Kimura’s Theory of Non-Adaptive Radiation and Peto’s Paradox: A Missing Link?
Abstract
:Simple Summary
Abstract
1. Introduction: Formulating the Question
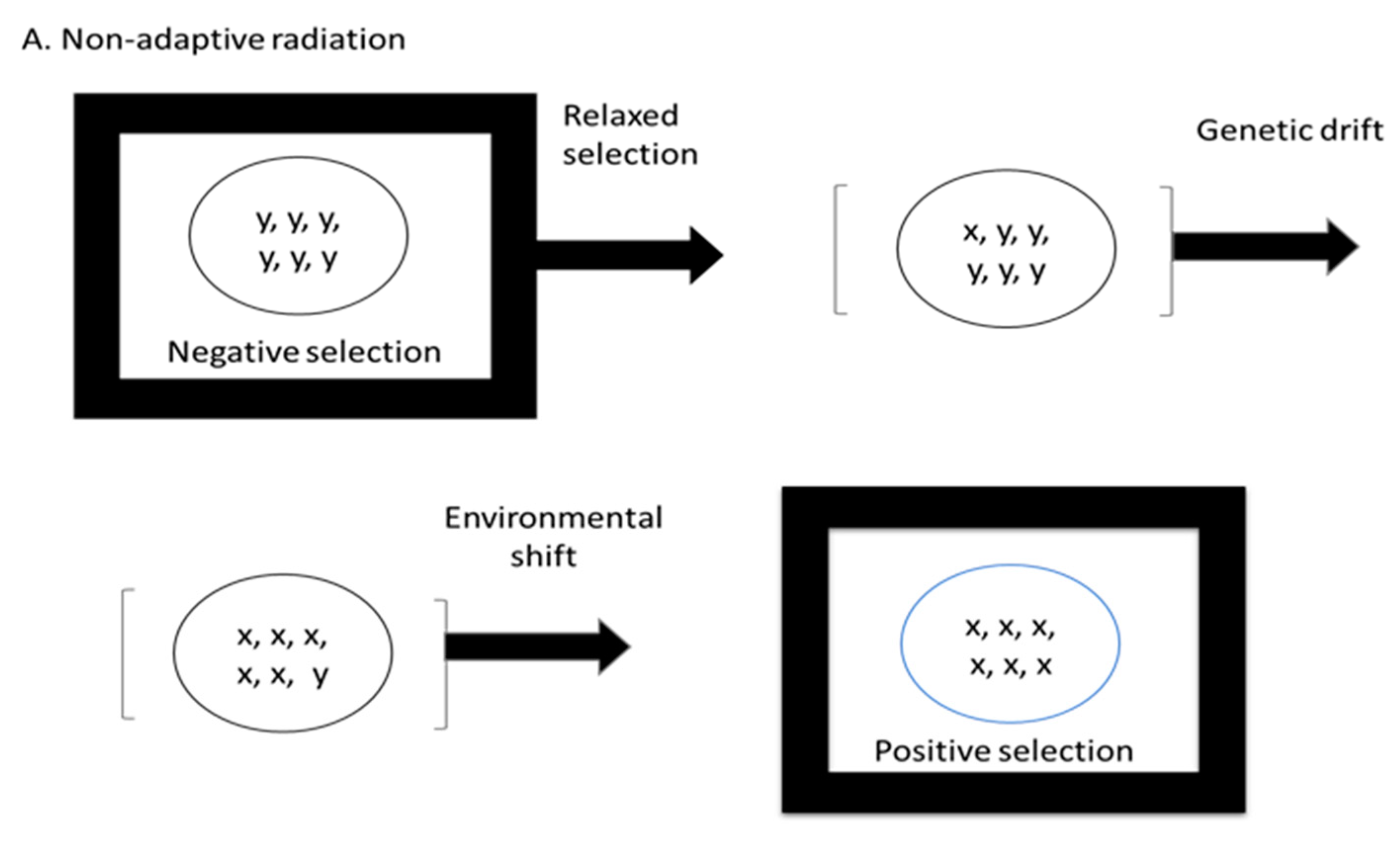
2. The Neutral Theory of Evolution and the Hypothesis of Non-Adaptive Radiation
Kimura’s Hypothesis
- (1)
- Liberation from the preexisting selective constraint;
- (2)
- Sudden increase, or boom, of neutral variants under relaxed selection that are then fixed in the population by random genetic drift;
- (3)
- Realization of latent selective potential: some of the accumulated neutral mutants become useful at the phenotypic level in a new environment, which the population is then able to exploit;
- (4)
- Intergroup competition, as well as individual selection, leads to extensive adaptive evolution creating a radically different taxonomic group adapted to a newly opened ecological niche.
3. Peto’s Paradox
4. The DNA Damage Response (DDR) System Mediates the Rate of Mutation Input
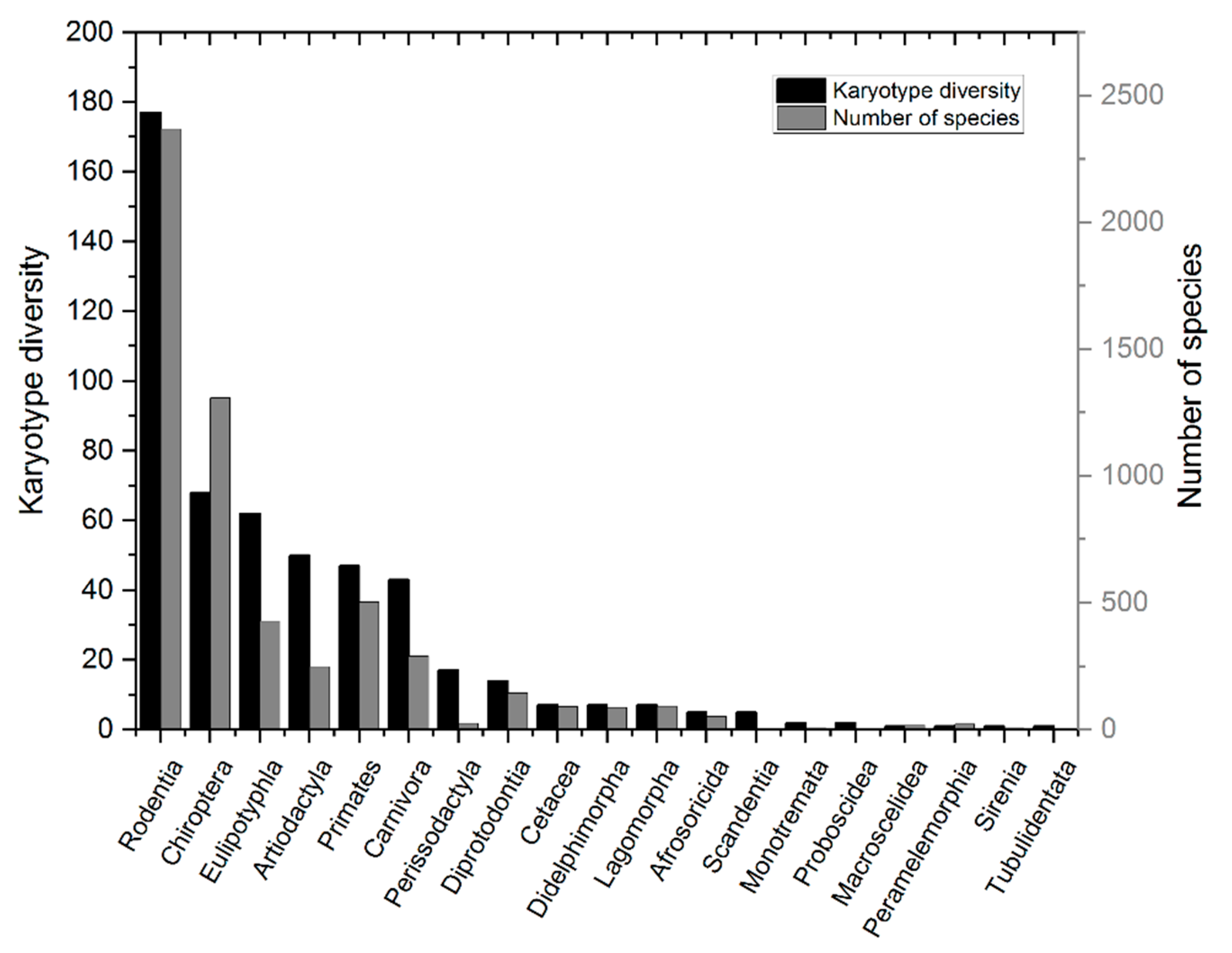
4.1. The Fidelity and Efficiency of the DDR Varies Significantly across Taxonomic Groups
4.2. Sirtuin 6 and the Naked Mole Rat (NMR): A More-Proficient DDR Promotes a Longer Life Span Independent of Body Size
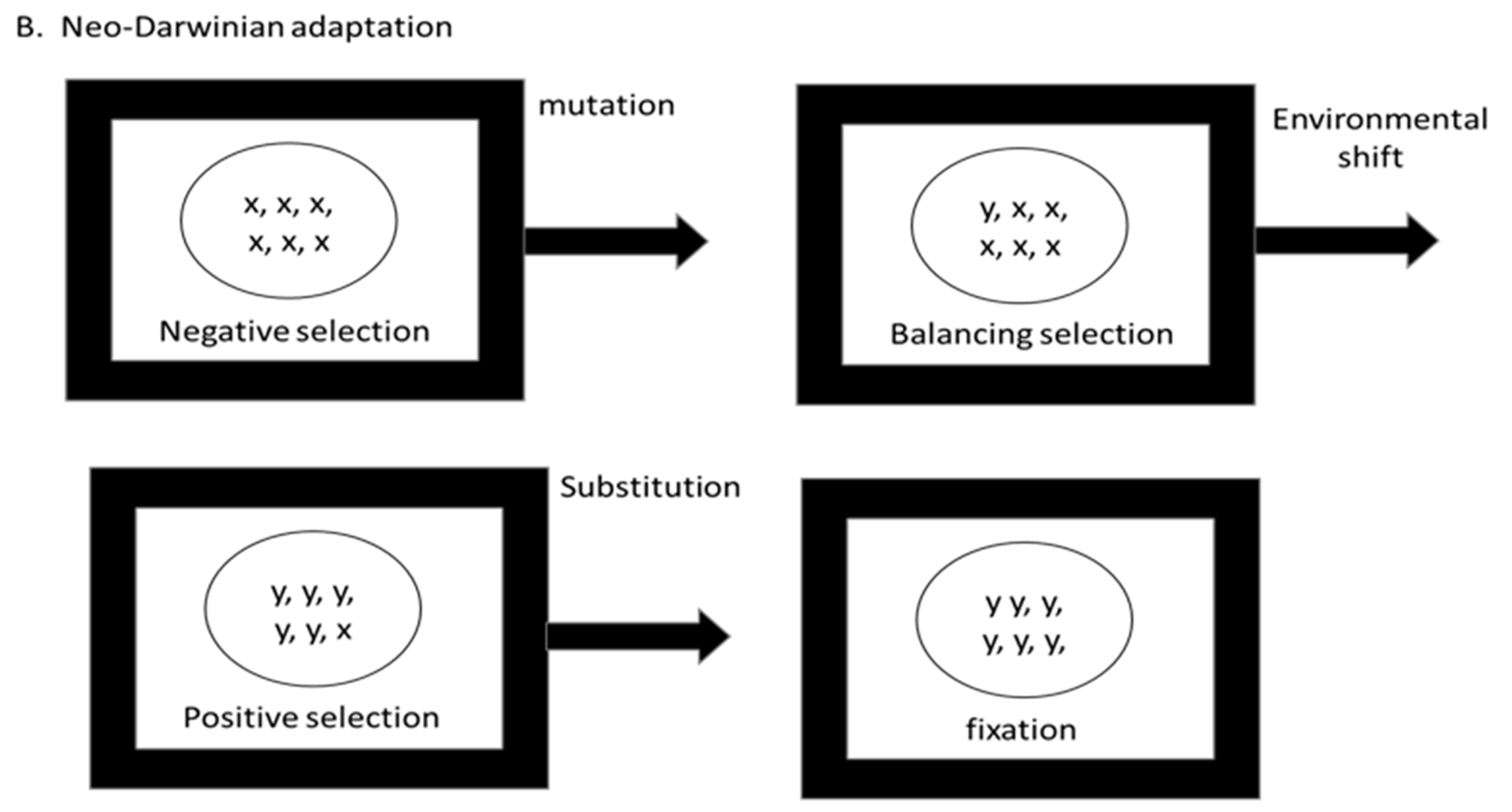
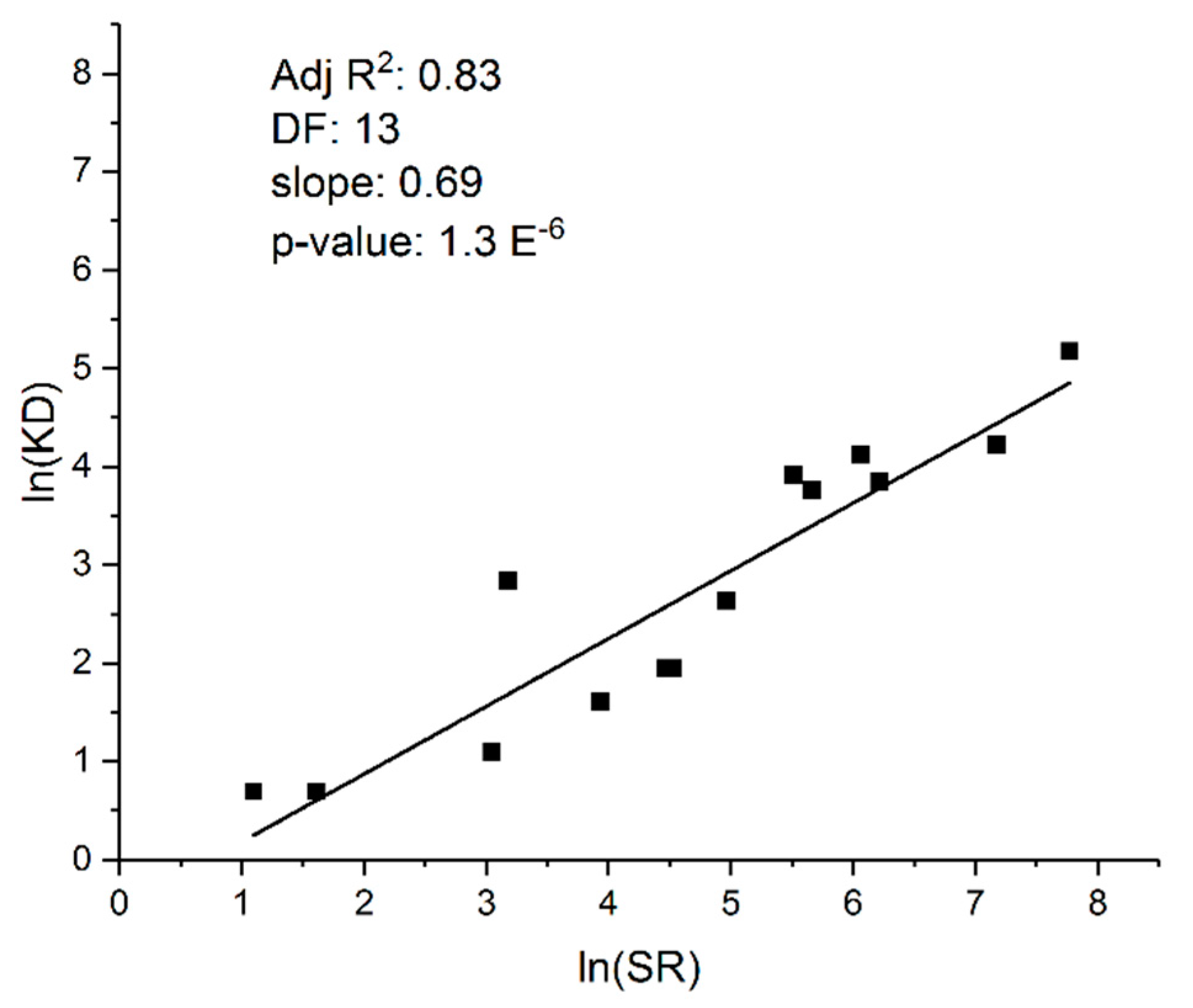
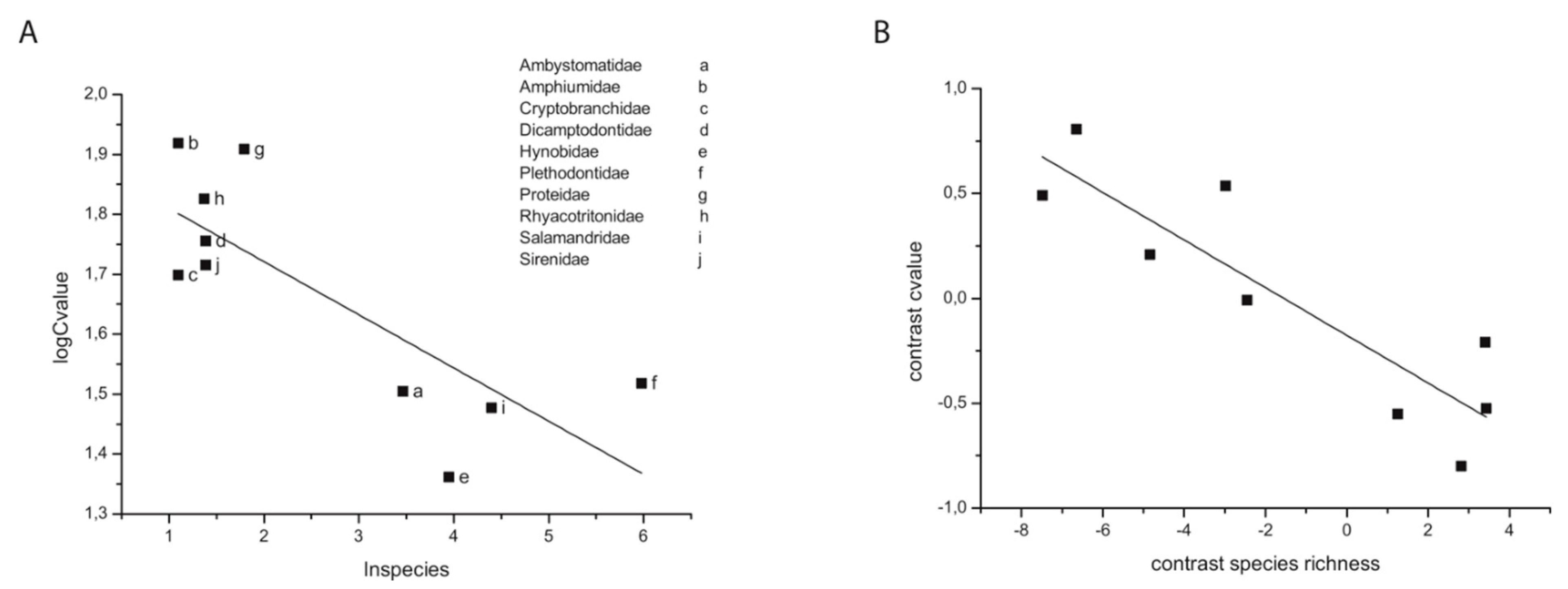
5. Adaptive Evolution: Mutation Limited, Selection Limited, or Both?
6. Other Questions
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ayala, F.J.; Coluzzi, M. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc. Nat. Acad. Sci. USA 2005, 102 (Suppl. S1), 6535–6542. [Google Scholar] [CrossRef] [PubMed]
- Presgraves, D.C. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 2010, 11, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Graphodatsky, A.S.; Trifonov, V.A.; Stanyon, R. The genome diversity and karyotype evolution of mammals. Mol. Cytogenet. 2011, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zhou, P.K. DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal. Transduct. Target Ther. 2021, 6, 254. [Google Scholar]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C. A brief history of the DNA repair field. Cell Res. 2008, 18, 3–7. [Google Scholar] [CrossRef]
- Kern, A.D.; Hahn, M.W. The Neutral Theory in Light of Natural Selection. Mol. Biol. Evol. 2018, 35, 1366–1371. [Google Scholar] [CrossRef]
- Jensen, J.D.; Payseur, B.A.; Stephan, W.; Aquadro, C.F.; Lynch, M.; Charlesworth, D.; Charlesworth, B. The importance of the Neutral Theory in 1968 and 50 years on: A response to Kern and Hahn 2018. Evolution 2019, 73, 111–114. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, J. Antagonistic pleiotropy conceals molecular adaptations in changing environments. Nat. Ecol. Evol. 2020, 4, 461–469. [Google Scholar] [CrossRef]
- Rousselle, M.; Simion, P.; Tilak, M.K.; Figuet, E.; Nabholz, B.; Galtier, N. Is adaptation limited by mutation? A timescale-dependent effect of genetic diversity on the adaptive substitution rate in animals. PLoS Genet. 2020, 16, e1008668. [Google Scholar] [CrossRef]
- Gerrish, P.J.; Colato, A.; Sniegowski, P.D. Genomic mutation rates that neutralize adaptive evolution and natural selection. J. R. Soc. Interface 2013, 10, 20130329. [Google Scholar] [CrossRef]
- Alvarez-Valin, F.; Jabbari, K.; Bernardi, G. Synonymous and nonsynonymous substitutions in mammalian genes: Intragenic correlations. J. Mol. Evol. 1998, 46, 37–44. [Google Scholar] [CrossRef]
- Cameron, J.M.; Kreitman, M. The correlation between synonymous and nonsynonymous substitutions in Drosphila: Mutation, selection or relaxed constraints? Genetics 1998, 150, 767–775. [Google Scholar] [CrossRef]
- Wyckoff, G.J.; Malcom, C.M.; Vallender, E.J.; Lahn, B.T. A highly unexpected strong correlation between fixation probability of nonsynonymous mutations and mutation rate. Genome Anal. 2005, 21, 381–385. [Google Scholar] [CrossRef]
- Kimura, M. The neutral theory of molecular evolution: A review of recent evidence. Jpn. J. Genet. 1991, 66, 367–386. [Google Scholar] [CrossRef]
- Jones, K.E.; Safi, K. Ecology and evolution of mammalian biodiversity. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 2451–2461. [Google Scholar] [CrossRef]
- Burgin, C.J.; Colella, J.P.; Kahn, P.L.; Upham, N.S. How many species of mammals are there? J. Mammal. 2018, 99, 1–14. [Google Scholar] [CrossRef]
- Caulin, S.F.; Maley, C.C. Peto’s Paradox: Evolution’s Prescription for Cancer Prevention. Trends Ecol. Evol. 2011, 26, 175–182. [Google Scholar] [CrossRef]
- Callier, V. Solving Peto’s Paradox to better understand cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 1825–1828. [Google Scholar] [CrossRef]
- Dart, A. Peto’s paradox put to the test. Nat. Rev. Cancer 2022, 22, 129. [Google Scholar] [CrossRef]
- Nery, M.F.; Rennó, M.; Picorelli, A.; Ramos, E. A phylogenetic review of cancer resistance highlights evolutionary solutions to Peto’s Paradox. Genet. Mol. Biol. 2022, 45 (Suppl. S1), e20220133. [Google Scholar] [CrossRef]
- Maciak, S. Cell size, body size and Peto’s paradox. BMC Ecol. Evol. 2022, 22, 142. [Google Scholar] [CrossRef]
- Moreno, A.; Carrington, J.T.; Albergante, L.; Al Mamun, M.; Haagensen, E.J.; Komseli, E.S.; Gorgoulis, V.G.; Newman, T.J.; Blow, J.J. Unreplicated DNA remaining from unperturbed S phases passes through mitosis for resolution in daughter cells. Proc. Natl. Acad. Sci. USA 2016, 113, E5757–E5764. [Google Scholar] [CrossRef]
- Feschotte, C.; Pritham, E.J. DNA Transposons and the Evolution of Eukaryotic Genomes. Ann. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef]
- Platt, R.N., 2nd; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 2018, 26, 25–43. [Google Scholar] [CrossRef]
- Uphoff, S. Real-time dynamics of mutagenesis reveal the chronology of DNA repair and damage tolerance responses in single cells. Proc. Natl. Acad. Sci. USA 2018, 115, E6516–E6525. [Google Scholar] [CrossRef]
- Bergeron, L.A.; Besenbacher, S.; Zheng, J.; Li, P.; Bertelsen, M.F.; Quintard, B.; Hoffman, J.I.; Li, Z.; St Leger, J.; Shao, C.; et al. Evolution of the germline mutation rate across vertebrates. Nature 2023, 615, 285–291. [Google Scholar] [CrossRef]
- Seluanov, A.; Gladyshev, V.N.; Vijg, J.; Gorbunova, V. Mechanisms of cancer resistance in long-lived mammals. Nat. Rev. Cancer 2018, 18, 433–441. [Google Scholar] [CrossRef]
- Abegglen, L.M.; Caulin, A.F.; Chan, A.; Lee, K.; Robinson, R.; Campbell, M.S.; Kiso, W.K.; Schmitt, D.L.; Waddell, P.J.; Bhaskara, S.; et al. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 2015, 314, 1850–1860. [Google Scholar] [CrossRef]
- Sulak, M.; Fong, L.; Mika, K.; Chigurupati, S.; Yon, L.; Mongan, N.P.; Emes, R.D.; Lynch, V.J. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife 2016, 5, e11994. [Google Scholar] [CrossRef]
- Callaway, E. How Elephants Avoid Cancer. Nature 2015, 1038, 18534. [Google Scholar] [CrossRef]
- Takemoto, K.; Ii, M.; Nishizuka, S.S. Importance of metabolic rate to the relationship between the number of genes in a functional category and body size in Peto’s paradox for cancer. R. Soc. Open Sci. 2016, 3, 160267. [Google Scholar] [CrossRef] [PubMed]
- Firsanov, D.; Zacher, M.; Tian, X.; Zhao, Y.; George, J.C.; Sformo, T.L.; Trombline, G.; Biashed, A.; Gilman, A.; Hamilton, N.; et al. DNA repair and anti-cancer mechanisms in the longest-living mammal: The bowhead whale. bioRxiv 2023. [Google Scholar] [CrossRef]
- Marra, N.J.; Richards, V.P.; Early, A.; Bogdanowicz, S.M.; Bitar, P.D.P.; Stanhope, M.J.; Shivji, M.S. Comparative transcriptomics of elasmobranchs and teleosts highlight important processes in adaptive immunity and regional endothermy. BMC Genom. 2017, 18, 87. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; Deelen, J.; Jones, O.R. Editorial: Mechanisms and Pathways Contributing to the Diversity of Aging across the Tree of Life. Front. Cell Dev. Biol. 2022, 10, 854700. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.H. Salamander Insights into Ageing and Rejuvenation. Front. Cell Dev. Biol. 2021, 9, 689062. [Google Scholar] [CrossRef]
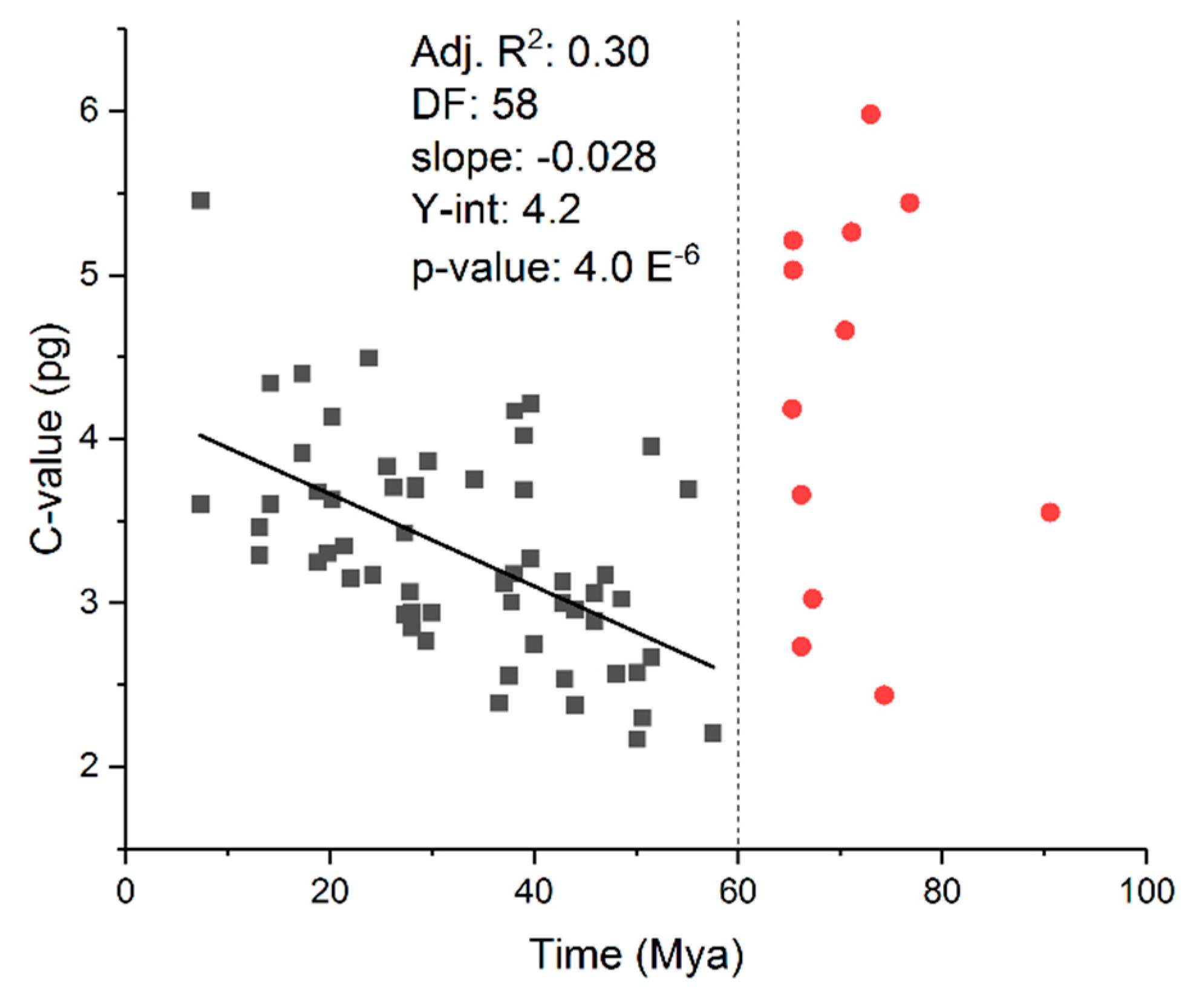
- Mohlhenrich, E.R.; Mueller, R.L. Genetic drift and mutational hazard in the evolution of salamander genomic gigantism. Evolution 2016, 70, 2865–2878. [Google Scholar] [CrossRef]
- Liedtke, H.C.; Gower, D.J.; Wilkinson, M.; Gomez-Mestre, I. Macroevolutionary shift in the size of amphibian genomes and the role of life history and climate. Nat. Ecol. Evol. 2018, 2, 1792–1799. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, X.; Tian, X.; Lee, M.; Ablaeva, J.; Firsanov, D.; Lee, S.G.; Maslov, A.Y.; Gladyshev, V.N.; Seluanov, A.; et al. Maintenance of genome sequence integrity in long- and short-lived rodent species. Sci. Adv. 2021, 7, eabj3284. [Google Scholar] [CrossRef]
- Lewis, K.N.; Mele, J.; Hornsby, P.J.; Buffenstein, R. Stress resistance in the naked mole-rat: The bare essentials—A mini-review. Gerontology 2012, 58, 453–462. [Google Scholar] [CrossRef]
- Munro, D.; Baldy, C.; Pamenter, M.E.; Treberg, J.R. The exceptional longevity of the naked mole-rat may be explained by mitochondrial antioxidant defenses. Aging Cell 2019, 18, e12916. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Firsanov, D.; Zhang, Z.; Cheng, Y.; Luo, L.; Tombline, G.; Tan, R.; Simon, M.; Henderson, S.; Steffan, J.; et al. SIRT6 Is Responsible for More Efficient DNA Double-Strand Break Repair in Long-Lived Species. Cell 2019, 177, 622–638.e22. [Google Scholar] [CrossRef]
- Maxson, L.E.R.; Wilson, A.C. Rates of molecular and chromosomal evolution in salamanders. Evolution 1978, 33, 734–740. [Google Scholar] [CrossRef]
- Gorbunova, V.; Seluanov, A.; Mita, P.; McKerrow, W.; Fenyö, D.; Boeke, J.D.; Linker, S.B.; Gage, F.H.; Kreiling, J.A.; Petrashen, A.P.; et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature 2021, 596, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Roscoe, S.; Stone, G.; Garcia, M.; Lutz, F. Reciprocal chromosome painting shows that squirrels, unlike murid rodents, have a highly conserved genome organization. Genomics 2003, 82, 745–749. [Google Scholar]
- Bengtsson, B.O. Rates of karyotype evolution in placental mammals. Hereditas 1980, 92, 37–47. [Google Scholar] [CrossRef]
- Bush, G.L.; Case, S.M.; Wilson, A.C.; Patton, J.L. Rapid speciation and chromosomal evolution in mammals. Proc. Natl. Acad. Sci. USA. 1977, 74, 3942–3946. [Google Scholar] [CrossRef]
- Herrick, J.; Sclavi, B. Genome diversity and species richness in mammals. bioRxiv 2019. [Google Scholar] [CrossRef]
- Pierce, B.A.; Mitton, J.B. The relationship between genome size and genetic variation. Am. Nat. 1980, 116, 850–861. [Google Scholar] [CrossRef]
- Nevo, E.; Beiles, A. Genetic diversity and ecological heterogeneity in amphibian evolution. Copeia 1991, 1991, 565–592. [Google Scholar] [CrossRef]
- Sclavi, B.; Herrick, J. Genome size variation and species diversity in salamanders. J. Evol. Biol. 2019, 32, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Leffler, E.M.; Bullaughey, K.; Matute, D.R.; Meyer, W.K.; Ségurel, L.; Venkat, A.; Andolfatto, P.; Przeworski, M. Revisiting an old riddle: What determines genetic diversity levels within species? PLoS Biol. 2012, 10, e1001388. [Google Scholar] [CrossRef] [PubMed]
- Vellend, M. Species diversity and genetic diversity: Parallel processes and correlated patterns. Am. Nat. 2005, 166, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Vellend, M.; Geber, M.A. Connections between species diversity and genetic diversity. Ecol. Lett. 2005, 8, 762–781. [Google Scholar] [CrossRef]
- Petersen, H.C.; Hansen, B.W.; Knott, K.E.; Banta, G.T. Species and genetic diversity relationships in benthic macroinvertebrate communities along a salinity gradient. BMC Ecol. Evol. 2022, 22, 125. [Google Scholar] [CrossRef]
- Marchesini, A.; Vernesi, C.; Battisti, A.; Ficetola, G.F. Deciphering the drivers of negative species-genetic diversity correlation in Alpine amphibians. Mol. Ecol. 2018, 27, 4916–4930. [Google Scholar] [CrossRef]
- Osmanski, A.B.; Paulat, N.S.; Korstian, J.; Grimshaw, J.R.; Halsey, M.; Sullivan, K.A.M.; Moreno-Santillán, D.D.; Crookshanks, C.; Roberts, J.; Garcia, C.; et al. Insights into mammalian TE diversity through the curation of 248 genome assemblies. Science 2023, 380, eabn1430. [Google Scholar] [CrossRef]
- Ricci, M.; Peona, V.; Guichard, E.; Taccioli, C.; Boattini, A. Transposable Elements Activity is Positively Related to Rate of Speciation in Mammals. J. Mol. Evol. 2018, 86, 303–310. [Google Scholar] [CrossRef]
- Wang, J.; Jia, S.T.; Jia, S. New Insights into the Regulation of Heterochromatin. Trends Genet. 2016, 32, 284–294. [Google Scholar] [CrossRef]
- Sung, W.; Ackerman, M.S.; Miller, S.F.; Doak, T.G.; Lynch, M. Drift-barrier hypothesis and mutation-rate evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 18488–18492. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.V. Non-adaptive evolution of genome complexity. Bioessays 2006, 28, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Serrato-Capuchina, A.; Matute, D.R. The Role of Transposable Elements in Speciation. Genes 2018, 9, 254. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J.; Bao, W.; Kojima, K.K. Families of transposable elements, population structure and the origin of species. Biol. Direct 2011, 6, 44. [Google Scholar] [CrossRef]
- Glazier, D.S. Genome Size Covaries More Positively with Propagule Size than Adult Size: New Insights into an Old Problem. Biology 2021, 10, 270. [Google Scholar] [CrossRef]
- Knight, C.A.; Molinari, N.A.; Petrov, D.A. The large genome constraint hypothesis: Evolution ecology and phenotype. Ann. Bot. 2005, 95, 177–190. [Google Scholar] [CrossRef]
- Francis, D.; Stuart Davies, M.; Barlow, P.W. A strong nucleotypic effect on the cell cycle regardless of ploidy level. Ann. Bot. 2008, 101, 747–757. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herrick, J. Kimura’s Theory of Non-Adaptive Radiation and Peto’s Paradox: A Missing Link? Biology 2023, 12, 1140. https://doi.org/10.3390/biology12081140
Herrick J. Kimura’s Theory of Non-Adaptive Radiation and Peto’s Paradox: A Missing Link? Biology. 2023; 12(8):1140. https://doi.org/10.3390/biology12081140
Chicago/Turabian StyleHerrick, John. 2023. "Kimura’s Theory of Non-Adaptive Radiation and Peto’s Paradox: A Missing Link?" Biology 12, no. 8: 1140. https://doi.org/10.3390/biology12081140
APA StyleHerrick, J. (2023). Kimura’s Theory of Non-Adaptive Radiation and Peto’s Paradox: A Missing Link? Biology, 12(8), 1140. https://doi.org/10.3390/biology12081140





