2. Materials and Methods
To study how cell-to-cell communication occurs, we must first study the motion of proteins inside the cell membrane; this motion of proteins gives rise to many bodily functions, from hormone production to hemostasis and disease prevention [
7]. Proteins are found in the cell membrane, which separates the interior of the cells from the outside environment. The cell membrane protects cells and provides a fixed environment within the cell; some of the main functions of the cell membrane are to transport nutrients into the cell, transport toxic substances out of the cells, and allow proteins to pass through the cell membrane [
8]. We shall study the motion of proteins inside the cell membrane, followed by the study of proteins when they diffuse through the cell membrane to the exterior of the cell so that they can bind with other proteins from other cells during cell communication. Fick’s law of diffusion describes the macroscopic diffusion of particles from a region of higher concentration to a level of lower concentration, with a diffusion rate being the concentration gradient [
9]; the macroscopic view of Fick’s law does not take into account the microscopic view of diffusion, especially when considering diffusion within a cell membrane. At a microscopic level, the diffusion of proteins in the cell membrane is highly stochastic [
9].
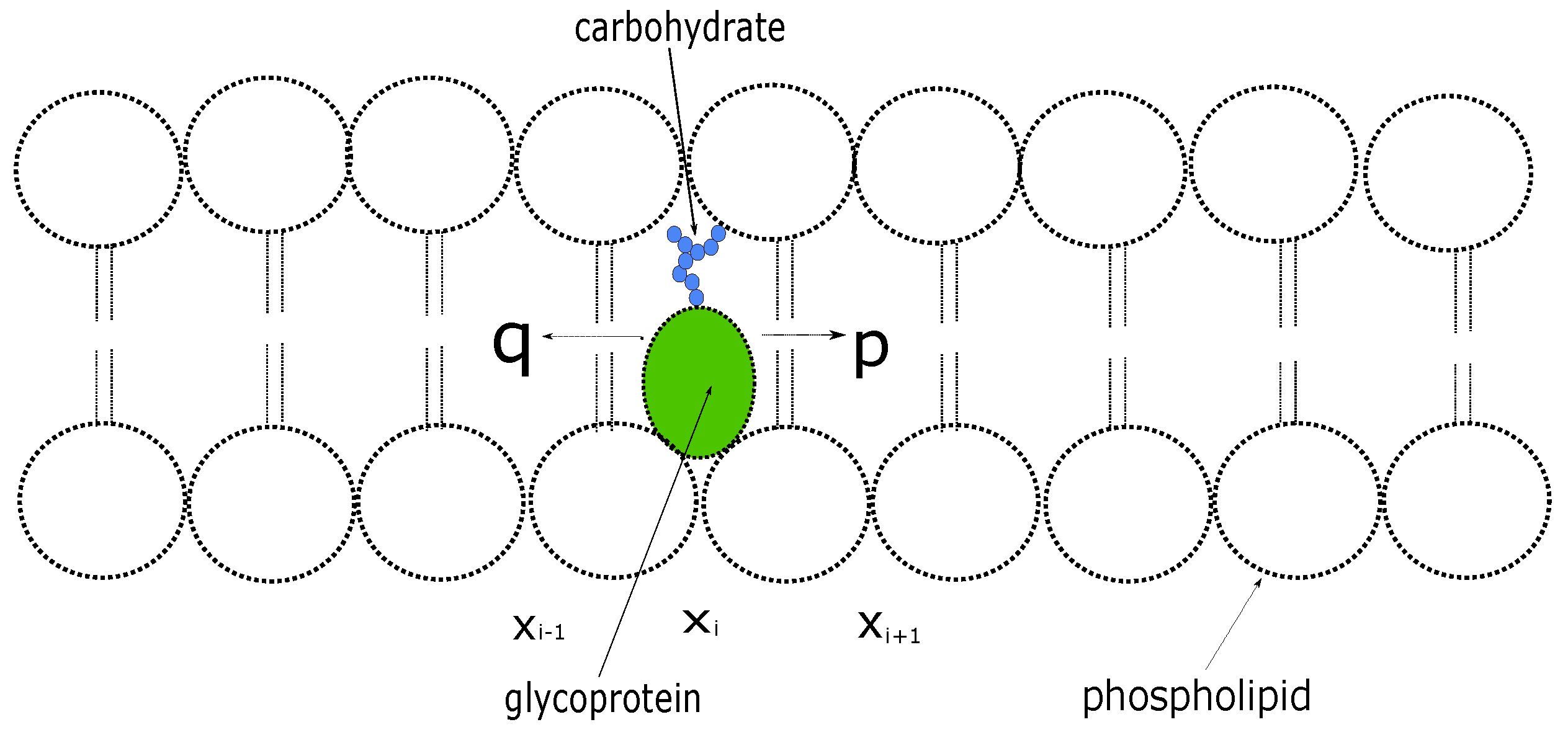
We consider a protein inside a cell membrane that can move at discrete times between neighboring sites on a one-dimensional lattice with unit spacing [
9] (see
Figure 1); At each step, the protein moves a unit distance to the right with probability
p or to the left with probability
. Let
denote the probability that the protein is at location
at the
n th time step. The evolution of the probability distribution is described by the discrete-time equation given by
We introduce the characteristic equation by taking the Discrete Fourier Transform on both sides of Equation (
1) for fixed
n.
To solve Equation (
2), we need to know what the value
is; let us assume, without loss of generality that
. Thus, then Equation (
2) becomes
Taking the Inverse Fourier Transform of Equation (
3),
from the binomial theorem, we can write Equation (
4) as
For infinitesimal step length
and time length
, where
denotes the number of points along the one-dimensional lattice, and
is the number of time points, we define the probability per unit length as the probability density function given by
We substitute Equation (
6) into Equation (
1),
using a Taylor Series expansion with a first order of
and second-order
[
10],
We divide Equation (
8) by
,
Due to the fact that proteins move extremely fast [
11], the step length
and the time
it takes for a protein to move from one point to another become significantly small; thus, we can take
,
we denote
and
.
Consider the case where the drift velocity
, and initial condition
,
applying the Fourier transform,
Multiplying by the integrating factor, Equation (
13) becomes
integration with respect to
t yields
The Inverse Fourier Transform yields
This is the probability density function describing the likelihood of the protein being located at point
x. Consider the random motion of a protein inside a cell membrane, subjected to external forces
F, with drag coefficient
. Let
be the position of the protein at time
t inside the cell membrane; we define the infinitesimal change in the position of the protein as it moves through the cell membrane in terms of a random variable that is normally distributed by the process:
where we have used the fact that
,
,
and
are random variables that are normally distributed with
and
=
[
12]. We introduced the diffusivity
D, which is the rate of diffusion that controls the volatility of the process, a measure of the rate at which proteins diffuse into and out of the cell membrane; we also introduced the drift term
.
We set the initial position of the protein at time
,
, to zero, consider the time interval
, and we divide the time
T into small
n equal interval
, since the position of the protein changes by the amount given by
, where
over the
ith time interval; the final position
is given by,
We now have a stochastic description of the motion of a protein in a cell membrane. The behavior and motion of the proteins determine the success of cell-to-cell communication; the exact solution of Equation (
17) is found by applying Ito’s formula and solving for
. The analytical solution is given by,
where
; let
be a function of the stochastic function
X and time
t.
From Ito’s Lemma, we can determine the process followed by the function
by applying Taylor expansion up to second order in variables
X and
t [
12].
where
.
We substitute Equation (
17) into Equation (
21), and take the limit
, in Equation (
21). The limit
is computed because the time it takes for a protein to move from one location to another becomes significantly small as proteins move at high speeds; we obtain the below equation:
This is the change in value for over the infinitesimal time interval . The stochastic behavior of proteins allows us to study the interaction of one protein with another protein; this mainly serves as the receiving end of chemical reactions, as they float freely inside and outside the cell membranes during cell communication, exhibiting Brownian motion behavior.
We transform Equation (
22) into a diffusion equation by taking the limit
[
12], where
is equal to 1.
As suggested by Paul Wolfgang [
13], in order to make Equation (
22) dimensionless, we make use of the following change in variables,
,
,
, where
K is a scaling factor, and
is the volatility of the process, we obtain the below equation:
Inside the cell membrane, we define the diffusivity rate to be proportional to the external forces acting on the protein
F and the drag coefficient
by a simple relationship given by
However, Dan V. Nicolau Jr. et al. [
14] suggested that the diffusivity inside a cell membrane, is spatially dependent and is given by
where
is the Gamma function. For maximum diffusivity, we set
,
This is the diffusion advection equation subjected to the Dirichlet boundary conditions and initial condition .
3. Results and Discussion
In this section, we present and discuss the numerical results of Equations (
17) and (
29); the results are computed in R using the
diffeqr,
ReacTran and
plotly libraries.
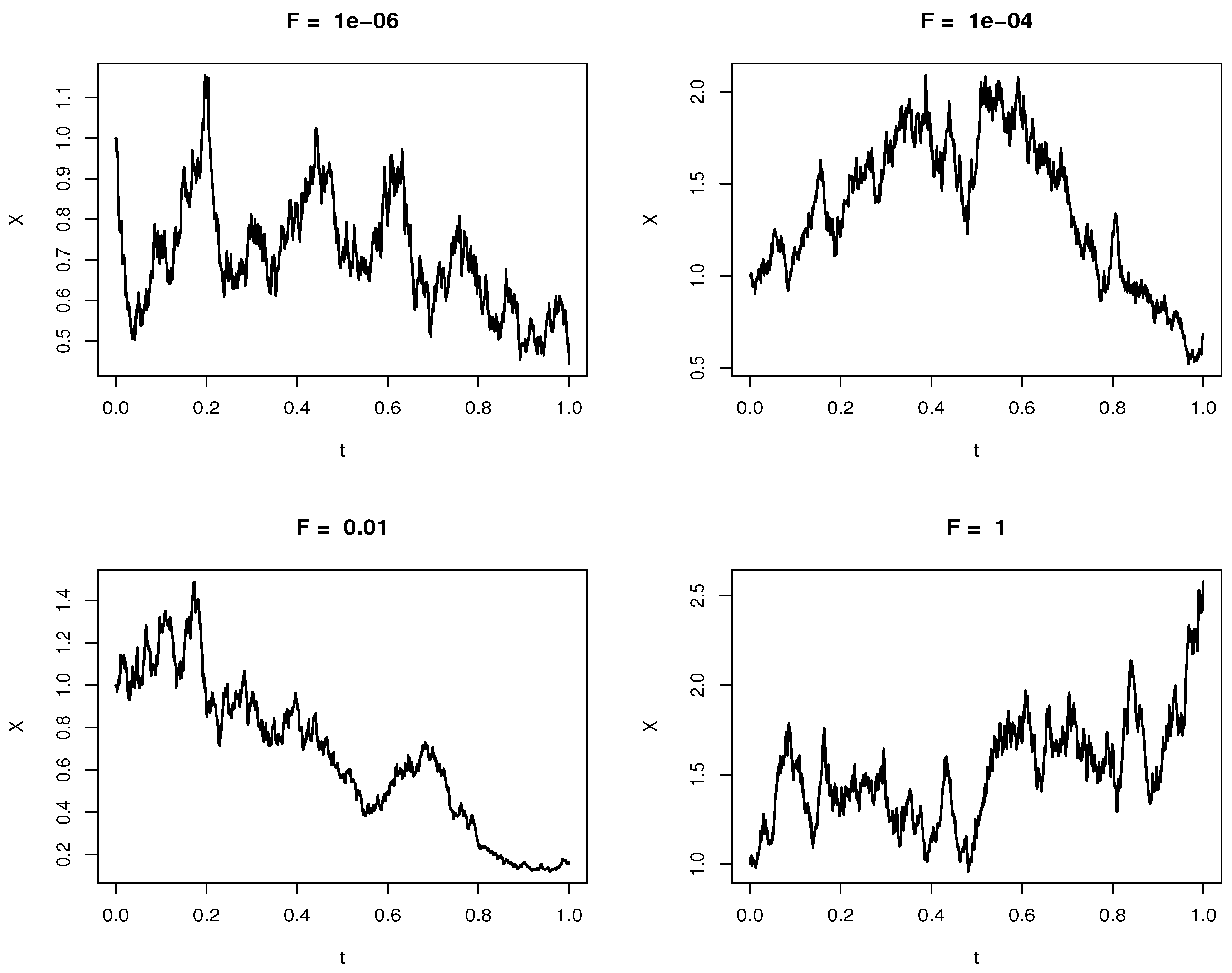
Figure 2 shows the numerical simulations of Equation (
17) with varying external forces
F; the force varies from
N to
N with all other factors kept constant [
15]. These external forces regulate the cellular membrane so that it adjusts to the changing cellular environment by controlling the cell shape. The numerical simulation shows that the random motion exhibits an exponential motion; this motion is highlighted in the analytical solution in Equation (
19). The movement of proteins is evident from the fluid mosaic model; this motion is important in cell communication; the external forces and the drag force facilitate this motion, and the physical forces at the plasma membrane trigger nanoscale membrane deformations that are then translated into chemical signals [
15]. The relationship of the external force with the drag coefficient is highlighted in Equation (
28). Equation (
29) is solved numerically subjected to the boundary and initial conditions stated in Equation (
29); the simulations are carried out for different values of
, and the solutions are presented in line plots of the diffusion advection equation. We start by varying the value of
from
to
; the diffusivity and advective terms in Equation (
29) are spatially dependent exponential functions implying that diffusion grows quicker with varying spatial values. The function
, which is a function of the stochastic process
X and time
t, is a function that describes the diffusion and advection of proteins into and out of the cell membrane as supported by the fluid mosaic model. This movement is essential in cell communication; it results in successful cell binding and also allows cells to recognize foreign cells such as cancerous cells.
Consider Equation (
17); this equation is solved numerically as depicted in
Figure 2 using the Euler–Maruyama method on the interval
, where
by assigning the grid points
with estimated x-values
, where
, we solve the initial value problem of Equation (
17) subjected to the initial condition
; we represent this equation as follows:
where
,
; recall that
where
,
, the set of the solutions
is the approximate solution to the stochastic differential equation
which depends on the random values
. The exact solution in Equation (
17) and the numerically simulated Equation (
29) are exponential; recall that each computed point
is only an approximation to the stochastic differential equation
. Each of these values results in a random variable
T, such that the difference between the exact solution and the approximated solution converges strongly if the expected value of the difference converges to zero, i.e.,
, if
; then, the solution converges strongly, where
is the exact solution of the stochastic differential equation [
16].
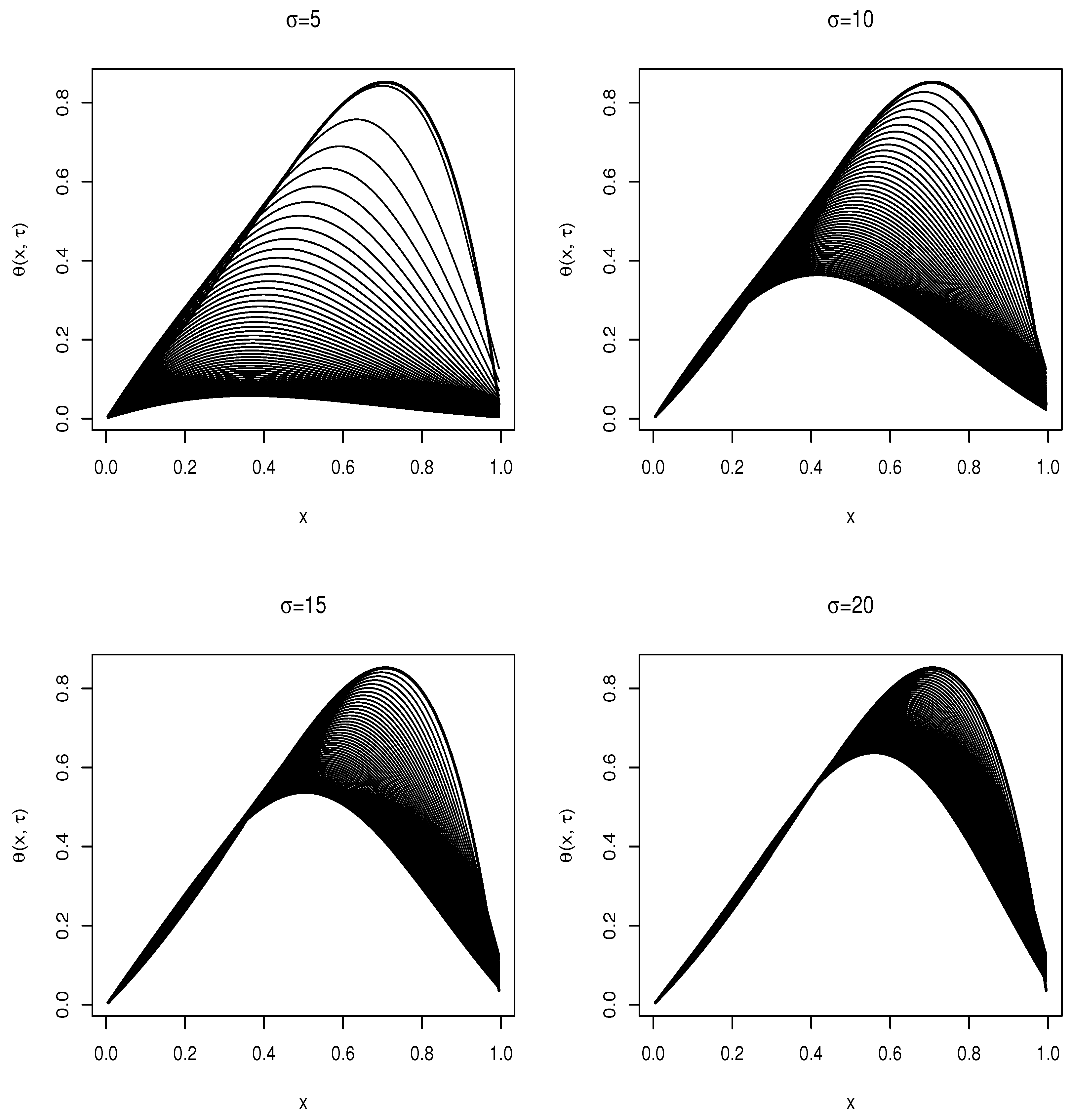
Consider the differential equation in Equation (
29); this is a convection–diffusion equation where the convection and diffusivity terms are spatially dependent; we simulate this equation for varying volatility
, and show the results as line plots in
Figure 3, where the dark bold line represents the initial condition. Consider the limit
; the diffusivity and convection term becomes significantly prominent in the equation; the solution rapidly takes on the values imposed by the boundary condition closer to the upstream value
x = 1. For the limit
, the solution becomes more consistent with the initial conditions imposed.
The function
is a function of the stochastic function
X and time
t; it is a function that describes the diffusion of proteins as a function of the protein’s stochastic motion for successful protein binding during cell communication. The movement of proteins allows the proteins to recognize each other and to also relay information with each other and recognize foreign cells; proteins travel through diffusion and convection. The concept of diffusion is not only limited to proteins, but it also allows other molecules to find each other and bind successfully, such as substrates locating enzymes and DNA binding to proteins; thus, it is clear that diffusion is essential in the overall functioning and well-being of cells. It is worth mentioning that cells are densely packed with proteins, ribosomes, and other biological molecules, such that the distance between two biological molecules is significantly minimal; due to thermal motion, proteins move extremely fast, reaching speeds of about 400 km/h [
11]. This means that delays in cell signaling are significantly minor; the proteins constantly collide with other proteins, thus exhibiting random motion. Because of this high-speed random motion and frequent molecular collision, it is complicated to simulate the reality of the motion exhibited by proteins inside the cell membrane.