Cuproptosis- and m6A-Related lncRNAs for Prognosis of Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets Retrieval
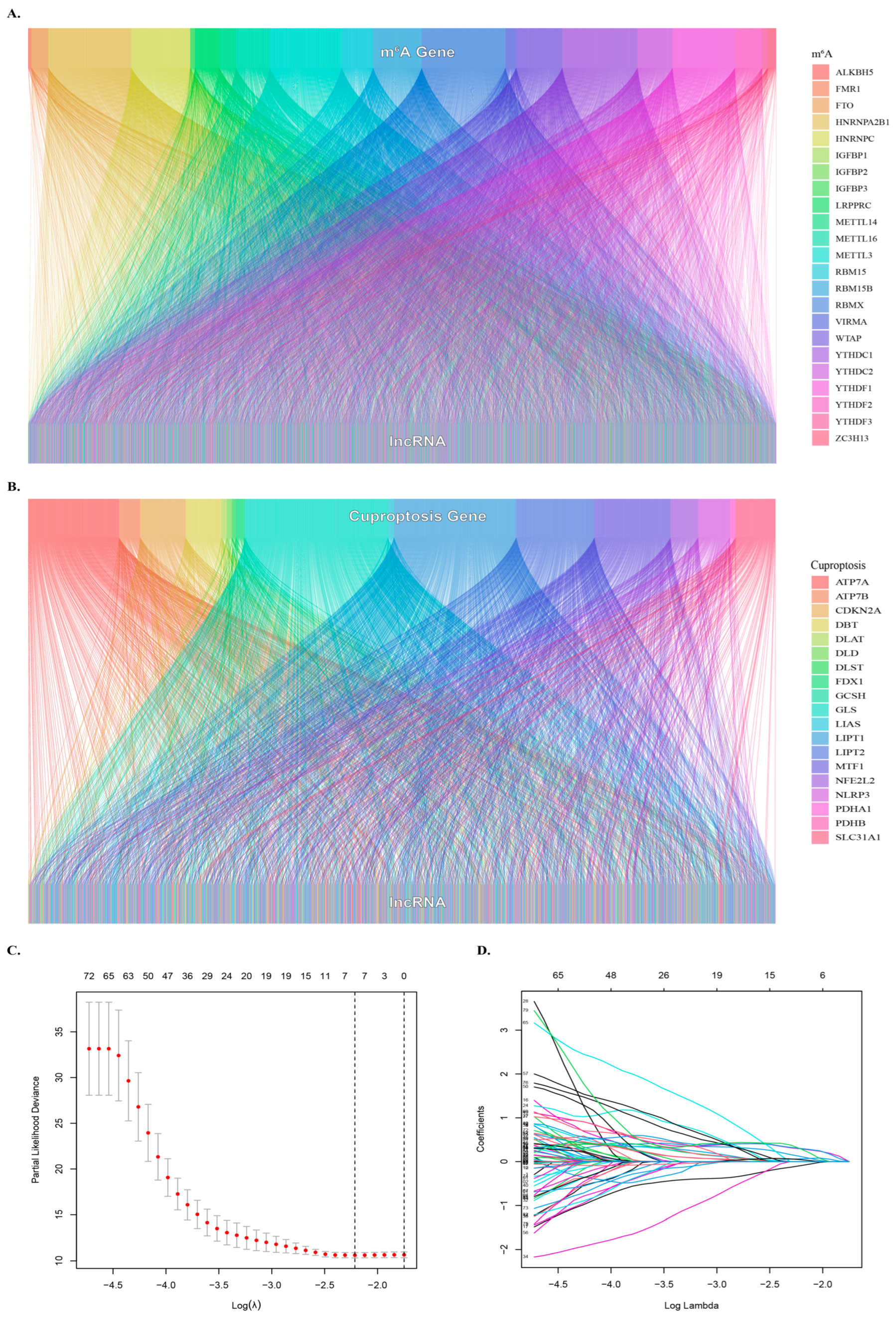
2.2. Identification of Cuproptosis-Related lncRNAs
2.3. Identify the m6A-Related lncRNAs
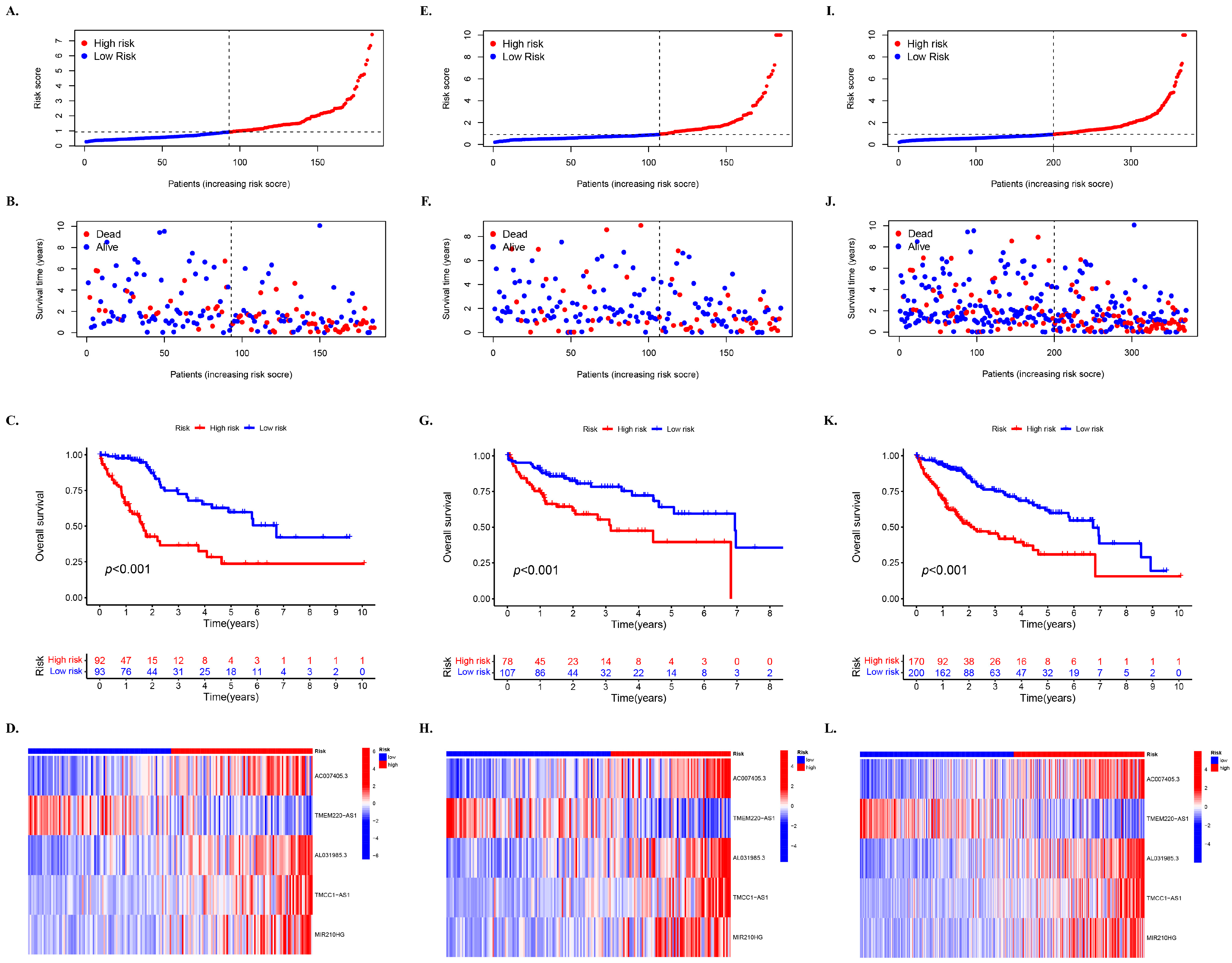
2.4. Construction of m6A and Cuproptosis-Related lncRNA Mortality Risk Model for HCC
2.5. Evaluation of the Grouping Ability of Risk Scores
2.6. Evaluation of the Validity of the Model
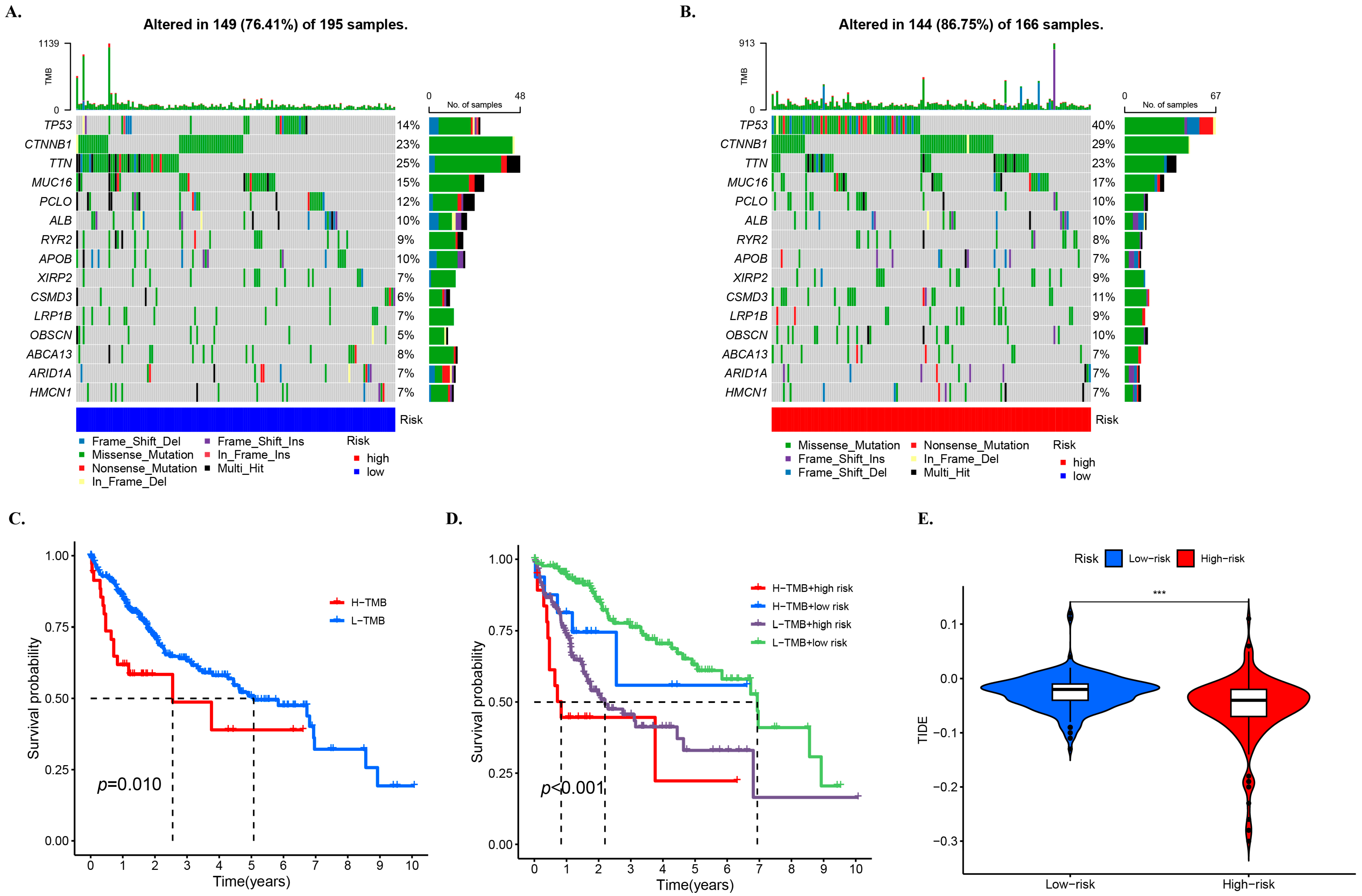
2.7. Oncoplots and Combined Analysis of Other Metrics
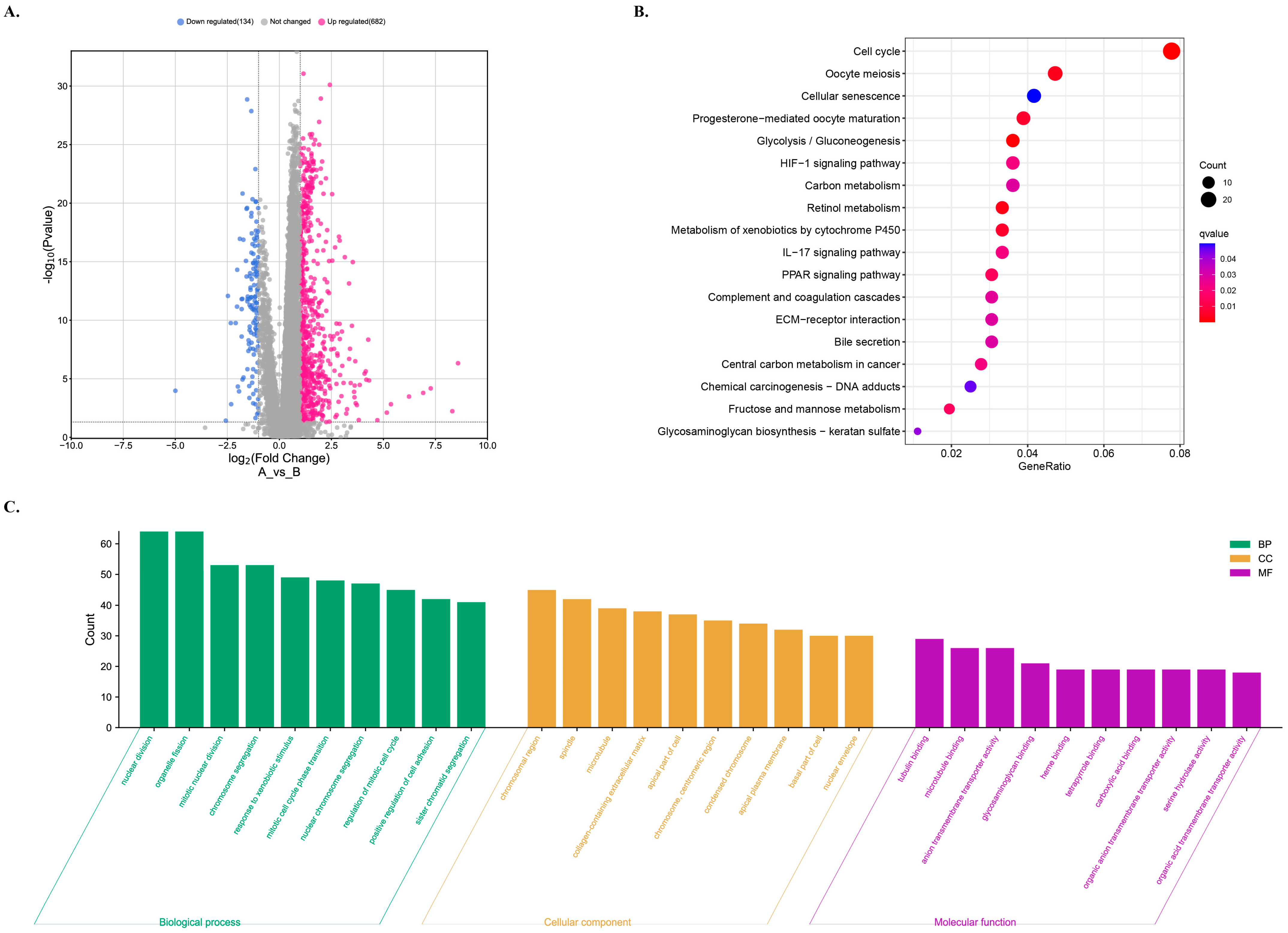
2.8. Exploring Meaningful Gene Function Pathways
2.9. Immunological Infiltration and Drug Screening
2.10. Statistical Analysis
3. Results
3.1. Clinical Sample Information
3.2. Identification of Cuproptosis- and m6A- Related lncRNAs
3.3. Prognostic Impact of Mcplncrnas at Different Risks on HCC
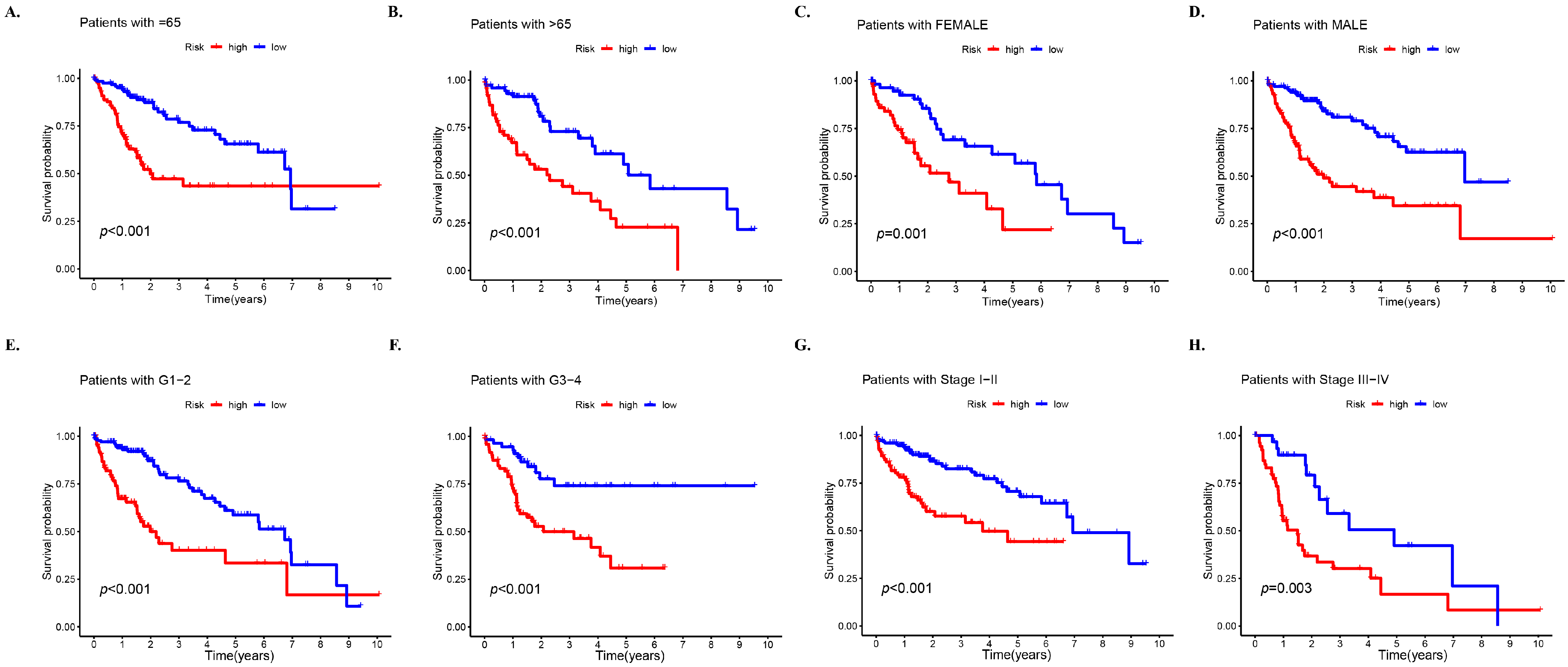
3.4. Prognosis Stratified by Various Clinicopathological Features in Different Risk Subgroups
3.5. PCA of Different Risk Groups Based on Selected Factors
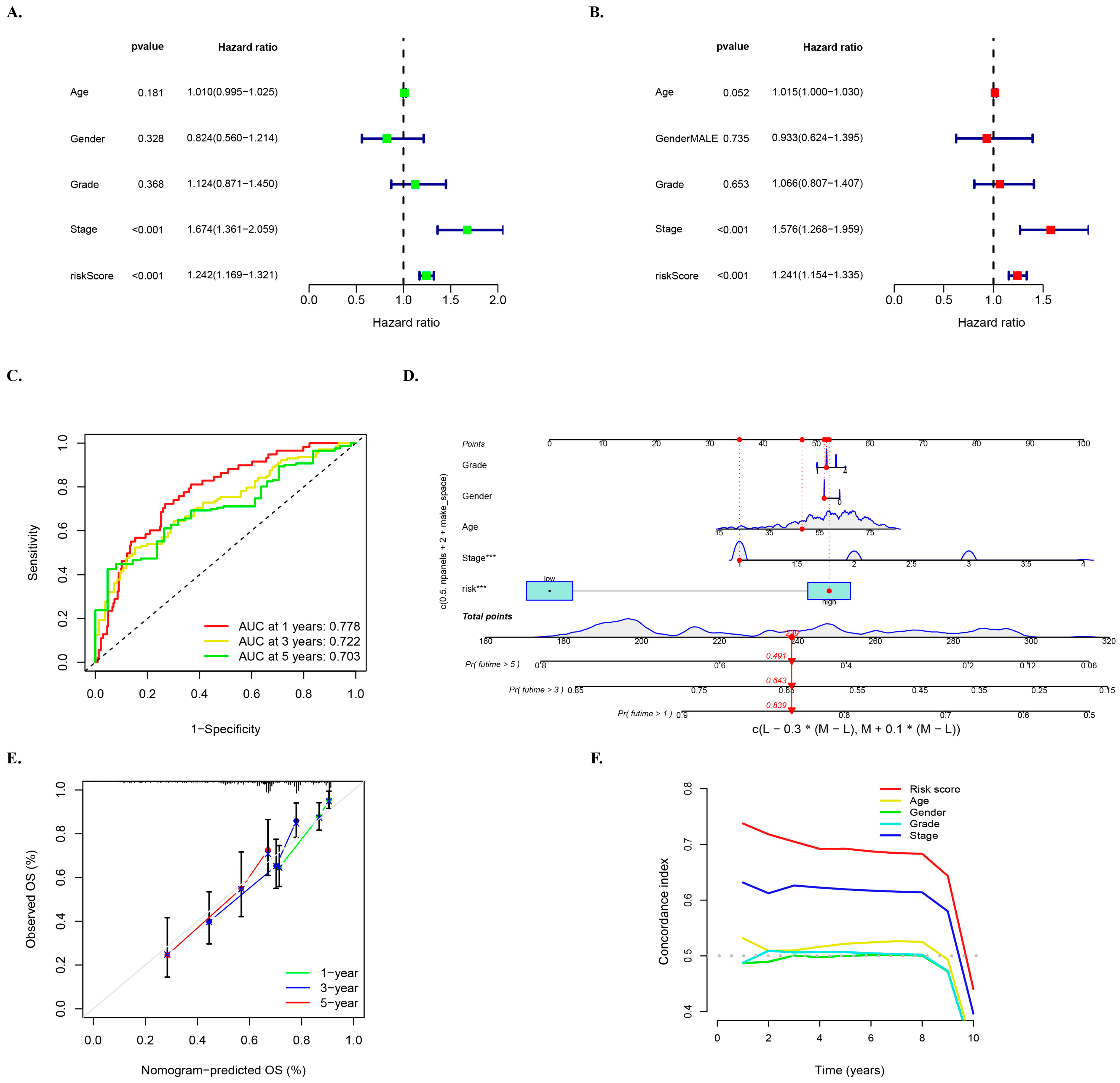
3.6. Multivariate Survival Regression Analyses for Verifying the Prognostic Ability of the Risk Model
3.7. TMB and Related Prognosis in High- and Low-Risk Groups
3.8. GO and KEGG Analyses for the Functional Enrichment of Differential Genes in Different Risk Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ganesan, P.; Kulik, L.M. Hepatocellular Carcinoma: New Developments. Clin. Liver Dis. 2023, 27, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.K.; Guan, X.Y.; Ma, S. Cancer stem cells in hepatocellular carcinoma—From origin to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 26–44. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Meyer, T.; Sapisochin, G.; Salem, R.; Saborowski, A. Hepatocellular carcinoma. Lancet 2022, 400, 1345–1362. [Google Scholar] [CrossRef]
- Liu, D.; Song, T. Changes in and challenges regarding the surgical treatment of hepatocellular carcinoma in China. Biosci. Trends 2021, 15, 142–147. [Google Scholar] [CrossRef]
- Wang, D.X.; Yang, X.; Lin, J.Z.; Bai, Y.; Long, J.Y.; Yang, X.B.; Seery, S.; Zhao, H.T. Efficacy and safety of lenvatinib for patients with advanced hepatocellular carcinoma: A retrospective, real-world study conducted in China. World J. Gastroenterol. 2020, 26, 4465–4478. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Song, T. Conversion therapy and maintenance therapy for primary hepatocellular carcinoma. Biosci. Trends 2021, 15, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Powell, E.E.; Wong, V.W.; Rinella, M. Non-alcoholic fatty liver disease. Lancet 2021, 397, 2212–2224. [Google Scholar] [CrossRef]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021, 73 (Suppl. 1), 4–13. [Google Scholar] [CrossRef]
- Da Silva, D.A.; De Luca, A.; Squitti, R.; Rongioletti, M.; Rossi, L.; Machado, C.; Cerchiaro, G. Copper in tumors and the use of copper-based compounds in cancer treatment. J. Inorg. Biochem. 2022, 226, 111634. [Google Scholar] [CrossRef]
- Ge, E.J.; Bush, A.I.; Casini, A.; Cobine, P.A.; Cross, J.R.; DeNicola, G.M.; Dou, Q.P.; Franz, K.J.; Gohil, V.M.; Gupta, S.; et al. Connecting copper and cancer: From transition metal signalling to metalloplasia. Nat. Rev. Cancer 2022, 22, 102–113. [Google Scholar] [CrossRef]
- Jiang, Y.; Huo, Z.; Qi, X.; Zuo, T.; Wu, Z. Copper-induced tumor cell death mechanisms and antitumor theragnostic applications of copper complexes. Nanomedicine 2022, 17, 303–324. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, J.; Wu, C.; Wang, L.; Chen, Z.S.; Cui, W. The combination of disulfiram and copper for cancer treatment. Drug Discov. Today 2020, 25, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tang, L.; Huang, W.; Abisola, F.H.; Zhang, Y.; Zhang, G.; Yao, L. Identification of a prognostic cuproptosis-related signature in hepatocellular carcinoma. Biol. Direct 2023, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Y.; Ye, S.; Feng, H.; Ma, L. Development and validation of cuproptosis-related gene signature in the prognostic prediction of liver cancer. Front. Oncol. 2022, 12, 985484. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Wang, L.; Zhang, N.; Xu, W.; Zhou, J.; Zhao, Y.; Zhu, W.; Zhang, T.; Wang, L. Development and experimental verification of a prognosis model for cuproptosis-related subtypes in HCC. Hepatol. Int. 2022, 16, 1435–1447. [Google Scholar] [CrossRef]
- Zhang, C.; Zeng, Y.; Guo, X.; Shen, H.; Zhang, J.; Wang, K.; Ji, M.; Huang, S. Pan-cancer analyses confirmed the cuproptosis-related gene FDX1 as an immunotherapy predictor and prognostic biomarker. Front. Genet. 2022, 13, 923737. [Google Scholar] [CrossRef]
- Zhang, Z.; Zeng, X.; Wu, Y.; Liu, Y.; Zhang, X.; Song, Z. Cuproptosis-Related Risk Score Predicts Prognosis and Characterizes the Tumor Microenvironment in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 925618. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, J.; Yin, S.; Shi, J.; Zheng, M.; He, C.; Meng, H.; Han, Y.; Han, J.; Guo, J.; et al. The expression of cuproptosis-related genes in hepatocellular carcinoma and their relationships with prognosis. Front. Oncol. 2022, 12, 992468. [Google Scholar] [CrossRef]
- Wang, X.X.; Wu, L.H.; Ji, H.; Liu, Q.Q.; Deng, S.Z.; Dou, Q.Y.; Ai, L.; Pan, W.; Zhang, H.M. A novel cuproptosis-related prognostic signature and potential value in HCC immunotherapy. Front. Mol. Biosci. 2022, 9, 1001788. [Google Scholar] [CrossRef]
- Lin, Z.; Niu, Y.; Wan, A.; Chen, D.; Liang, H.; Chen, X.; Sun, L.; Zhan, S.; Chen, L.; Cheng, C.; et al. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020, 39, e103181. [Google Scholar] [CrossRef]
- Ally, A.; Balasundaram, M.; Carlsen, R.; Chuah, E.; Clarke, A.; Dhalla, N.; Holt, R.A.; Jones, S.J.M.; Lee, D.; Ma, Y.; et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341.e23. [Google Scholar] [CrossRef] [PubMed]
- Montironi, C.; Castet, F.; Haber, P.K.; Pinyol, R.; Torres-Martin, M.; Torrens, L.; Mesropian, A.; Wang, H.; Puigvehi, M.; Maeda, M.; et al. Inflamed and non-inflamed classes of HCC: A revised immunogenomic classification. Gut 2023, 72, 129–140. [Google Scholar] [CrossRef]
- Hon, K.W.; Abu, N.; Ab Mutalib, N.S.; Jamal, R. miRNAs and lncRNAs as Predictive Biomarkers of Response to FOLFOX Therapy in Colorectal Cancer. Front. Pharmacol. 2018, 9, 846. [Google Scholar] [CrossRef] [PubMed]
- Muhammad Yusuf, A.N.; Raja Ali, R.A.; Muhammad Nawawi, K.N.; Mokhtar, N.M. Potential biomarkers in NASH-induced liver cirrhosis with hepatocellular carcinoma: A preliminary work on roles of exosomal miR-182, miR-301a, and miR-373. Malays. J. Pathol. 2020, 42, 377–384. [Google Scholar]
- Azit, N.A.; Sahran, S.; Voon Meng, L.; Subramaniam, M.; Mokhtar, S.; Mohammed Nawi, A. Risk factors of hepatocellular carcinoma in type 2 diabetes patients: A two-centre study in a developing country. PLoS ONE 2021, 16, e0260675. [Google Scholar] [CrossRef]
- Chen, B.; Yang, Z.; Lang, Z.; Tao, Q.; Zhang, R.; Zhan, Y.; Xu, X.; Zhu, K.; Zheng, J.; Yu, Z.; et al. M6A-related lncRNAs predict clinical outcome and regulate the tumor immune microenvironment in hepatocellular carcinoma. BMC Cancer 2022, 22, 867. [Google Scholar] [CrossRef]
- Hao, K.; Li, J.; Zhang, Y.; Zhao, W.; Chen, X.; Xu, J.; Tian, Y.; Li, X.; Fen, J.; He, X. Expression and prognostic signatures of m6A-related lncRNAs in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 4429–4441. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef]
- Chakraborty, E.; Sarkar, D. Emerging Therapies for Hepatocellular Carcinoma (HCC). Cancers 2022, 14, 2798. [Google Scholar] [CrossRef]
- Wu, Y.; Hao, X.; Wei, H.; Sun, R.; Chen, Y.; Tian, Z. Blockade of T-cell receptor with Ig and ITIM domains elicits potent antitumor immunity in naturally occurring HBV-related HCC in mice. Hepatology 2023, 77, 965–981. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, S.; Tümen, D.; Volz, B.; Neumeyer, K.; Egler, N.; Kunst, C.; Tews, H.C.; Schmid, S.; Kandulski, A.; Müller, M.; et al. HCC biomarkers—State of the old and outlook to future promising biomarkers and their potential in everyday clinical practice. Front. Oncol. 2022, 12, 1016952. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wang, H.; Chen, J.; Zhu, H. Sinomenine improve diabetic nephropathy by inhibiting fibrosis and regulating the JAK2/STAT3/SOCS1 pathway in streptozotocin-induced diabetic rats. Life Sci. 2021, 265, 118855. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Shen, J.; Qiao, X.; Gao, Y.; Su, H.B.; Zhang, S. Long Non-Coding RNA Signatures Associated with Ferroptosis Predict Prognosis in Colorectal Cancer. Int. J. Gen. Med. 2022, 15, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Tang, Y.; Lu, J.; Zhuang, Y.; Wang, J. MIR210HG promotes breast cancer progression by IGF2BP1 mediated m6A modification. Cell Biosci. 2022, 12, 38. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, Y.; Huang, W. Identification of immune-related lncRNA signature for predicting immune checkpoint blockade and prognosis in hepatocellular carcinoma. Int. Immunopharmacol. 2021, 92, 107333. [Google Scholar] [CrossRef]
- Zhu, H.; Mao, F.; Wang, K.; Feng, J.; Cheng, S. Cuproptosis-related lncRNAs predict the clinical outcome and immune characteristics of hepatocellular carcinoma. Front. Genet. 2022, 13, 972212. [Google Scholar] [CrossRef]

| Covariates | Type | Total | Test | Train | p Value |
|---|---|---|---|---|---|
| Age | ≤65 | 232 (62.7%) | 116 (62.7%) | 116 (62.7%) | 1 |
| >65 | 138 (37.3%) | 69 (37.3%) | 69 (37.3%) | ||
| Gender | FEMALE | 121 (32.7%) | 59 (31.89%) | 62 (33.51%) | 0.82 |
| MALE | 249 (67.3%) | 126 (68.11%) | 123 (66.49%) | ||
| Grade | G1 | 55 (14.86%) | 23 (12.43%) | 32 (17.3%) | 0.37 |
| G2 | 177 (47.84%) | 92 (49.73%) | 85 (45.95%) | ||
| G3 | 121 (32.7%) | 59 (31.89%) | 62 (33.51%) | ||
| G4 | 12 (3.24%) | 8 (4.32%) | 4 (2.16%) | ||
| unknow | 5 (1.35%) | 3 (1.62%) | 2 (1.08%) | ||
| Stage | Stage I | 171 (46.22%) | 87 (47.03%) | 84 (45.41%) | 0.24 |
| Stage II | 85 (22.97%) | 49 (26.49%) | 36 (19.46%) | ||
| Stage III | 85 (22.97%) | 36 (19.46%) | 49 (26.49%) | ||
| Stage IV | 5 (1.35%) | 2 (1.08%) | 3 (1.62%) | ||
| unknow | 24 (6.49%) | 11 (5.95%) | 13 (7.03%) | ||
| T | T1 | 181 (48.92%) | 91 (49.19%) | 90 (48.65%) | 0.1 |
| T2 | 93 (25.14%) | 55 (29.73%) | 38 (20.54%) | ||
| T3 | 80 (21.62%) | 33 (17.84%) | 47 (25.41%) | ||
| T4 | 13 (3.51%) | 5 (2.7%) | 8 (4.32%) | ||
| unknow | 3 (0.81%) | 1 (0.54%) | 2 (1.08%) | ||
| M | M0 | 266 (71.89%) | 129 (69.73%) | 137 (74.05%) | 1 |
| M1 | 4 (1.08%) | 2 (1.08%) | 2 (1.08%) | ||
| unknow | 100 (27.03%) | 54 (29.19%) | 46 (24.86%) | ||
| N | N0 | 252 (68.11%) | 121 (65.41%) | 131 (70.81%) | 0.57 |
| N1 | 4 (1.08%) | 3 (1.62%) | 1 (0.54%) | ||
| unknow | 114 (30.81%) | 61 (32.97%) | 53 (28.65%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Tan, J.K.; Goon, J.A. Cuproptosis- and m6A-Related lncRNAs for Prognosis of Hepatocellular Carcinoma. Biology 2023, 12, 1101. https://doi.org/10.3390/biology12081101
Zhu Y, Tan JK, Goon JA. Cuproptosis- and m6A-Related lncRNAs for Prognosis of Hepatocellular Carcinoma. Biology. 2023; 12(8):1101. https://doi.org/10.3390/biology12081101
Chicago/Turabian StyleZhu, Yuezhi, Jen Kit Tan, and Jo Aan Goon. 2023. "Cuproptosis- and m6A-Related lncRNAs for Prognosis of Hepatocellular Carcinoma" Biology 12, no. 8: 1101. https://doi.org/10.3390/biology12081101
APA StyleZhu, Y., Tan, J. K., & Goon, J. A. (2023). Cuproptosis- and m6A-Related lncRNAs for Prognosis of Hepatocellular Carcinoma. Biology, 12(8), 1101. https://doi.org/10.3390/biology12081101






