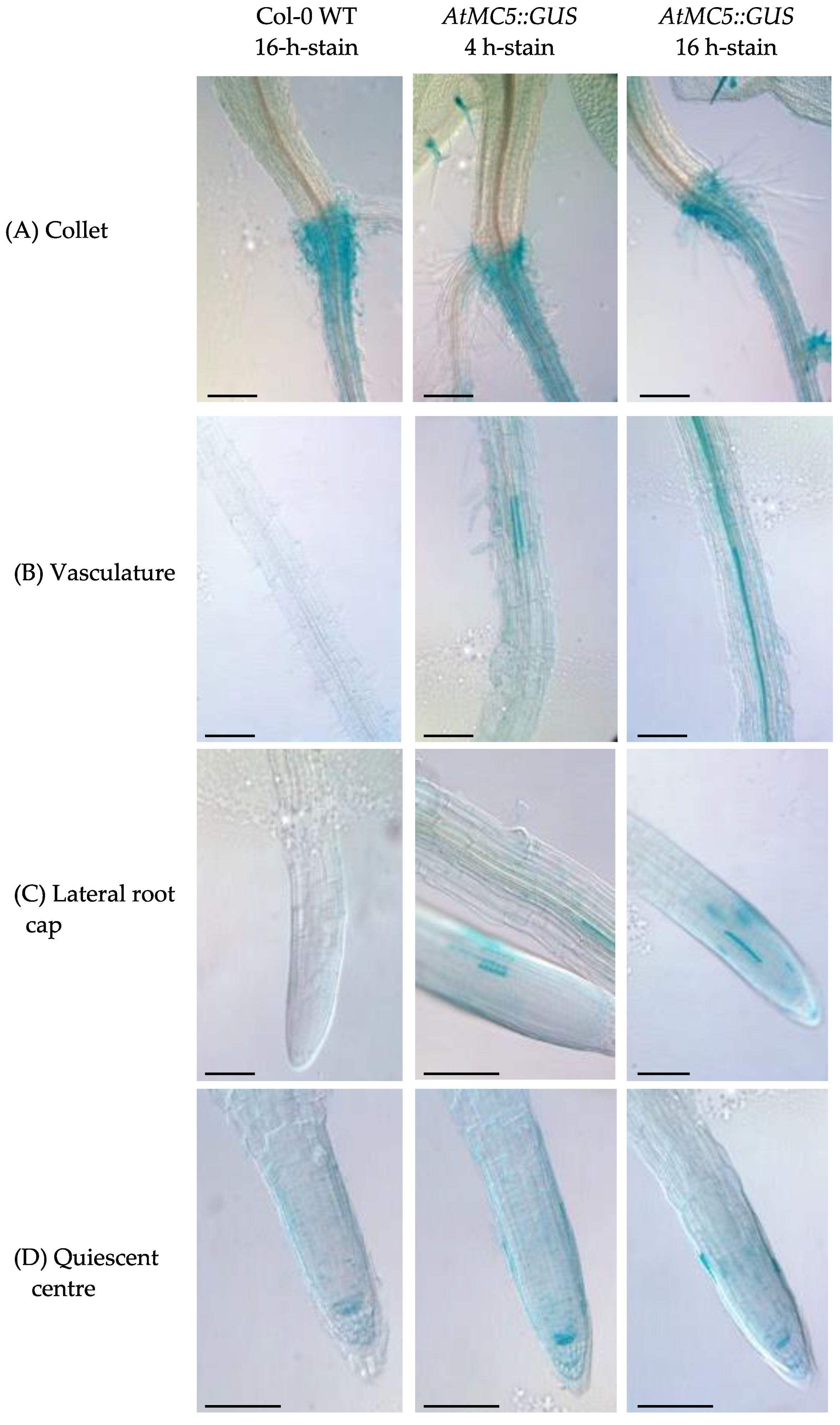
3.1. Expression of AtMC5::GUS in A. thaliana Tissues
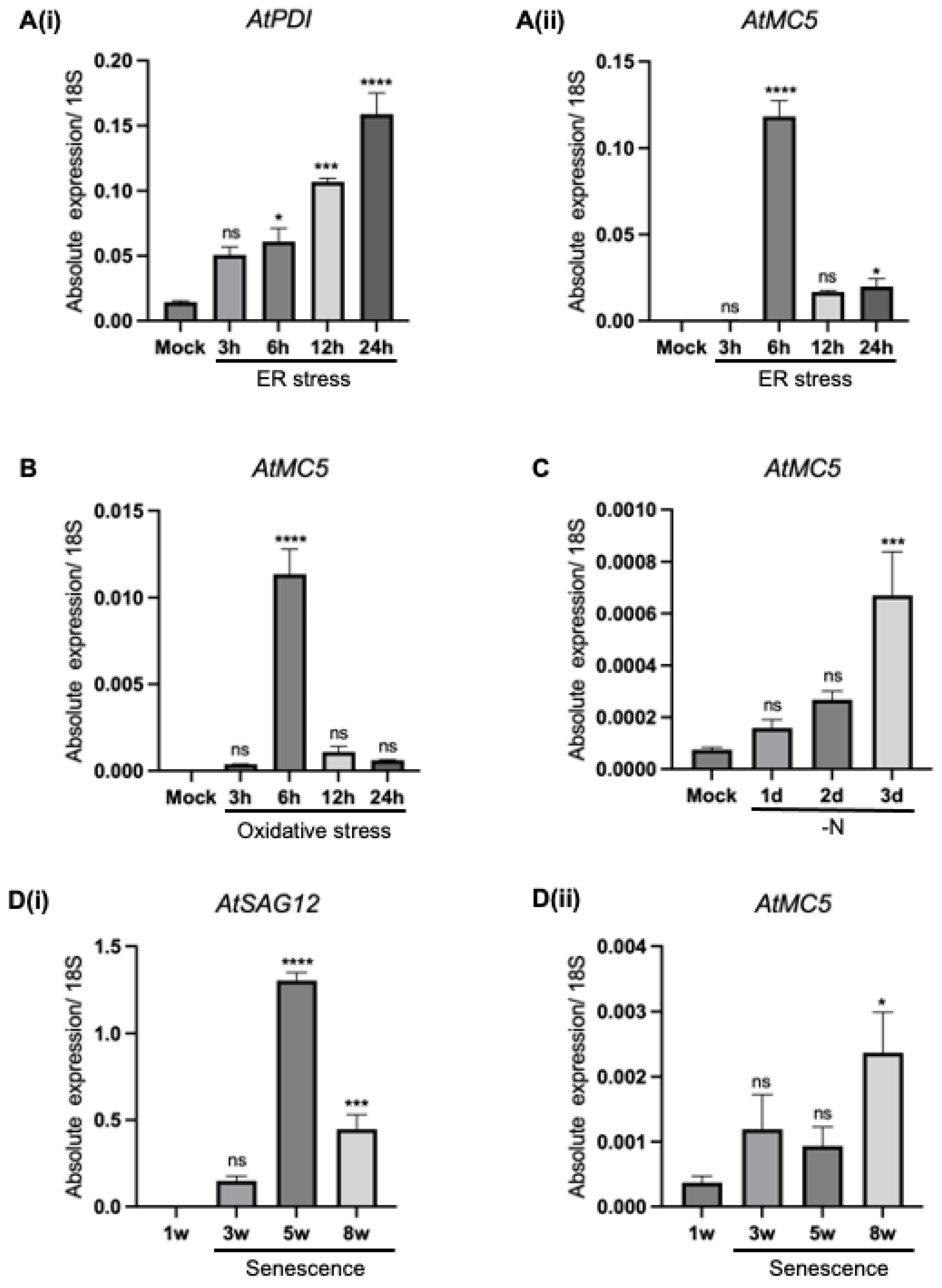
3.2. AtMC5 Responses in Various Stress Inductions
3.2.1. Endoplasmic Reticulum Stress
3.2.2. Oxidative Stress
3.2.3. Nitrogen Starvation
3.2.4. Leaf Senescence
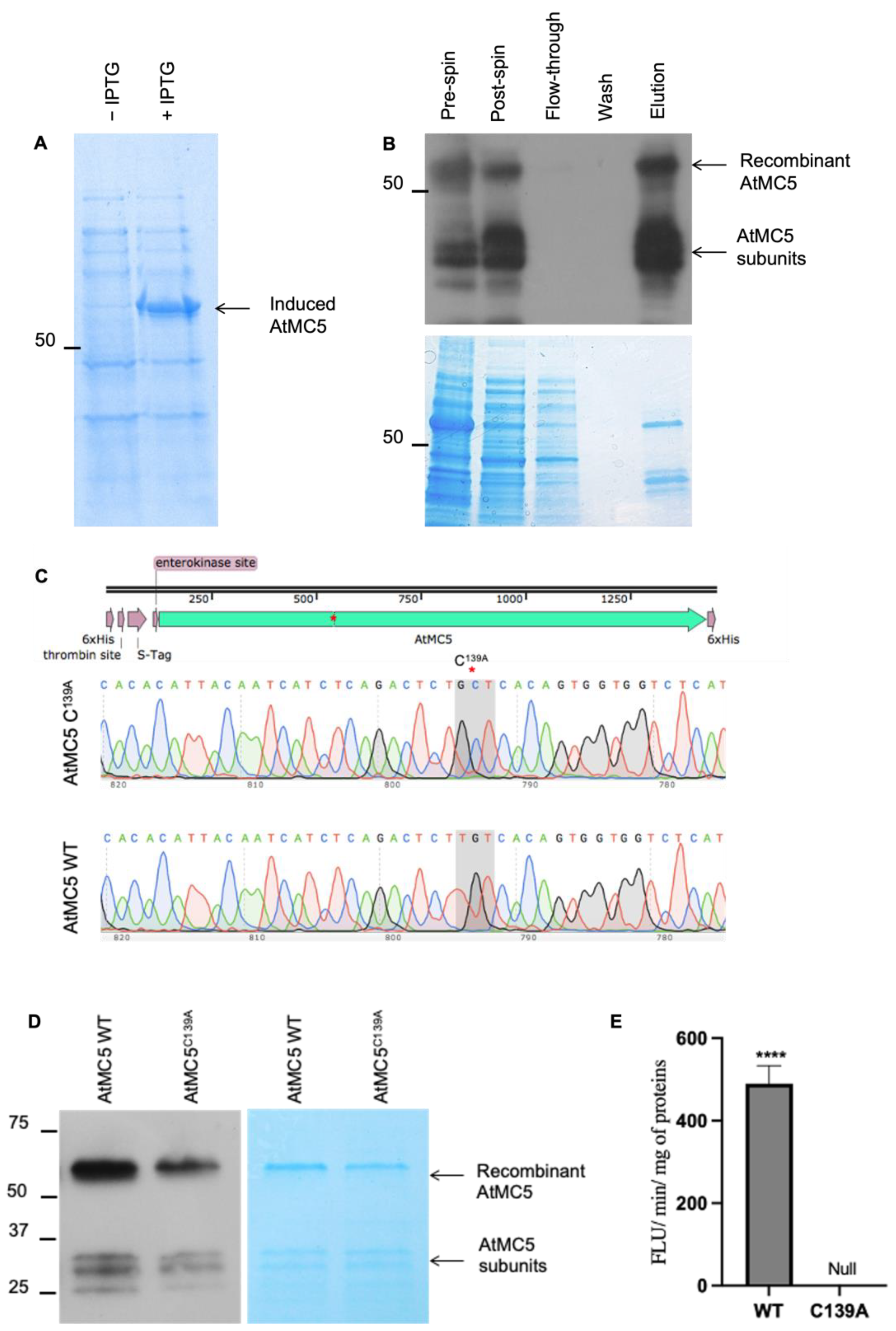
3.3. Purification of Recombinant AtMC5 Expressed in E. coli
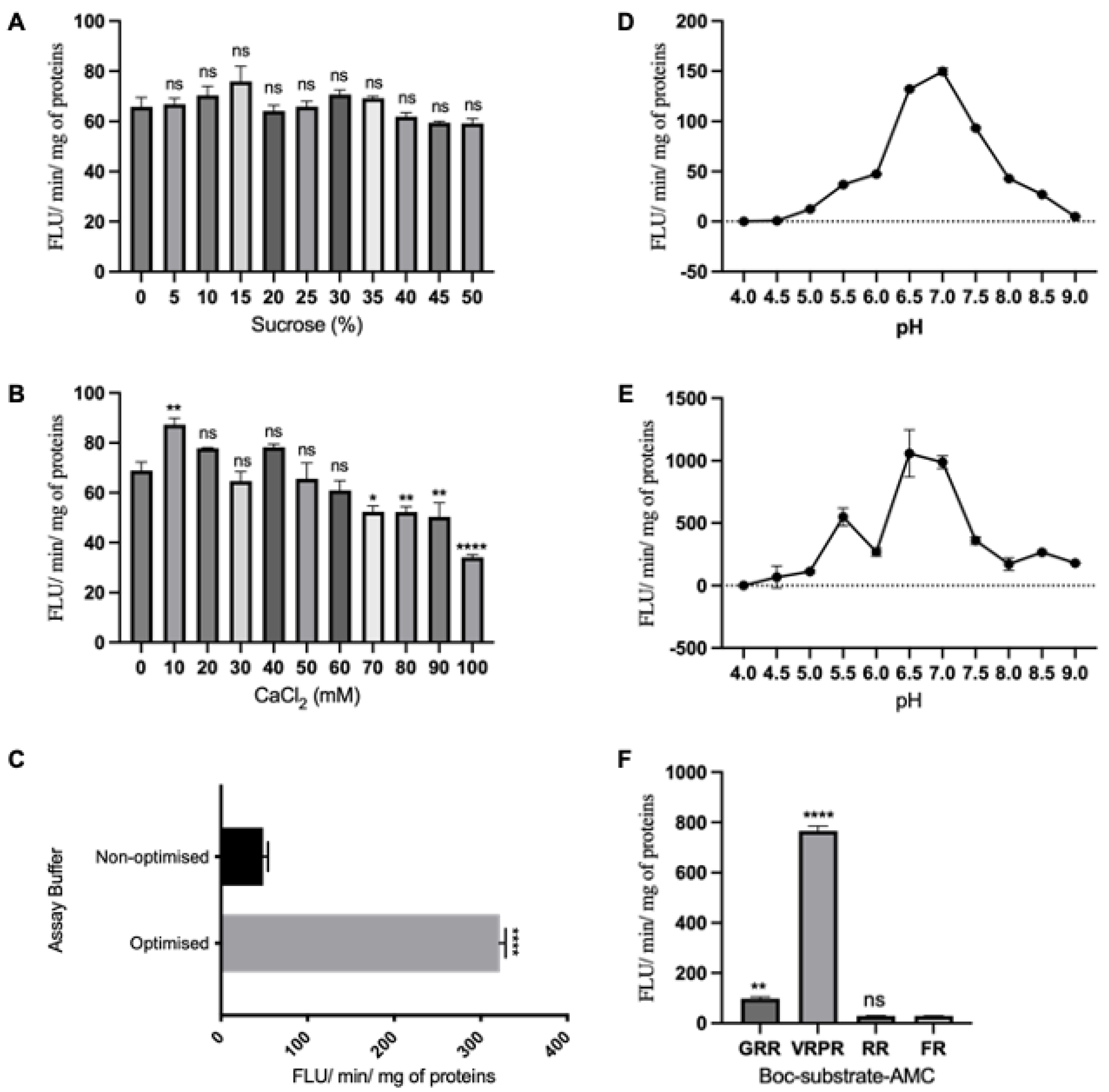
3.4. Enzymatic Activities of AtMC5
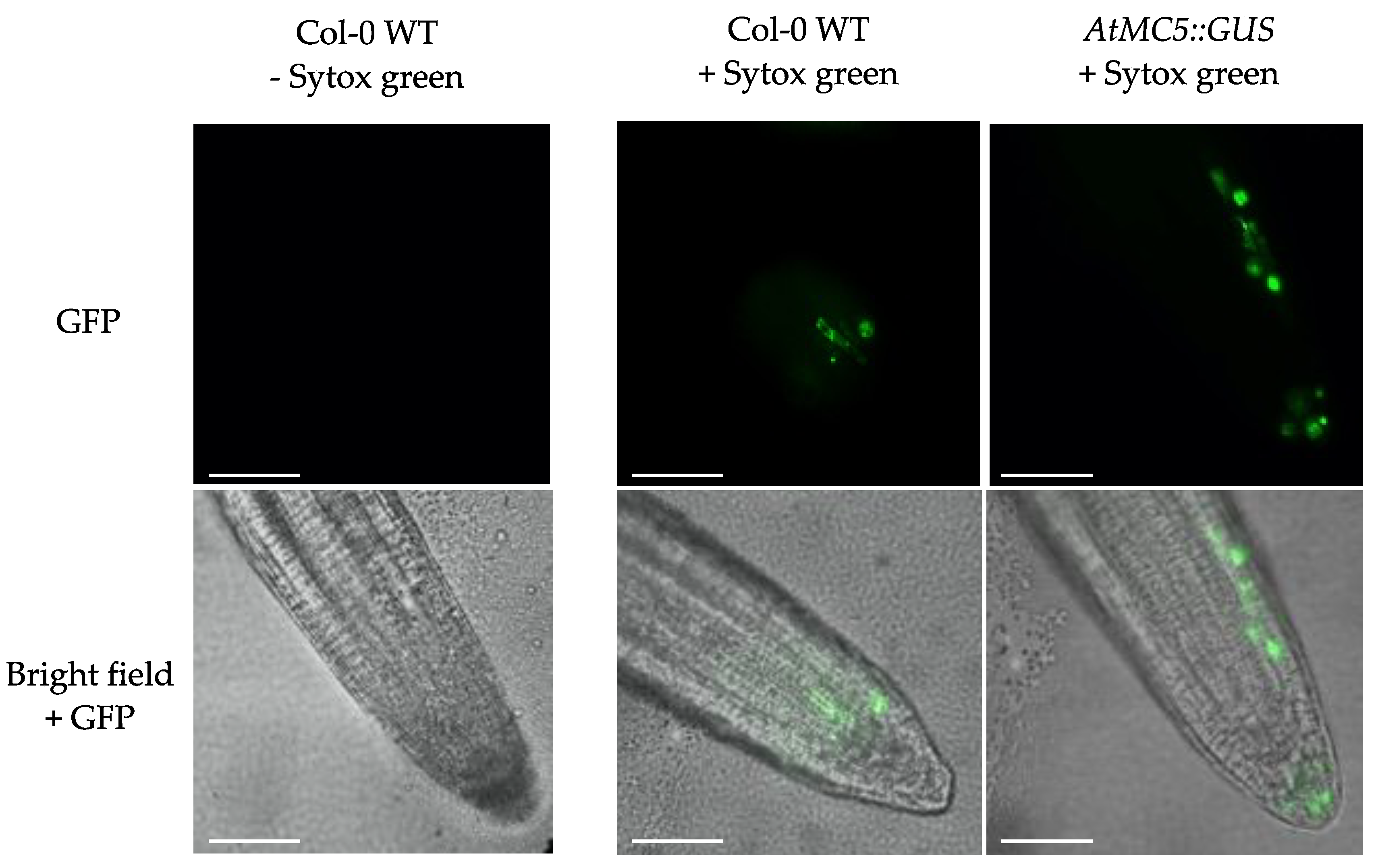
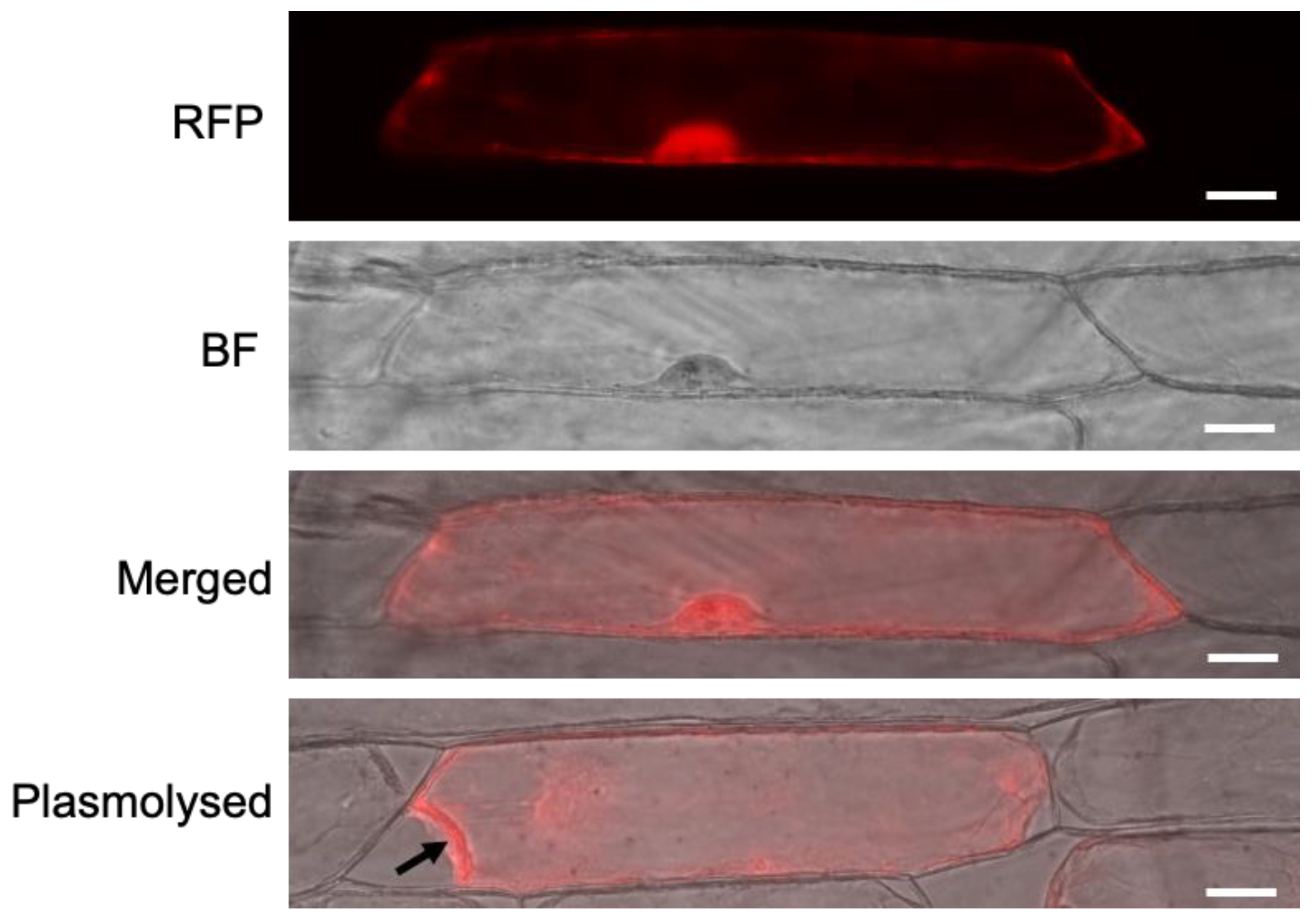
3.5. Subcellular Localisation of AtMC5-RFP
4. Discussion
4.1. AtMC5 Is Only Expressed in Specific Cells of A. thaliana without a Strong PCD Association
4.2. AtMC5 Expression Is Inducible by ER Stress and Oxidative Stress
4.3. AtMC5 Enzymatic Activity Is Enhanced by Sucrose and Calcium
4.4. AtMC5-RFP Is Localised in the Cytosol and Nucleus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vercammen, D.; van de Cotte, B.; De Jaeger, G.; Eeckhout, D.; Casteels, P.; Vandepoele, K.; Vandenberghe, I.; Van Beeumen, J.; Inzé, D.; Van Breusegem, F. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J. Biol. Chem. 2004, 279, 45329–45336. [Google Scholar] [CrossRef]
- Bollhöner, B.; Zhang, B.; Stael, S.; Denancé, N.; Overmyer, K.; Goffner, D.; Van Breusegem, F.; Tuominen, H. Post mortem function of AtMC9 in xylem vessel elements. New Phytol. 2013, 200, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Stael, S.; Sabljić, I.; Audenaert, D.; Andersson, T.; Tsiatsiani, L.; Kumpf, R.P.; Vidal-Albalat, A.; Lindgren, C.; Vercammen, D.; Jacques, S.; et al. Structure-function study of a Ca(2+)-independent metacaspase involved in lateral root emergence. Proc. Natl. Acad. Sci. USA 2023, 120, e2303480120. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; Van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Lam, E. Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 2011, 66, 969–982. [Google Scholar] [CrossRef]
- He, R.; Drury, G.E.; Rotari, V.I.; Gordon, A.; Willer, M.; Farzaneh, T.; Woltering, E.J.; Gallois, P. Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J. Biol. Chem. 2008, 283, 774–783. [Google Scholar] [CrossRef]
- Coll, N.S.; Smidler, A.; Puigvert, M.; Popa, C.; Valls, M.; Dangl, J.L. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: Functional linkage with autophagy. Cell Death Differ. 2014, 21, 1399–1408. [Google Scholar] [CrossRef]
- Sundström, J.F.; Vaculova, A.; Smertenko, A.P.; Savenkov, E.I.; Golovko, A.; Minina, E.; Tiwari, B.S.; Rodriguez-Nieto, S.; Zamyatnin, A.A.; Välineva, T.; et al. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat. Cell Biol. 2009, 11, 1347–1354. [Google Scholar] [CrossRef] [PubMed]
- Wrzaczek, M.; Vainonen, J.P.; Stael, S.; Tsiatsiani, L.; Help-Rinta-Rahko, H.; Gauthier, A.; Kaufholdt, D.; Bollhöner, B.; Lamminmäki, A.; Staes, A.; et al. GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 2015, 34, 55–66. [Google Scholar] [CrossRef]
- Hander, T.; Fernández-Fernández, D.; Kumpf, R.P.; Willems, P.; Schatowitz, H.; Rombaut, D.; Staes, A.; Nolf, J.; Pottie, R.; Yao, P.; et al. Damage on plants activates Ca2+ -dependent metacaspases for release of immunomodulatory peptides. Science 2019, 363, aar7486. [Google Scholar] [CrossRef]
- Shen, W.; Liu, J.; Li, J.-F. Type-II Metacaspases Mediate the Processing of Plant Elicitor Peptides in Arabidopsis. Mol. Plant. 2019, 12, 1524–1533. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Cui, Y.; Hou, X.; Huang, T. The AtMC4 regulates the stem cell homeostasis in Arabidopsis by catalyzing the cleavage of AtLa1 protein in response to environmental hazards. Plant Sci. 2018, 266, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-T.; Wang, M.-J.; Sun, L.; Lu, S.-J.; Bi, D.-L.; Sun, L.; Song, Z.-T.; Zhang, S.-S.; Zhou, S.-F.; Liu, J.-X. The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants. PLoS Genet. 2014, 10, e1004243. [Google Scholar] [CrossRef]
- Wilkinson, B.; Gilbert, H.F. Protein disulfide isomerase. Biochim. Biophys. Acta. 2004, 1699, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, B.C.; Oelmüller, R. Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 2012, 7, 1621–1633. [Google Scholar] [CrossRef]
- Martin, T.; Oswald, O.; Graham, I.A. Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 2002, 128, 472–481. [Google Scholar] [CrossRef]
- Thomas, H. Senescence, ageing and death of the whole plant. N. Phytol. 2013, 197, 696–711. [Google Scholar] [CrossRef]
- Hamann, A.; Brust, D.; Osiewacz, H.D. Deletion of putative apoptosis factors leads to lifespan extension in the fungal ageing model Podospora anserina. Mol. Microbiol. 2007, 65, 948–958. [Google Scholar] [CrossRef]
- Kwon SIl Hwang, D.J. Expression analysis of the metacaspase gene family in Arabidopsis. J. Plant Biol. 2013, 56, 391–398. [Google Scholar] [CrossRef]
- Lee, J.C.; Timasheff, S.N. The stabilization of proteins by sucrose. J. Biol. Chem. 1981, 256, 7193–7201. [Google Scholar] [CrossRef]
- Watanabe, N.; Lam, E. Calcium-dependent activation and autolysis of Arabidopsis metacaspase 2d. J. Biol. Chem. 2011, 286, 10027–10040. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; Timmerman, E.; De Bock, P.-J.; Vercammen, D.; Stael, S.; van de Cotte, B.; Staes, A.; Goethals, M.; Beunens, T.; Van Damme, P.; et al. The Arabidopsis metacaspase9 degradome. Plant Cell. 2013, 25, 2831–2847. [Google Scholar] [CrossRef] [PubMed]
- Escamez, S.; Tuominen, H. Programmes of cell death and autolysis in tracheary elements: When a suicidal cell arranges its own corpse removal. J. Exp. Bot. 2014, 65, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Huysmans, M.; Buono, R.A.; Skorzinski, N.; Radio, M.C.; De Winter, F.; Parizot, B.; Mertens, J.; Karimi, M.; Fendrych, M.; Nowack, M.K. NAC Transcription Factors ANAC087 and ANAC046 Control Distinct Aspects of Programmed Cell Death in the Arabidopsis Columella and Lateral Root Cap. Plant Cell. 2018, 30, 2197–2213. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.; Phillips, A.R.; Vierstra, R.D. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 2010, 62, 483–493. [Google Scholar] [CrossRef]
- Herker, E.; Jungwirth, H.; Lehmann, K.A.; Maldener, C.; Frohlich, K.-U.; Wissing, S.; Büttner, S.; Fehr, M.; Sigrist, S.; Madeo, F. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 2004, 164, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Moss, C.X.; Westrop, G.D.; Juliano, L.; Coombs, G.H.; Mottram, J.C. Metacaspase 2 of Trypanosoma brucei is a calcium-dependent cysteine peptidase active without processing. FEBS Lett. 2007, 581, 5635–5639. [Google Scholar] [CrossRef] [PubMed]
- Thibaud, M.C.; Gineste, S.; Nussaume, L.; Robaglia, C. Sucrose increases pathogenesis-related PR-2 gene expression in Arabidopsis thaliana through an SA-dependent but NPR1-independent signaling pathway. Plant Physiol. Biochem. 2004, 42, 81–88. [Google Scholar] [CrossRef]
- Moghaddam, M.R.B.; Van Den Ende, W. Sugars and plant innate immunity. J. Exp. Bot. 2012, 63, 3989–3998. [Google Scholar] [CrossRef]
- Belenghi, B.; Romero-Puertas, M.C.; Vercammen, D.; Brackenier, A.; Inzé, D.; Delledonne, M.; Van Breusegem, F. Metacaspase Activity of Arabidopsis thaliana Is Regulated by S-Nitrosylation of a Critical Cysteine Residue. J Biol Chem. 2007, 282, 1352–1358. [Google Scholar] [CrossRef]

| Primer | Sequences (5′-3′) |
|---|---|
| Bip2_F | TCAGCACCAAGTCCGTGTAG |
| Bip2_R | CTTCACAGGTCCCATGGTCT |
| PDI_F | TGAGAAATGGAGGGAAGTCG |
| PDI_R | CAACAACCTCAGTGGCAGAA |
| 18S_29F | GGTCTGTGATGCCCTTAGATGTT |
| 18S_102R | GGCAAGGTGTGAACTCGTTGA |
| MC5_580F | ATCTAAAGGGATCGCCATTCC |
| MC5_640R | AGCTTAAACTAGAAGATGGGGCAA |
| MC5_811F | AAGCTGCAAGAAGGTAAAACTGAAG |
| MC5_904R | TTAAGCATAAACTAAACGATGACGAAG |
| MC5v3_F | CTAACAAAGCTGCAAGAAGG |
| MCFv3_R | GGGAATGATTGGGAAACTAG |
| ATG5_F | TCTCAACAAGTTGTGCCTGAG |
| ATG5_R | GTACGAGATGTCATCCCAGGT |
| GPX8_F | GATGAAGCTTTGTTGCTGAATCG |
| GPX8_R | TCCAAGTCAATGTCATGGCTCTT |
| SAG12_F | CAGCTGCGGATGTTGTTG |
| SAG12_R | CCACTTTCTCCCCATTTTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobri, Z.M.; Gallois, P. Characterising the Gene Expression, Enzymatic Activity and Subcellular Localisation of Arabidopsis thaliana Metacaspase 5 (AtMCA-IIb). Biology 2023, 12, 1155. https://doi.org/10.3390/biology12091155
Sobri ZM, Gallois P. Characterising the Gene Expression, Enzymatic Activity and Subcellular Localisation of Arabidopsis thaliana Metacaspase 5 (AtMCA-IIb). Biology. 2023; 12(9):1155. https://doi.org/10.3390/biology12091155
Chicago/Turabian StyleSobri, Zulfazli M., and Patrick Gallois. 2023. "Characterising the Gene Expression, Enzymatic Activity and Subcellular Localisation of Arabidopsis thaliana Metacaspase 5 (AtMCA-IIb)" Biology 12, no. 9: 1155. https://doi.org/10.3390/biology12091155
APA StyleSobri, Z. M., & Gallois, P. (2023). Characterising the Gene Expression, Enzymatic Activity and Subcellular Localisation of Arabidopsis thaliana Metacaspase 5 (AtMCA-IIb). Biology, 12(9), 1155. https://doi.org/10.3390/biology12091155






