Mid-Pleistocene Transitions Forced Himalayan ibex to Evolve Independently after Split into an Allopatric Refugium
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Sequencing, Variant Calling, and Phylogeny Based on Nuclear Variants
2.2. Assembling Mitogenomes and Phylogeny
2.3. Population Genetic Assignment, Admixture, and Gene Flow
2.4. Demographic Estimations and Population Divergence
2.5. Genetic Affinity and Gene Flow
2.6. Modeling Geo-Climatic Changes
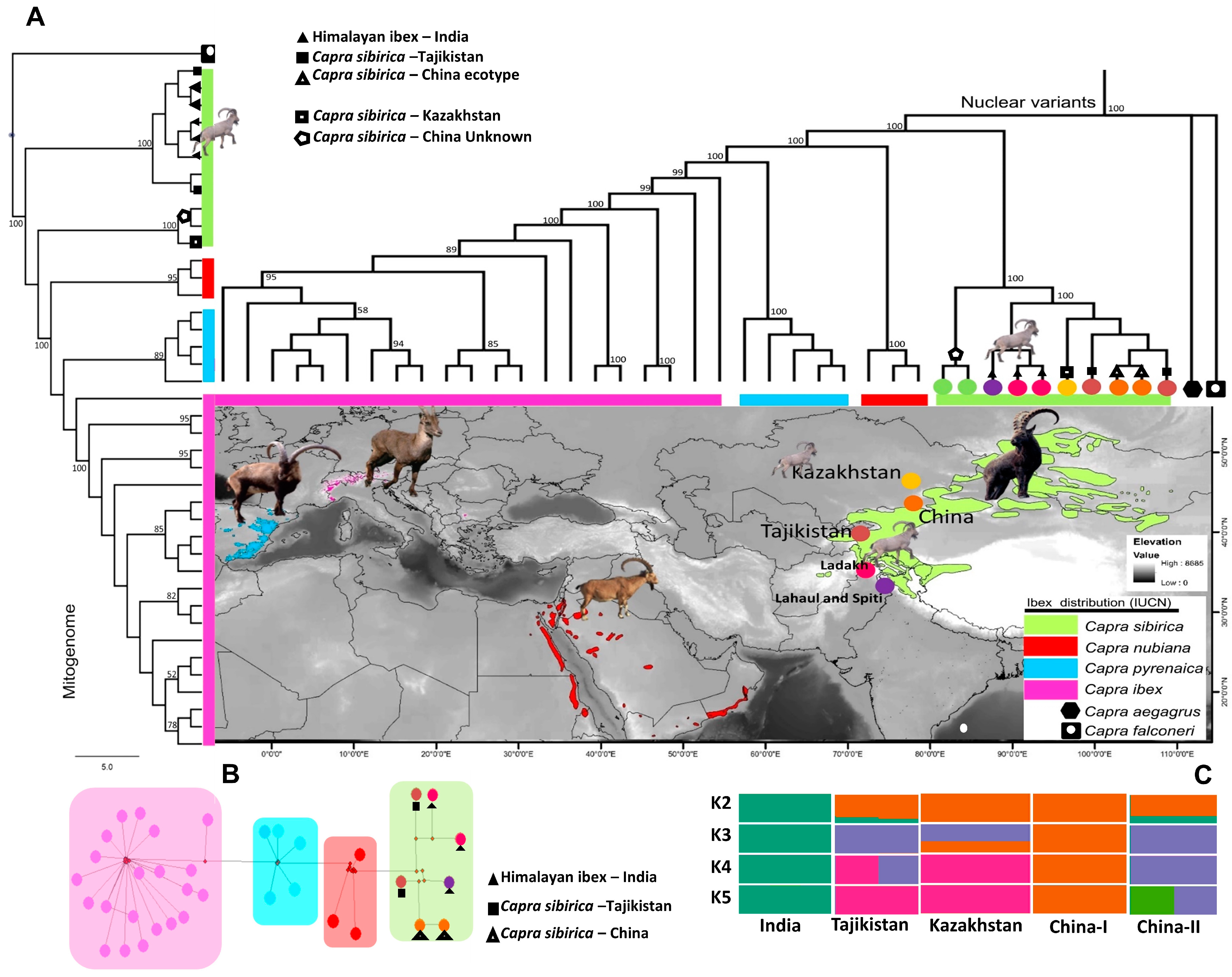
3. Results
3.1. Phylogeny and Population Genetic Assignment of Himalayan ibex
3.2. Demographic History, Genetic Divergence, and Gene Flow
3.3. Paleo-Geo Climatic Modeling
4. Discussion
4.1. Phylogeny and Population Genetic Assignment of Himalayan ibex
4.2. Demographic History, Genetic Divergence, and Gene Flow
4.3. Paleo-Geo Climatic Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jablonski, D. Species Selection: Theory and Data. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 501–524. [Google Scholar] [CrossRef]
- Bibi, F.; Kiessling, W. Continuous Evolutionary Change in Plio-Pleistocene Mammals of Eastern Africa. Proc. Natl. Acad. Sci. USA 2015, 112, 10623–10628. [Google Scholar] [CrossRef]
- Luo, S.; Wu, Y.; Chang, Q.; Liu, Y.; Yang, X.; Zhang, Z.; Zhang, M.; Zhang, Q.; Zou, F. Deep Phylogeographic Divergence of a Migratory Passerine in Sino-Himalayan and Siberian Forests: The Red-Flanked Bluetail (Tarsiger cyanurus) Complex. Ecol. Evol. 2014, 4, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Wilting, A.; Courtiol, A.; Christiansen, P.; Niedballa, J.; Scharf, A.K.; Orlando, L.; Balkenhol, N.; Hofer, H.; Kramer-Schadt, S.; Fickel, J.; et al. Planning Tiger Recovery: Understanding Intraspecific Variation for Effective Conservation. Sci. Adv. 2015, 1, e1400175. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Schättin, E.W.; McShea, W.J. Globally Common, Locally Rare: Revisiting Disregarded Genetic Diversity for Conservation Planning of Widespread Species. Biodivers. Conserv. 2018, 27, 3031–3035. [Google Scholar] [CrossRef]
- Roca, A.L.; Georgiadis, N.; Pecon-Slattery, J.; O’Brien, S.J. Genetic Evidence for Two Species of Elephant in Africa. Science 2001, 293, 1473–1477. [Google Scholar] [CrossRef]
- Janecka, J.E.; Zhang, Y.; Li, D.; Munkhtsog, B.; Bayaraa, M.; Galsandorj, N.; Wangchuk, T.R.; Karmacharya, D.; Li, J.; Lu, Z.; et al. Range-Wide Snow Leopard Phylogeography Supports Three Subspecies. J. Hered. 2017, 108, 597–607. [Google Scholar] [CrossRef]
- Lee, Y.S.; Markov, N.; Argunov, A.; Voloshina, I.; Bayarlkhagva, D.; Kim, B.J.; Min, M.S.; Lee, H.; Kim, K.S. Genetic Diversity and Phylogeography of Siberian Roe Deer, Caproulus Pygargus, in Central and Peripheral Populations. Ecol. Evol. 2016, 6, 7286–7297. [Google Scholar] [CrossRef]
- Hu, Y.; Thapa, A.; Fan, H.; Ma, T.; Wu, Q.; Ma, S.; Zhang, D.; Wang, B.; Li, M.; Yan, L.; et al. Genomic Evidence for Two Phylogenetic Species and Long-Term Population Bottlenecks in Red Pandas. Sci. Adv. 2020, 6, eaax5751. [Google Scholar] [CrossRef]
- Frantz, L.A.F.; Schraiber, J.G.; Madsen, O.; Megens, H.J.; Bosse, M.; Paudel, Y.; Semiadi, G.; Meijaard, E.; Li, N.; Crooijmans, R.P.M.A.; et al. Genome Sequencing Reveals Fine Scale Diversification and Reticulation History during Speciation in Sus. Genome Biol. 2013, 14, R107. [Google Scholar] [CrossRef] [PubMed]
- Muellner-Riehl, A.N. Mountains as Evolutionary Arenas: Patterns, Emerging Approaches, Paradigm Shifts, and Their Implications for Plant Phylogeographic Research in the Tibeto-Himalayan Region. Front. Plant Sci. 2019, 10, 195. [Google Scholar] [CrossRef]
- de Jong, M.J.; Li, Z.; Qin, Y.; Quéméré, E.; Baker, K.; Wang, W.; Hoelzel, A.R. Demography and Adaptation Promoting Evolutionary Transitions in a Mammalian Genus That Diversified during the Pleistocene. Mol. Ecol. 2020, 29, 2777–2792. [Google Scholar] [CrossRef]
- Kaya, S.; Kabasakal, B.; Erdoğan, A. Geographic Genetic Structure of Alectoris Chukar in Türkiye: Post-LGM-Induced Hybridization and Human-Mediated Contaminations. Biology 2023, 12, 401. [Google Scholar] [CrossRef] [PubMed]
- Sharief, A.; Joshi, B.D.; Kumar, V.; Singh, H.; Singh, V.K.; Dar, S.A.; Graham, C.; Ramesh, C.; Quyoom, I.; Thakur, M.; et al. Empirical Data Suggest That the Kashmir Musk Deer (Moschus Cupreus, Grubb 1982) Is the One Musk Deer Distributed in the Western Himalayas: An Integration of Ecology, Genetics and Geospatial Modelling Approaches. Biology 2023, 12, 786. [Google Scholar] [CrossRef]
- Fedosenko, A.K.; Blank, D.A. Capra Sibirica. Mamm. Species 2001, 675, 1–13. [Google Scholar] [CrossRef]
- Reading, R.; Michel, S.; Suryawanshi, K.; Bhatnagar, Y. Capra Sibirica, Siberian Ibex. Available online: https://doi.org/10.2305/IUCN.UK.2020-2.RLTS.T42398A22148720.en (accessed on 13 October 2021).
- Smith, A.T.; Xie, Y. A Field Guide to the Mammals of China; Princeton University Press: Princeton, NJ, USA, 2009. [Google Scholar]
- Wilson, D.E.; Reeder, D.M. Mammal Species of the World: A Taxonomic and Geographic Reference; JHU Press: Baltimore, MD, USA, 2005; Volume 1. [Google Scholar]
- Joshi, B.D.; Jabin, G.; Sharief, A.; Kumar, V.; Mukherjee, T.; Kumar, M.; Singh, A.; Singh, S.K.; Chandra, K.; Sharma, L.K.; et al. Genetic Evidence for Allopatric Speciation of the Siberian Ibex Capra Sibirica in India. Endanger. Species Res. 2020, 42, 1–5. [Google Scholar] [CrossRef]
- Carstens, B.C.; Knowles, L.L. Estimating Species Phylogeny from Gene-Tree Probabilities despite Incomplete Lineage Sorting: An Example from Melanoplus Grasshoppers. Syst. Biol. 2007, 56, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Degnan, J.H.; Rosenberg, N.A. Gene Tree Discordance, Phylogenetic Inference and the Multispecies Coalescent. Trends Ecol. Evol. 2009, 24, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.S.; Wang, S.; Li, Y.; Jhala, Y.; Thakur, M.; Otecko, N.O.; Si, J.F.; Chen, H.M.; Shapiro, B.; Nielsen, R.; et al. Ancient Hybridization with an Unknown Population Facilitated High-Altitude Adaptation of Canids. Mol. Biol. Evol. 2020, 37, 2616–2629. [Google Scholar] [CrossRef]
- Coimbra, R.T.F.; Winter, S.; Kumar, V.; Koepfli, K.P.; Gooley, R.M.; Dobrynin, P.; Fennessy, J.; Janke, A. Whole-Genome Analysis of Giraffe Supports Four Distinct Species. Curr. Biol. 2021, 31, 2929–2938.e5. [Google Scholar] [CrossRef]
- Grossen, C.; Guillaume, F.; Keller, L.F.; Croll, D. Purging of Highly Deleterious Mutations through Severe Bottlenecks in Alpine Ibex. Nat. Commun. 2020, 11, 1001. [Google Scholar] [CrossRef]
- Head, M.J.; Gibbard, P.L. Early-Middle Pleistocene Transitions: Linking Terrestrial and Marine Realms. Quat. Int. 2015, 389, 7–46. [Google Scholar] [CrossRef]
- Bishop, M.P.; Björnsson, H.; Haeberli, W.; Oerlemans, J.; Shroder, J.F.; Tranter, M. Encyclopedia of Snow, Ice and Glaciers; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Otecko, N.O.; Wang, S.; Wu, D.; Yang, M.; Xu, Y.; Murphy, R.W.; Peng, M.; Zhang, Y. An Evolutionary Genomic Perspective on the Breeding of Dwarf Chickens. Mol. Biol. Evol. 2017, 34, 3081–3088. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Korneliussen, T.S.; Albrechtsen, A.; Nielsen, R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinform. 2014, 15, 356. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-Wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE Algorithm for Individual Ancestry Estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nadachowska-Brzyska, K.; Burri, R.; Smeds, L.; Ellegren, H. PSMC Analysis of Effective Population Sizes in Molecular Ecology and Its Application to Black-and-White Ficedula Flycatchers. Mol. Ecol. 2016, 25, 1058–1072. [Google Scholar] [CrossRef]
- Brambilla, A.; Biebach, I.; Bassano, B.; Bogliani, G.; von Hardenberg, A. Direct and Indirect Causal Effects of Heterozygosity on Fitness-Related Traits in Alpine Ibex. Proc. R. Soc. B Biol. Sci. 2014, 282, 20141873. [Google Scholar] [CrossRef]
- Chen, L.; Qiu, Q.; Jiang, Y.; Wang, K.; Lin, Z.; Li, Z.; Bibi, F.; Yang, Y.; Wang, J.; Nie, W.; et al. Large-Scale Ruminant Genome Sequencing Provides Insights into Their Evolution and Distinct Traits. Science 2019, 364, eaav6202. [Google Scholar] [CrossRef] [PubMed]
- Schiffels, S.; Durbin, R. Inferring Human Population Size and Separation History from Multiple Genome Sequences. Nat. Genet. 2014, 46, 919–925. [Google Scholar] [CrossRef]
- Browning, B.L.; Browning, S.R. Genotype Imputation with Millions of Reference Samples. Am. J. Hum. Genet. 2016, 98, 116–126. [Google Scholar] [CrossRef]
- Malaspinas, A.S.; Westaway, M.C.; Muller, C.; Sousa, V.C.; Lao, O.; Alves, I.; Bergström, A.; Athanasiadis, G.; Cheng, J.Y.; Crawford, J.E.; et al. A Genomic History of Aboriginal Australia. Nature 2016, 538, 207–214. [Google Scholar] [CrossRef]
- Pickrell, J.; Pritchard, J. Inference of population splits and mixtures from genome-wide allele frequency data. Nat. Preced. 2012. [Google Scholar] [CrossRef]
- Green, R.E.; Krause, J.; Briggs, A.W.; Maricic, T.; Stenzel, U.; Kircher, M.; Patterson, N.; Li, H.; Zhai, W.; Fritz, M.H.Y.; et al. A Draft Sequence of the Neandertal Genome. Science 2010, 328, 710–722. [Google Scholar] [CrossRef]
- Scotese, C.R.; Wright, N. PALEOMAP Paleodigital Elevation Models (PaleoDEMS) for the Phanerozoic. Available online: https://www.earthbyte.org/webdav/ftp/Data_Collections/Scotese_Wright_2018_PaleoDEM/Scotese_Wright2018_PALEOMAP_PaleoDEMs.pdf (accessed on 22 September 2021).
- Evans, J.S.; Oakleaf, J.; Cushman, S.A.; Theobald, D. An ArcGIS Toolbox for Surface Gradient and Geomorphometric Modeling, Version 2.0-0. 2014. Available online: https://evansmurphy.wixsite.com/evansspatialevansspatial (accessed on 25 June 2019).
- Ellegren, H.; Galtier, N. Determinants of Genetic Diversity. Nat. Rev. Genet. 2016, 17, 422–433. [Google Scholar] [CrossRef]
- Clark, P.U.; Archer, D.; Pollard, D.; Blum, J.D.; Rial, J.A.; Brovkin, V.; Mix, A.C.; Pisias, N.G.; Roy, M. The Middle Pleistocene Transition: Characteristics, Mechanisms, and Implications for Long-Term Changes in Atmospheric PCO2. Quat. Sci. Rev. 2006, 25, 3150–3184. [Google Scholar] [CrossRef]
- Hewitt, G. The Genetic Legacy of the Quaternary Ice Ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Qiu, Q.; Zhang, G.; Ma, T.; Qian, W.; Ye, Z.; Cao, C.; Hu, Q.; Kim, J.; Larkin, D.M.; Auvil, L.; et al. The Yak Genome and Adaptation to Life at High Altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef]
- Past Interglacials Working Group of PAGES. Interglacials of the Last 800,000 Years. Rev. Geophys. 2016, 54, 162–219. [Google Scholar] [CrossRef]
- Zheng, B.; Xu, Q.; Shen, Y. The Relationship between Climate Change and Quaternary Glacial Cycles on the Qinghai-Tibetan Plateau: Review and Speculation. Quat. Int. 2002, 97–98, 93–101. [Google Scholar] [CrossRef]
- Bird, M.I.; Taylor, D.; Hunt, C. Palaeoenvironments of Insular Southeast Asia during the Last Glacial Period: A Savanna Corridor in Sundaland? Quat. Sci. Rev. 2005, 24, 2228–2242. [Google Scholar] [CrossRef]
- Wurster, C.M.; Bird, M.I.; Bull, I.D.; Creed, F.; Bryant, C.; Dungait, J.A.J.; Paz, V. Forest Contraction in North Equatorial Southeast Asia during the Last Glacial Period. Proc. Natl. Acad. Sci. USA 2010, 107, 15508–15511. [Google Scholar] [CrossRef] [PubMed]
- Owen, L.A.; Caffee, M.W.; Finkel, R.C.; Seong, Y.B. Quaternary Glaciation of the Himalayan--Tibetan Orogen. J. Quat. Sci. Publ. Quat. Res. Assoc. 2008, 23, 513–531. [Google Scholar] [CrossRef]
- Shi, Y.; Ren, B.; Wang, J.; Derbyshire, E. Quaternary Glaciation in China. Quat. Sci. Rev. 1986, 5, 503–507. [Google Scholar] [CrossRef]
- Zhao, S.; Zheng, P.; Dong, S.; Zhan, X.; Wu, Q.; Guo, X.; Hu, Y.; He, W.; Zhang, S.; Fan, W.; et al. Whole-Genome Sequencing of Giant Pandas Provides Insights into Demographic History and Local Adaptation. Nat. Genet. 2013, 45, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Wang, L.; Wang, K.; Yang, Y.; Ma, T.; Wang, Z.; Zhang, X.; Ni, Z.; Hou, F.; Long, R.; et al. Yak Whole-Genome Resequencing Reveals Domestication Signatures and Prehistoric Population Expansions. Nat. Commun. 2015, 6, 10283. [Google Scholar] [CrossRef] [PubMed]
- Saijo, K.; Tanaka, S. Paleosols of Middle Holocene Age in the Thakkhola Basin, Central Nepal, and Their Paleoclimatic Significance. J. Asian Earth Sci. 2002, 21, 323–329. [Google Scholar] [CrossRef]
- Miehe, G.; Miehe, S.; Schlütz, F. Early Human Impact in the Forest Ecotone of Southern High Asia (Hindu Kush, Himalaya). Quat. Res. 2009, 71, 255–265. [Google Scholar] [CrossRef]
- Abramowski, U.; Bergau, A.; Seebach, D.; Zech, R.; Glaser, B.; Sosin, P.; Kubik, P.W.; Zech, W. Pleistocene Glaciations of Central Asia: Results from 10Be Surface Exposure Ages of Erratic Boulders from the Pamir (Tajikistan), and the Alay-Turkestan Range (Kyrgyzstan). Quat. Sci. Rev. 2006, 25, 1080–1096. [Google Scholar] [CrossRef]
- Jin, H.; Vandenberghe, J.; Luo, D.; Harris, S.A.; He, R.; Chen, X.; Jin, X.; Wang, Q.; Zhang, Z.; Spektor, V.; et al. Quaternary Permafrost in China: Framework and Discussions. Quaternary 2020, 3, 32. [Google Scholar] [CrossRef]
- Wang, M.S.; Thakur, M.; Peng, M.S.; Jiang, Y.; Frantz, L.A.F.; Li, M.; Zhang, J.J.; Wang, S.; Peters, J.; Otecko, N.O.; et al. 863 Genomes Reveal the Origin and Domestication of Chicken. Cell Res. 2020, 30, 693–701. [Google Scholar] [CrossRef]
- Wang, M.S.; Zhang, J.J.; Guo, X.; Li, M.; Meyer, R.; Ashari, H.; Zheng, Z.Q.; Wang, S.; Peng, M.S.; Jiang, Y.; et al. Large-Scale Genomic Analysis Reveals the Genetic Cost of Chicken Domestication. BMC Biol. 2021, 19, 118. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Sinding, M.H.S.; Ramos-Madrigal, J.; Niemann, J.; Samaniego Castruita, J.A.; Vieira, F.G.; Carøe, C.; de Manuel Montero, M.; Kuderna, L.; Serres, A.; et al. Interspecific Gene Flow Shaped the Evolution of the Genus Canis. Curr. Biol. 2018, 28, 3441–3449.e5. [Google Scholar] [CrossRef]
- Galov, A.; Fabbri2, E.; Caniglia, R.; Arbanasić, H.; Lapalombella, S.; Florijančić, T.; Bošković, I.; Randi, E.; Randi, E. First Evidence of Hybridization between Golden Jackal (Canis aureus) and Domestic Dog (Canis familiaris) as Revealed by Genetic Markers. R. Soc. Open Sci. 2015, 2, 150450. [Google Scholar] [CrossRef]
- Wu, D.D.; Ding, X.D.; Wang, S.; Wójcik, J.M.; Zhang, Y.; Tokarska, M.; Li, Y.; Wang, M.S.; Faruque, O.; Nielsen, R.; et al. Pervasive Introgression Facilitated Domestication and Adaptation in the Bos Species Complex. Nat. Ecol. Evol. 2018, 2, 1139–1145. [Google Scholar] [CrossRef]
- Li, G.; Davis, B.W.; Eizirik, E.; Murphy, W.J. Phylogenomic Evidence for Ancient Hybridization in the Genomes of Living Cats (Felidae). Genome Res. 2016, 26, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Knoche, M.; Merz, R.; Lindner, M.; Weise, S.M. Bridging Glaciological and Hydrological Trends in the Pamir Mountains, Central Asia. Water 2017, 9, 422. [Google Scholar] [CrossRef]
- van der Geer, A. Capra Sibrica, the Asiatic Ibex. In Animals in Stone; Brill: Leiden, The Netherlands, 2008; pp. 205–210. [Google Scholar]
- Ahmad, I.; Zhang, F.; Tayyab, M.; Anjum, M.N.; Zaman, M.; Liu, J.; Farid, H.U.; Saddique, Q. Spatiotemporal Analysis of Precipitation Variability in Annual, Seasonal and Extreme Values over Upper Indus River Basin. Atmos. Res. 2018, 213, 346–360. [Google Scholar] [CrossRef]
- Glantz, M.H. Water, Climate, and Development Issues in the Amu Darya Basin. Mitig. Adapt. Strateg. Glob. Chang. 2005, 10, 23–50. [Google Scholar] [CrossRef]
- Namgail, T. Mountain Ungulates of the Trans-Himalayan Region of Ladakh, India. Int. J. Wilderness 2009, 15, 35–40. [Google Scholar]
- Khanyari, M.; Luecke, S.; Mishra, C.; Suryawanshi, K.R. Understanding Population Baselines: Status of Mountain Ungulate Populations in the Central Tien Shan Mountains, Kyrgyzstan. Mammalia 2021, 85, 16–23. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jabin, G.; Joshi, B.D.; Wang, M.-S.; Mukherjee, T.; Dolker, S.; Wang, S.; Chandra, K.; Chinnadurai, V.; Sharma, L.K.; Thakur, M. Mid-Pleistocene Transitions Forced Himalayan ibex to Evolve Independently after Split into an Allopatric Refugium. Biology 2023, 12, 1097. https://doi.org/10.3390/biology12081097
Jabin G, Joshi BD, Wang M-S, Mukherjee T, Dolker S, Wang S, Chandra K, Chinnadurai V, Sharma LK, Thakur M. Mid-Pleistocene Transitions Forced Himalayan ibex to Evolve Independently after Split into an Allopatric Refugium. Biology. 2023; 12(8):1097. https://doi.org/10.3390/biology12081097
Chicago/Turabian StyleJabin, Gul, Bheem Dutt Joshi, Ming-Shan Wang, Tanoy Mukherjee, Stanzin Dolker, Sheng Wang, Kailash Chandra, Venkatraman Chinnadurai, Lalit Kumar Sharma, and Mukesh Thakur. 2023. "Mid-Pleistocene Transitions Forced Himalayan ibex to Evolve Independently after Split into an Allopatric Refugium" Biology 12, no. 8: 1097. https://doi.org/10.3390/biology12081097
APA StyleJabin, G., Joshi, B. D., Wang, M.-S., Mukherjee, T., Dolker, S., Wang, S., Chandra, K., Chinnadurai, V., Sharma, L. K., & Thakur, M. (2023). Mid-Pleistocene Transitions Forced Himalayan ibex to Evolve Independently after Split into an Allopatric Refugium. Biology, 12(8), 1097. https://doi.org/10.3390/biology12081097





