Influence of Metabolic, Transporter, and Pathogenic Genes on Pharmacogenetics and DNA Methylation in Neurological Disorders
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Sample Collection and Analysis
2.3. DNA Extraction
2.4. Quantification of Global DNA Methylation (5mC)
2.5. Genotyping
2.6. Statistical Analysis
3. Results
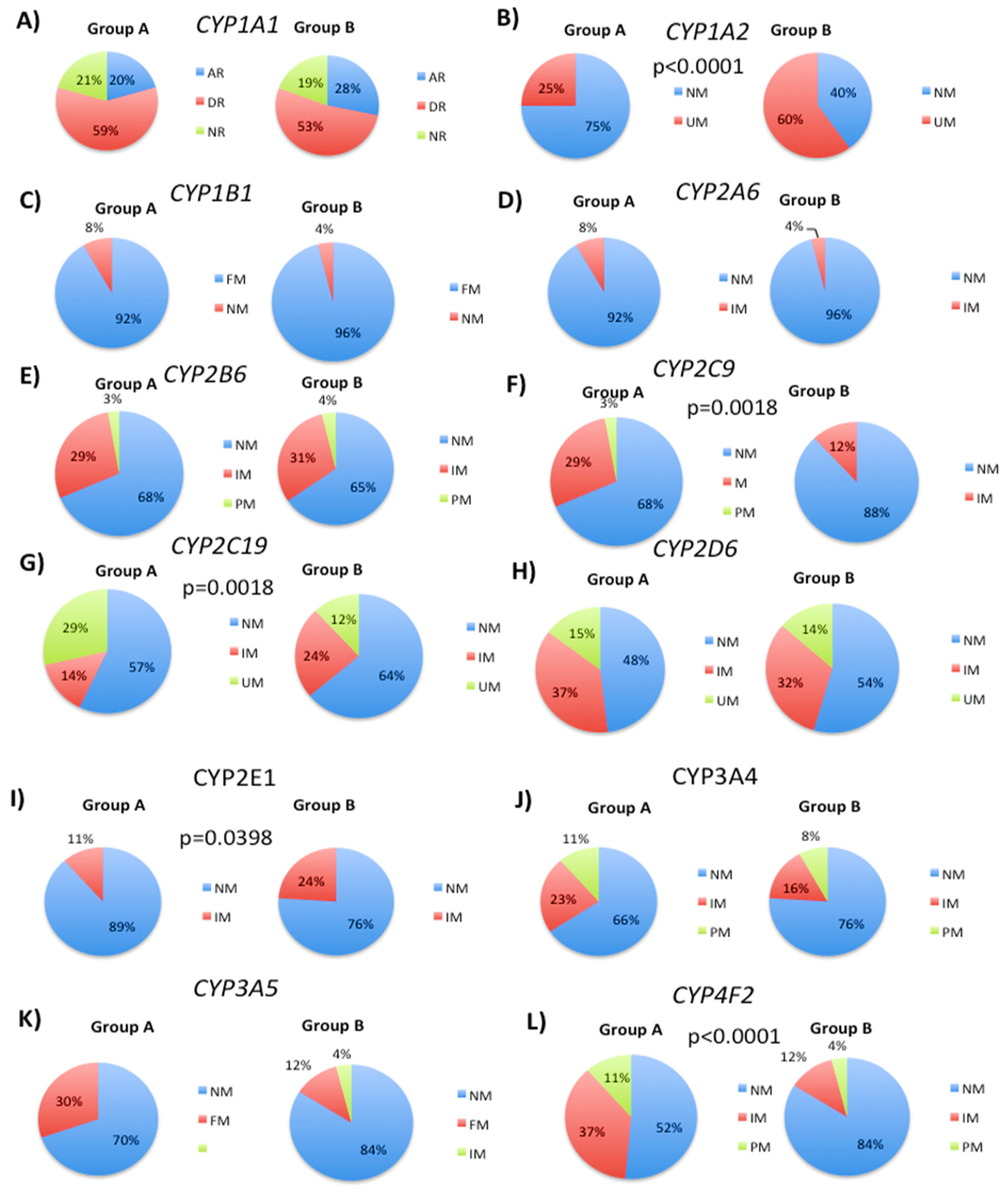
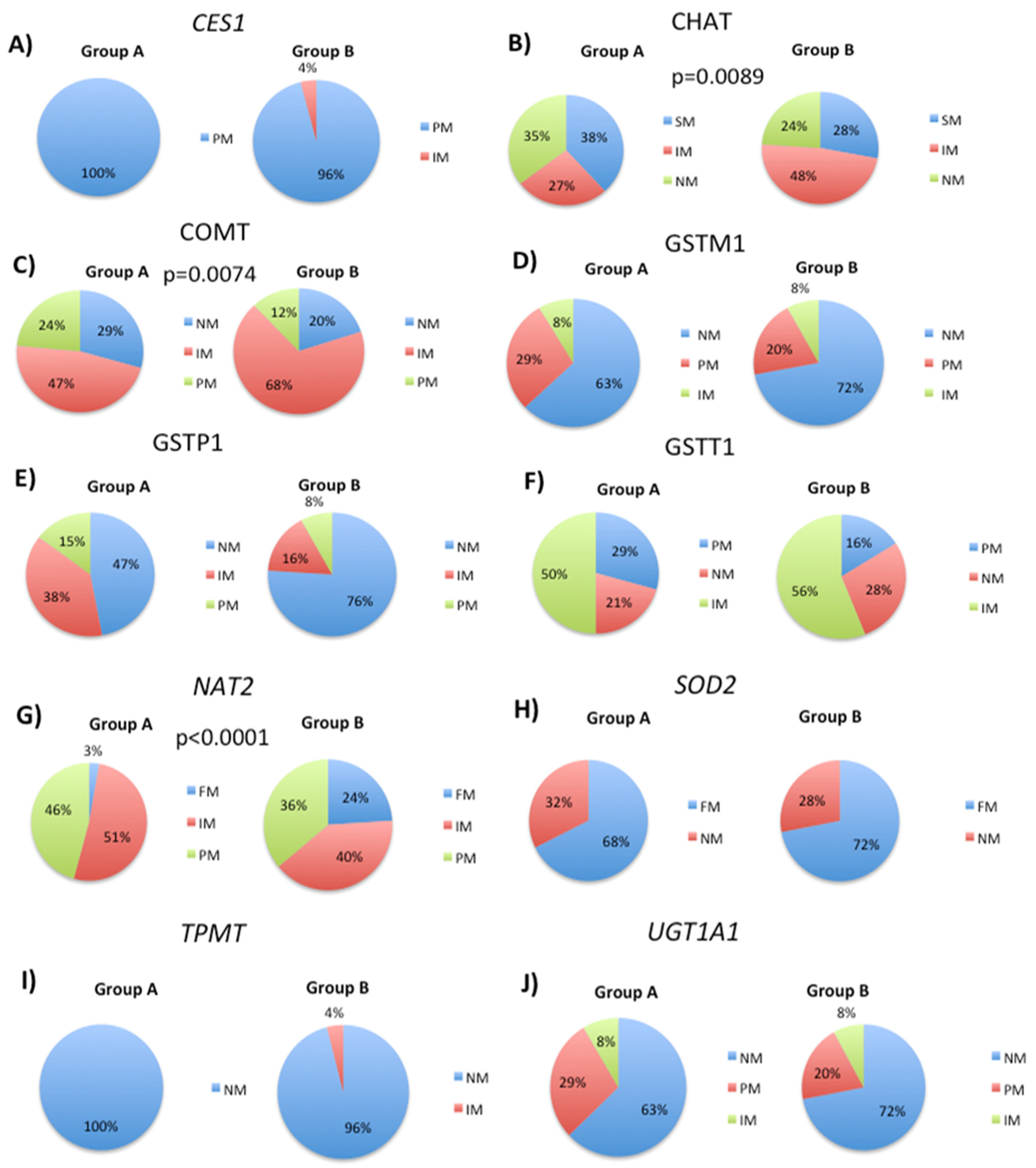
3.1. Genetic Variability in Drug Metabolizing Enzymes Influences 5mC Levels
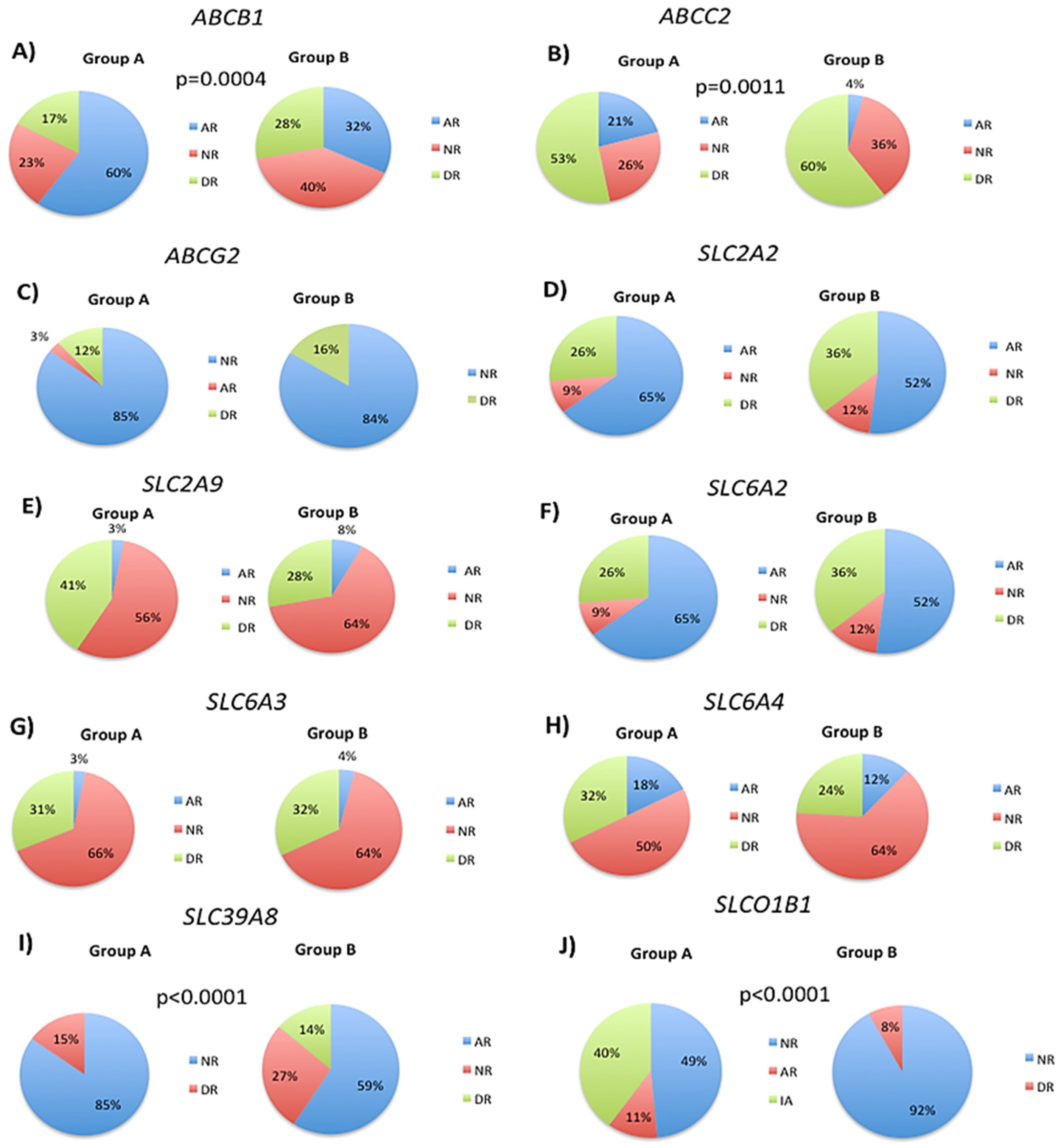
3.2. Modification of DNA Methylation Levels by Transport Gene Phenotypes
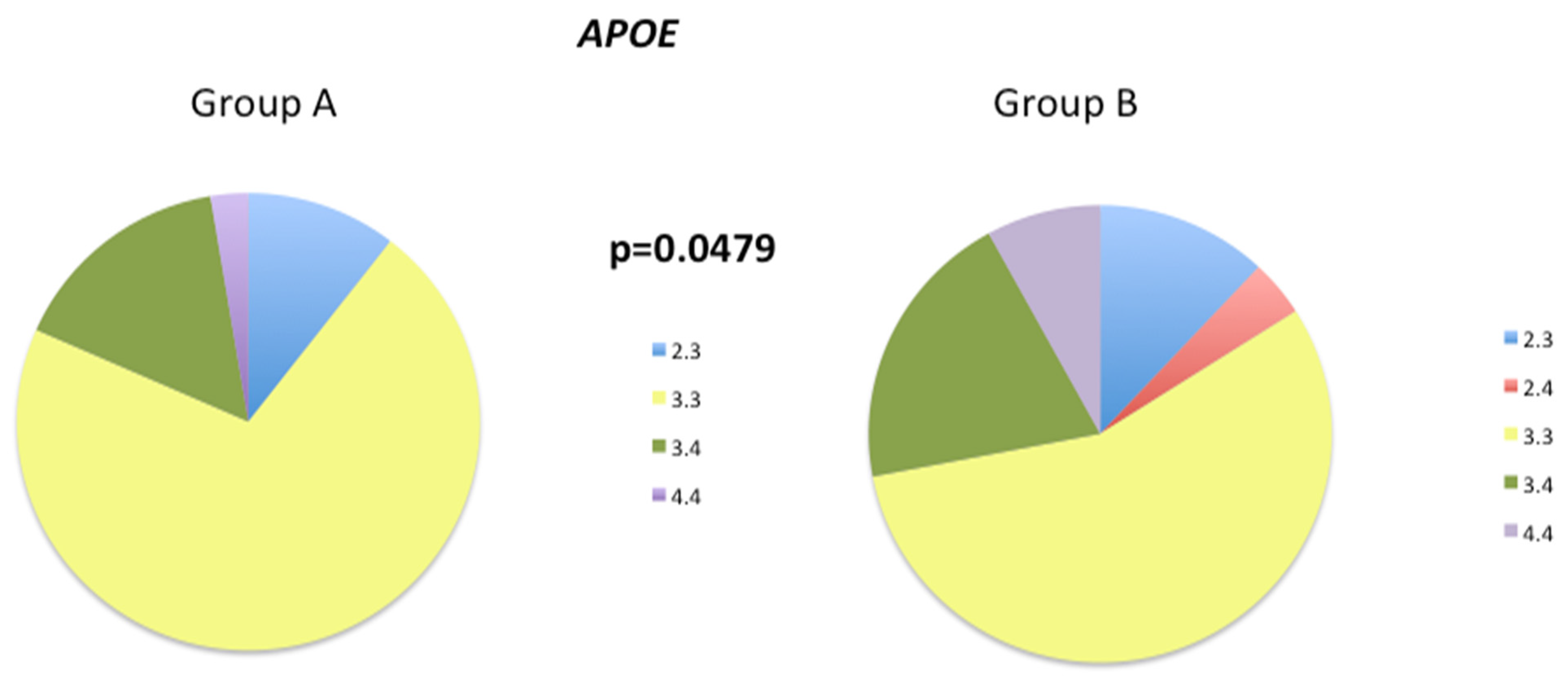
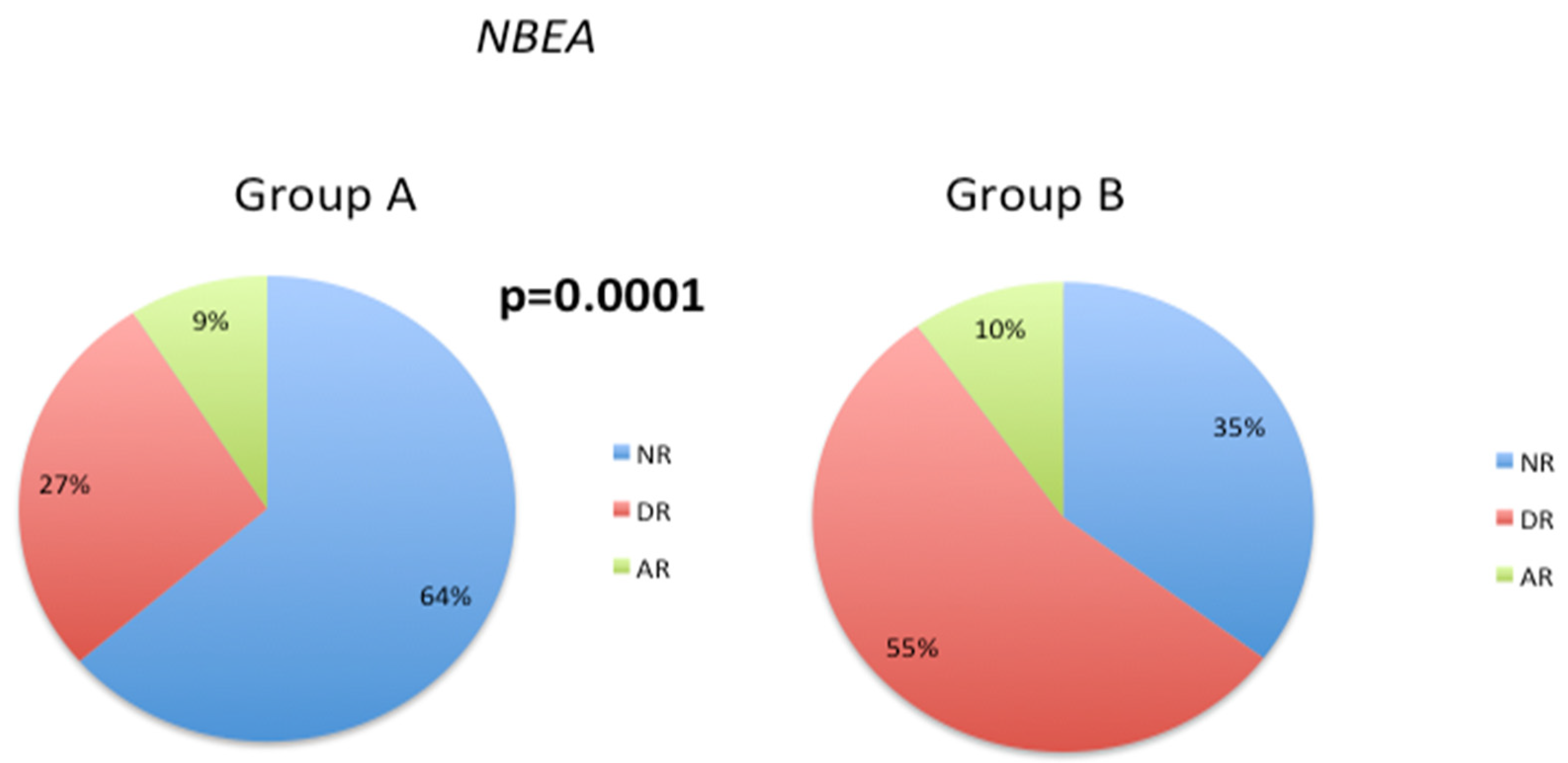
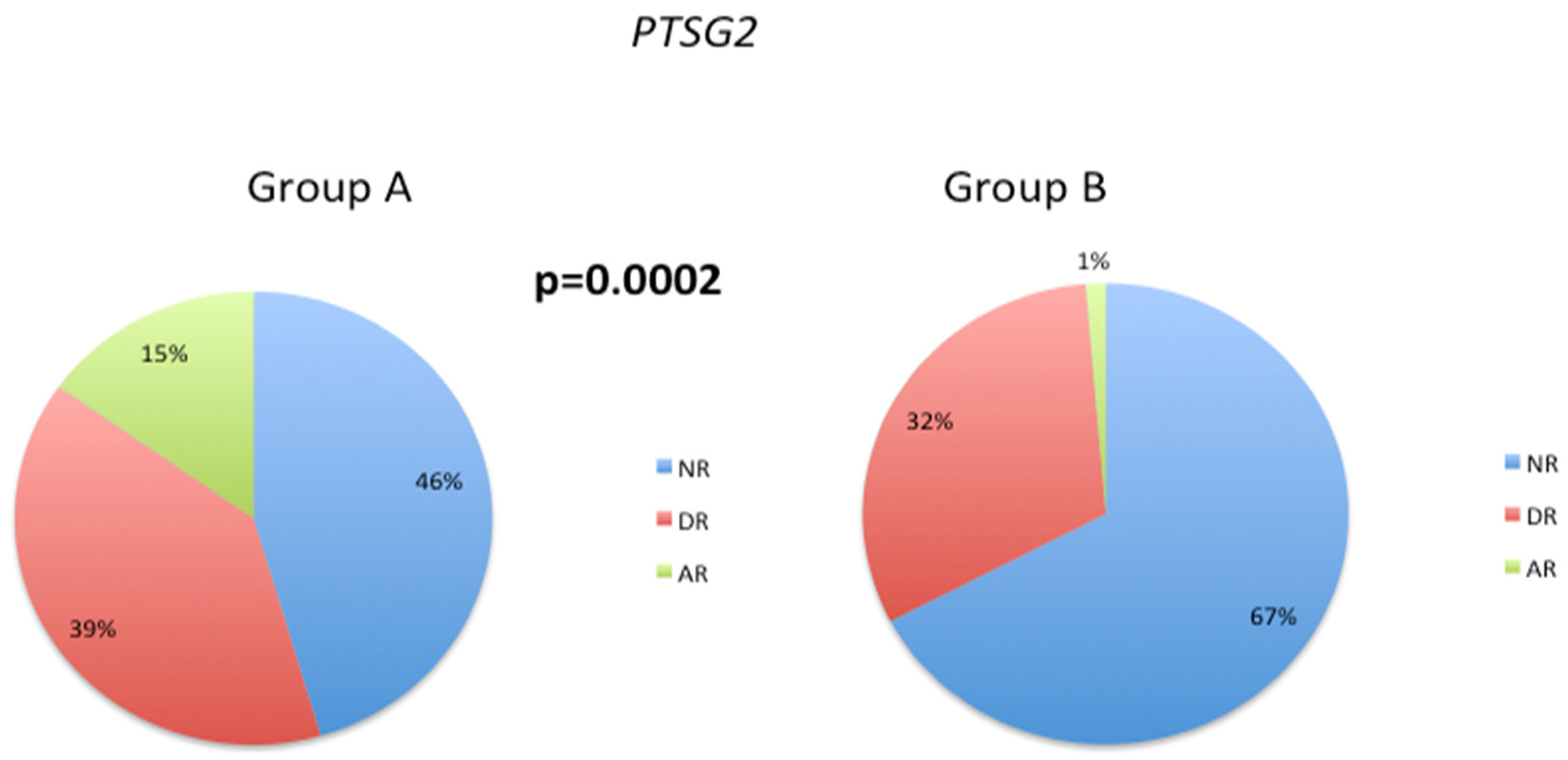
3.3. Pathological Gene Phenotypes Alter DNA Methylation Levels
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdur, R.; Mominur, R. Potential therapeutics against neurological disorders: Natural products-based drugs. Front. Pharmacol. 2022, 13, 950457. [Google Scholar]
- GBD 2019 Mental Disorders Collaborators. Global, Regional, and Netional burden of 12 Mental Disorders in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study. Lancet Psychiatry 2022, 9, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Sander, J.W. The global burden of epilepsy report: Implications for low- and middle-income countries. Epilepsy Behav. 2020, 105, 106949. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Naidoo, V.; Martínez-Iglesias, O.; Corzo, L.; Cacabelos, N.; Pego, R.; Carril, J.C. Pharmacogenomics of Alzheimer’s Disease: Novel strategies for drug utilization and development. Pharmacogenom. Drug Discov. Dev. 2022, 2547, 275–387. [Google Scholar]
- Osanlou, O.; Pirmohamed, M.; Daly, A.K. Pharmacogenetics of adverse drug reactions. Adv. Pharmacol. 2018, 83, 155–190. [Google Scholar]
- Cacabelos, R.; Naidoo, V.; Corzo, L.; Cacbelos, N.; Carril, J.C. Genophenotypoc factors and Pharmacogenomics in Adverse Drug Reactions. Int. J. Mol. Sci. 2021, 22, 13302. [Google Scholar] [CrossRef] [PubMed]
- Eissenberg, J.C.; Aurora, R. Pharmacogenomics: What the Doctor ordered? Mo. Med. 2019, 116, 217–225. [Google Scholar]
- Becquemont, L. Pharmacogenomics of adverse drug reactions: Practical applications and prespectives. Pharmacogenomics 2009, 10, 961–969. [Google Scholar] [CrossRef]
- Cacabelos, R. Pharmacogenomi of drugs to treat brain disorders. Expert Rev. Prec. Med. Drug. Dev. 2020, 5, 181–234. [Google Scholar] [CrossRef]
- Hong, J.Y.; Kim, J.H. PG-path: Modeling and personalizing phramacogenomis-based pathways. PLoS ONE 2020, 15, e0230950. [Google Scholar] [CrossRef]
- Wilke, R.A.; Reif, D.M.; Moore, J.H. Combinatorial pharmacogenetics. Nat. Rev. Drug Discov. 2005, 4, 911–918. [Google Scholar] [CrossRef]
- Kozyra, M.; Ingelman-Sundberg, M.; Lauschke, V.M. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet. Med. 2017, 19, 20–29. [Google Scholar] [CrossRef]
- Cacabelos, R.; Cacabelos, N.; Carril, J.C. The role of pharmacogenomics in adverse drug reactions. Expert Rev. Clin. Pharmacol. 2019, 12, 407–442. [Google Scholar] [CrossRef]
- Desta, Z.; Flockhart, D. Pharmacogenetics of drug metabolism. Clin. Transl. Sci. 2017, 2017, 1563–1570. [Google Scholar]
- Lee, I.-S.; Kim, D. Polymorphic metabolism by functional alterations of human cytochrome P450 enzymes. Arch. Pharm. Res. 2011, 34, 1799–1816. [Google Scholar] [CrossRef] [PubMed]
- Sweatt, J.D. The emerging field of neuroepigenetics. Neuron 2013, 80, 624–632. [Google Scholar] [CrossRef]
- Maloney, B.; Lahiri, D. Epigenetics of dementia: Understanding the disease as a transformation rather than a state. Lancet Neurol. 2016, 15, 760–774. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. The essentials of DNA methylation. Cell 1992, 70, 5–8. [Google Scholar] [CrossRef]
- Hermann, A.; Gowher, H.; Jeltsch, A. Biochemistry and biology of mammal DNA methyltransferases. Cell Mol. Life Sci. 2004, 61, 2571–2587. [Google Scholar] [CrossRef]
- Gräff, J.; Mansuy, I.M. Epigenetic codes in cognition and behaviour. Behav. Brain Res. 2008, 192, 70–87. [Google Scholar] [CrossRef]
- Nan, X.; Campoy, F.J.; Bird, A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 1997, 88, 4471–4481. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G.; et al. Distribution recognition and regulation of non-CpG methylation in the adult mamamlian brain. Nat. Neurosci. 2013, 17, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Hermann, A.; Jeltsch, A. The Dnmt1 DNA-cytosine-c5-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004, 279, 48350–48359. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Xie, S.; Li, E. Cloning and characterization of a family of novel mammalian DNMA (cytosine-5) methyltransferases. Nat. Genet. 1998, 19, 219–220. [Google Scholar] [CrossRef]
- Martínez-Iglesias, O.; Carrera, I.; Carril, J.C.; Fernández-Novoa, L.; Cacabelos, N.; Cacabelos, R. DNA methylation in neurodegenrative and cerebrovascular disorders. Int. J. Mol. Sci. 2020, 21, 2220. [Google Scholar] [CrossRef]
- Martínez-Iglesias, O.; Naidoo, V.; Cacabelos, N.; Cacabelos, R. Epigenetic biomarkers as diagnostic tools for Neurodegenrative Disorders. Int. J. Mol. Sci. 2022, 23, 13. [Google Scholar] [CrossRef]
- Masliah, E.; Dumaop, W.; Galasko, D.; Desplats, P. Distinctive patterns of DNA methylation associated with Parkinson disease: Identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics 2013, 8, 1030–1038. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; Arosio, B.; Falconi, A.; Micioni Di Bonaventura, M.V.; Karimi, M.; Mari, D.; Casati, M.; Maccarrone, M.; D’Addario, C. Global changes in DNA methylation in Alzheimer’s Disease peripheral blood mononuclear cells. Brain. Behav. Immun. 2015, 45, 139–144. [Google Scholar] [CrossRef]
- Martínez-Iglesias, O.; Naidoo, V.; Corzo, L.; Pego, R.; Seoane, S.; Rodríguez, S.; Alcaraz, M.; Muñiz, A.; Cacabelos, N.; Cacabelos, R. DNA methylation as a biomarker for monitoring disease outcome in patients with hypovitaminosis and Neurological Disorders. Genes 2023, 14, 365. [Google Scholar] [CrossRef]
- Crettol, S.; Petrovic, N.; Murray, M. Pharmaciogenetics of pase I and phase II drug metabolism. Curr. Pharm. Des. 2010, 16, 204–219. [Google Scholar] [CrossRef]
- Murray, M. Role of CYO pharmacogenetics and drug-drug interactions in the efficacy and safety of atypical and other antipsychotic agents. J. Pharm. Pharmacol. 2010, 58, 871–885. [Google Scholar] [CrossRef] [PubMed]
- Phang-Lyn, S.; Llerena, V. Biochemistry, Biotransformation; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Preissner, S.C.; Hoffmann, M.F.; Preissner, R.; Dunkel, M.; Gewiess, A. Polymorphic Citochrome P450 enzymes (CYPs) and their role in personalized therapy. PLoS ONE 2013, 8, e82562. [Google Scholar] [CrossRef] [PubMed]
- Siokas, V.; Stamati, P.; Pateraki, G.; Liampas, I.; Aloizou, A.-M.; Tsirelis, D.; Nousia, A.; Sgantzos, M.; Nasios, G.; Bogdanos, D.P.; et al. Analysis of SOD2 rs4880 Genetic Variant in Patients with Alzheimer’s Disease. Curr. Issues Mol. Biol. 2022, 44, 4406–4414. [Google Scholar] [CrossRef] [PubMed]
- Abaji, R.; Krajinovic, M. Thiopurine S-methyltransferase polymorphisms in acute lymphoblastic leukemia, inflammatory bowel disease and autoimmune disorders: Influence on treatment response. Pharmacogenom. Pers. Med. 2017, 10, 143–156. [Google Scholar] [CrossRef]
- Relling, M.V.; Schwab, M.; Whirl-Carrillo, M.; Suarez-Kurtz, G.; Pui, C.H.; Stein, C.M.; Moyer, A.M.; Evans, W.E.; Klein, T.E.; Antillon-Klussmann, F.G.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT15 Genotypes: 2018 Update. Clin. Pharmacol. Ther. 2019, 105, 1095–1105. [Google Scholar] [CrossRef]
- Pereira, E.E.B.; Leitão, L.P.C.; Andrade, R.B.; Modesto, A.A.C.; Fernandes, B.M.; Burbano, R.M.R.; Assumpção, P.P.; Fernandes, M.R.; Guerreiro, J.F.; Santos, S.E.B.D.; et al. UGT1A1 Gene Polymorphism Contributes as a Risk Factor for Lung Cancer: A Pilot Study with Patients from the Amazon. Genes 2022, 11, 493. [Google Scholar] [CrossRef]
- Zhu, X.; Ma, R.; Ma, X.; Yang, G. Association of UGT1A1*6 polymorphism with irinotecan-based chemotherapy reaction in colorectal cancer patients: A systematic review and a meta-analysis. Biosci. Rep. 2020, 40, BSR20200576. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, M.; Hu, M.; Cui, Y.; Zhong, Q.; Liang, L.; Huang, F. UGT1A1*6 and UGT1A1*28 polymorphisms are correlated with irinotecan-induced toxicity: A meta-analysis. Asia Pac. J. Clin. Oncol. 2018, 14, e479–e489. [Google Scholar] [CrossRef]
- Yu, J.T.; Tan, L.; Hardy, J. Apolipoprotein E in Alzheiemer’s Disease: An update. Ann. Rev. Neurosci. 2014, 37, 79–100. [Google Scholar] [CrossRef]
- Angelopoulou, E.; Paudel, Y.N.; Papageorgiou, S.G.; Piperi, C. APOE genotype and Alzheimer’s disease: The influence of lifestyle and environmental factors. ACS Chem. Neurosci. 2021, 12, 2749–2764. [Google Scholar] [CrossRef]
- Martinelli-Boneschi, F.; Giacalone, G.; Magnani, G.; Biella, G.; Coppi, E.; Santangelo, R.; Brambilla, P.; Esposito, F.; Lupoli, S.; Clerici, F.; et al. Pharmacogenomics in Alzheimer’s disease: A genome-wide association study of response to cholinesterase inhibitors. Neurobiol. Aging 2013, 4, e7–e13. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R. Pharmacogenomics of cognitive and neuropsychiatric disorders in Dementia. Int. J. Mol. Sci. 2020, 21, 3059. [Google Scholar] [CrossRef] [PubMed]
- Kuring, J.K.; Mathias, J.L.; Ward, L. 28 Prevalence of Depression, Anxiety and PTSD in People with Dementia: A Systematic Review and Meta-Analysis. Neuropsychol. Rev. 2018, 28, 393–416. [Google Scholar] [CrossRef]
- Zhang, C.; Jiang, X.; Chen, W.; Li, Q.; Yun, F.; Yang, X.; Dai, R.; Cheng, Y. Population genetic differnece of pharmacigenomic VIP gene variants in the Lisu population from Yunnan Province. Medicine 2018, 97, e13674. [Google Scholar] [CrossRef] [PubMed]
- Bialek, K.; Czarny, P.; Watala, C.; Wigner, P.; Talarowska, M.; Galecki, P.; Szemraj, J.; Sliwinski, T. Novel association between TGFA, TGFB1, IRF1, PTSG2 and IKBKB single-nucleotide polymorphisms and occurrence, severity and treatment response of major depressive disorder. PeerJ 2020, 8, e8676. [Google Scholar] [CrossRef] [PubMed]
- Salemeh, Y.; Bejaoui, Y.; El Hajj, N. DNA methylation biomarkers in aging and age-related diseases. Front. Genet. 2020, 11, 171. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Chen, S. Epigenetic regulation of Cytochrome P450 enzymed and clinical implication. Curr. Drug Metab. 2015, 16, 86–96. [Google Scholar] [CrossRef]
- Song, Y.; Li, C.; Liu, G.; Liu, R.; Chen, Y.; Li, W.; Cao, Z.; Zhao, B.; Lu, C.; Liu, Y. Drug-metabolizing cytochrome P450 enzymes have multifarious influences on treatment outcomes. Clin. Pharm. 2021, 60, 585–601. [Google Scholar] [CrossRef]
- Meyer, U.A. Pharmacogenetics and adverse drug reactions. Lancet 2000, 356, 1667–1671. [Google Scholar] [CrossRef]
- Guo, J.; Zhu, X.; Badawy, S.; Ihsan, A.; Liu, Z.; Xie, C.; Wang, X. Metabolism and mechanism of human Cytochrome P450 Enzyme 1A2. Curr. Drug Metab. 2021, 22, 40–49. [Google Scholar]
- Popat, R.A.; Van Den Eeden, S.K.; Tanner, C.M.; Kamel, F.; Umbach, D.M.; Marder, K.; Mayeux, R.; Ritz, B.; Ross, G.W.; Petrovitch, H.; et al. Coffee, ADORA2A, and CYP1A2, the caffeine connection in Parkinson’s disease. Eur. J. Neurol. 2011, 18, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Doude van Troostwijk, L.J.; Koopmans, R.P.; Vermeulen, H.D.; Guchelaar, H.J. CYP1A2 activity is an important determinant of clozapine dosage in schizophrenic patients. Eur. J. Pharm. Sci. 2003, 20, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Dean, L.; Kane, M. Clozapine therapy and CYP genotype. In Medical Genetics Summaries; Publisher: Bethesda, USA, 2021. [Google Scholar]
- Jeyarasasingam, G.; Yeluashvili, M.; Quik, M. Tacrine, a reversible acetylcholinesterase inhibitor, induces myopathy. NeuroReport 2000, 27, 1173–1176. [Google Scholar] [CrossRef]
- Cacabelos, R. Pharmacogenetic considerations when prescribing cholinesterase inhibitors for the treatment of Alzheimer’s disease. Expert Opin. Drug Metab. Toxicol. 2020, 16, 673–701. [Google Scholar] [CrossRef] [PubMed]
- Linde, K.; Rossnagel, K. Propanolol for migraine prophylasis. Cochrane Datanase Syst. Rev. 2017, 2017, CD003225. [Google Scholar]
- Steenen, S.A.; van Wijk, A.J.; van der Heijden, G.J.; van Westrhenen, R.; de Lange, J.; de Jongh, A. Propanolol for the treatment of anxiety disorders: Systematic review and meta-analysis. J. Phsychopharmacol. 2016, 30, 128–139. [Google Scholar] [CrossRef]
- Thorn, C.F.; Aklillu, E.; Klein, T.E.; Altman, R.B. Pharma GKB summary: Very important pharmacogene information for CYP1A2. Pharmacogenet Genom. 2013, 22, 73–77. [Google Scholar] [CrossRef]
- Lieber, C.S. Microsomal ethanol-oxidizing system (MEOS): The first 30 years (1968–1998)—A review. Alcohol. Clin. Exp. Res. 1999, 23, 991–1007. [Google Scholar] [CrossRef]
- García-Suástegui, W.A.; Ramos-Chávez, L.A.; Rubio-Osornio, M.; Calvillo-Velasco, M.; Atzin-Méndez, J.A.; Guevara, J.; Silva-Adaya, D. The role of CYP2E1 in the drug metabolism or bioactivation in the brain. Oxid. Med. Cell. Longev. 2017, 2017, 4680732. [Google Scholar] [CrossRef]
- Bai, Y.; Ma, X. Chorzoxazone exhibits neuroprotection against AD by attenuating neuroinflammation and neurodegenration in vitro and in vivo. Int. Immunopharmacol. 2020, 88, 106790. [Google Scholar] [CrossRef]
- Kumar, S.; Singla, B.; Singh, A.K.; Thomas-Gooch, S.M.; Zhi, K.; Singh, U.P. Hepatic. extrahepatic and extracellular vesicle Cytochrome P450 2E1 in alcohol and acetaminophen-mediated adverse intreactions and potential treatment options. Cells 2022, 11, 2620. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, S.; Barone, J.; Malone, B. Pharmacogenomics of Warfarin. Stratif. Med. Chapter 2014, 14, 497–507. [Google Scholar]
- Beetch, M.; Harandi-Zadeh, S.; Shen, K.; Lubecka, K.; Kitts, D.D.; O’Hagan, H.M.; Stefanska, B. Dietary antioxidants remodel DNA methylation patterns in chronid disease. Br. J. Pharmacol. 2019, 177, 1382–1408. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Myung, W.; Lim, S.W.; Kang, H.S.; Kim, S.; Won, H.H.; Carroll, B.J.; Kim, D.K. Association of the cjoline acetyltransferase gene with responsiveness to acetylcholinesterase inhibitors in Alzheimer’s disease. Pharmacopsychiatry 2015, 48, 111–117. [Google Scholar]
- Axelrod, J.; Tomchick, R. Enzymmatic O-methylation of epinephrine and other catechol. J. Biol. Chem. 1958, 233, 702–705. [Google Scholar] [CrossRef]
- Hall, K.T.; Loscalzo, J.; Kaptchuk, T.J. System pharmacogenomics-gene, disease, drug and placebo interactions: A case study in COMT. Pharmacogenomics 2019, 20, 529–551. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.; Bell, P.; Jonovska, V.; Budge, M.; Sim, E. NAT gene polymorphisms and susceptibility to Alzheimer’s disease: Identification of a novel NAT1 allelic variant. BMC Med. Genet. 2004, 5, 6. [Google Scholar] [CrossRef][Green Version]
- Borlak, J.; Reamon-Buettner, S.M. N-acetyltransferase 2 (NAT2) gene polymorphisms in Parkinson’s disease. BMC Med. Genet. 2006, 7, 30. [Google Scholar] [CrossRef][Green Version]
- Cacabelos, R.; Fernández-Novoa, L.; Alejo, R.; Corzo, L.; Rodríguez, S.; Alcaraz, M.; Nebril, L.; Cacabelos, P.; Fraile, C.; Carrera, I.; et al. EPodoFavalin-15999 (Atremorine®)-induced dopamine response in Parkinson’s Disease: Pharmacogenetics-related effects. J. Genom. Med. Pharmacogenom. 2016, 1, 450–452. [Google Scholar]
- Martínez-Iglesias, O.; Naidoo, V.; Carril, J.C.; Carrera, I.; Corzo, L.; Susana, R.; Ramón, A.; Natalia, C.; Cacabelos, R. AtreMorine Treatment Regulates DNA Methylation in Neurodegenerative Disorders: Epigenetic and Pharmacogenetic Studies. Curr. Pharmacogen. Pers. Med. 2021, 17, 159–171. [Google Scholar] [CrossRef]
- Ahmed, S.; Zhou, Z.; Zhou, J.; Chen, S.Q. Pharmacogenomics of drug metabolizing enzymes and transporters: Relevance to precision medicine. Genom. Proteom. Bioinform. 2016, 14, 298–313. [Google Scholar] [CrossRef]
- Cacabelos, R.; Tarrellas, C. Tellado Opportinities in pharmacogenomics for the treatment of Alzheimer’s disease. Future Neurol. 2015, 10, 229–252. [Google Scholar] [CrossRef]
- Cacabelos, R.; Carrera, I.; Martínez, O.; Alejo, R.; Fernández-Novoa, L.; Cacabelos, P.; Corzo, L.; Rodríguez, S.; Alcaraz, M.; Nebril, L.; et al. Atremorine in Parkisnon’s disease: From dopaminergic neuroprotection to pharmacogenomics. Med. Res. Rev. 2021, 41, 2841–2886. [Google Scholar] [CrossRef]
- Cacabelos, R.; Carril, J.C.; Corzo, L.; Fernández-Novoa, L.; Pego, R.; Cacabelos, N.; Cacabelos, P.; Alcaraz, M.; Tellado, I.; Naidoo, V. Influence of pathogenic and metabolic genes on the pharmacogenetic of mood disorders in Alzheimer’s disease. Pharmaceuticals 2021, 14, 366. [Google Scholar] [CrossRef]
- Fernández-Calle, R.; Konings, S.C.; Frontiñán-Rubio, J.; García-Revilla, J.; Camprubí-Ferrer, L.; Svensson, M.; Martinson, I.; Boza-Serrano, A.; Venero, J.L.; Nielsen, H.M.; et al. APOE in the bullseye of neurodegenerative diseases: Impact of the APOE genotype in Alzheimer’s disease pathology and brain diseases. Mol. Neurodegener. 2022, 24, 17–62. [Google Scholar] [CrossRef]
- Zhao, N.; Attrebi, O.N.; Ren, Y.; Qiao, W.; Sonustun, B.; Martens, Y.A.; Meneses, A.D.; Li, F.; Shue, F.; Zheng, J.; et al. APOE4 exacerbates a-synuclein pathology and related toxicity independent of amyloid. Sci. Transl. Med. 2020, 12, eaay1809. [Google Scholar] [CrossRef]
- Werden, E.; Khlif, M.S.; Bird, L.J.; Cumming, T.; Bradshaw, J.; Khan, W.; Pase, M.; Restrepo, C.; Veldsman, M.; Egorova, N.; et al. APOE varepsilon4 carriers show delayed recovery of verbal memory and smaller enthrinal volume in the fisrt year after ischemic stroke. J. Alzheimer’s Dis. 2019, 71, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Pendlebury, S.; Poole, D.D.; Burgess, A.; Duerden, J.; Rothwell, P.M.; Oxford Vascular Study. APOE-epsilon4 genotype and dementia before and after transient ischemic attack and stroke: Population based cohort study. Stroke 2020, 51, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Iglesias, O.; Naidoo, V.; Carril, J.C.; Seoane, S.; Cacabelos, N.; Cacabelos, R. Gene expression profiling as a novel diagnostic tool for Neurodegenrative Disorders. Int. J. Mol. Sci. 2023, 24, 5746. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.L.; Tybjaerg-Hansen, A.; Nordestgaard, B.G.; Frikke-Schmidt, R. Plasma levels of apolipoprotein E and risk of dementia in general population. Ann. Neurol. 2014, 77, 301–311. [Google Scholar] [CrossRef]
- Pozzi, F.E.; Conti, E.; Appollonio, I.; Ferrarese, C.; Tremolizzo, L. Predictors of response to acetylcholinesterase inhibitors: A systematic review. Front. Neurosci. 2022, 16, 998224. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Carril, J.C.; Corzo, L.; Pego, R.; Cacabelos, N.; Alcaraz, M.; Muñiz, A.; Martínez-Iglesias, O.; Naidoo, V. Pharmacogenetics of anxiety and depression in Alzheimer’s disease. Future Med. 2023, 24, 27–57. [Google Scholar] [CrossRef] [PubMed]
- McMillan, M.; Gomez, N.; Hsieh, C.; Bekier, M.; Li, X.; Miguez, R.; Tank, E.M.H.; Barmada, S.J. RNA methylation influences TDP43 binding and disease pathogenesis in models of amyotrophic lateral sclerosis and frontotemporal dementia. Mol. Cell 2023, 83, 219–236.e7. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dou, X.; Liu, J.; Xiao, Y.; Zhang, Z.; Hayes, L.; Wu, R.; Fu, X.; Ye, Y.; Yang, B.; et al. Globally reduced N6-methyladenosine (m6A) in C9ORF72-ALS/FTD dysregulates RNA metabolism and contributes to neurodegeneration. Nat. Neurosci. 2023, 26, 1328–1338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, Y.; Guo, F.; Li, S.; Cui, H. RNA N6-Methyladenosine Modifications and Its Roles in Alzheimer’s Disease. Front. Cell. Neurosci. 2022, 16, 820378. [Google Scholar] [CrossRef]
| Metabolic (Phase I) | Metabolic (Phase II) | Transporter | Pathogenic |
|---|---|---|---|
| CYP1A2 | GSTP1 | ABCB1 | APOE |
| CYP2C9 | NAT2 | ABCC2 | NBEA |
| CYP4F2 | SLC2A9 | PTSG2 | |
| SLC39A8 | |||
| SLCO1B1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Iglesias, O.; Naidoo, V.; Carrera, I.; Carril, J.C.; Cacabelos, N.; Cacabelos, R. Influence of Metabolic, Transporter, and Pathogenic Genes on Pharmacogenetics and DNA Methylation in Neurological Disorders. Biology 2023, 12, 1156. https://doi.org/10.3390/biology12091156
Martínez-Iglesias O, Naidoo V, Carrera I, Carril JC, Cacabelos N, Cacabelos R. Influence of Metabolic, Transporter, and Pathogenic Genes on Pharmacogenetics and DNA Methylation in Neurological Disorders. Biology. 2023; 12(9):1156. https://doi.org/10.3390/biology12091156
Chicago/Turabian StyleMartínez-Iglesias, Olaia, Vinogran Naidoo, Iván Carrera, Juan Carlos Carril, Natalia Cacabelos, and Ramón Cacabelos. 2023. "Influence of Metabolic, Transporter, and Pathogenic Genes on Pharmacogenetics and DNA Methylation in Neurological Disorders" Biology 12, no. 9: 1156. https://doi.org/10.3390/biology12091156
APA StyleMartínez-Iglesias, O., Naidoo, V., Carrera, I., Carril, J. C., Cacabelos, N., & Cacabelos, R. (2023). Influence of Metabolic, Transporter, and Pathogenic Genes on Pharmacogenetics and DNA Methylation in Neurological Disorders. Biology, 12(9), 1156. https://doi.org/10.3390/biology12091156








