1. Introduction
Narcissus, a genus of flowering plants belonging to Amaryllidaceae, encompasses a diverse group of ornamental flowers renowned for their beauty and fragrance [
1]. Chinese narcissus, also known as Chinese daffodil (
Narcissus tazetta), is a widely cultivated ornamental flower known for its delicate allure [
2]. It is culturally and economically important as an ornamental plant in China and other parts of the world. However, like many other plant species, this plant is susceptible to viral infections that can severely affect its growth, development, and aesthetic value [
3]. Viral diseases affecting
Narcissus tazetta can reduce flower quality, hinder growth, and cause significant economic losses to the horticultural industry [
4].
Narcissus is primarily affected by various plant viruses, including members of families such as
Potyviridae,
Alphaflexiviridae, and
Carlaviridae [
3,
5,
6,
7,
8]. The most common viruses infecting the
Narcissus species from the
Potyviridae family include narcissus late season yellows virus (NLSYV) [
9], narcissus yellow stripe virus (NYSV) [
5], narcissus degeneration virus (NDV) [
6], and narcissus latent virus (NLV). Additionally, narcissus common latent virus (NCLV) and narcissus symptomless virus (NSV) in the
Carlaviridae family have been identified [
7]. Several previous studies have demonstrated that infection of narcissus plants by multiple viruses is widespread [
3,
4,
10,
11]. These viruses can be transmitted through various means, including insect vectors, contaminated tools, and vegetative propagation. When the narcissus is infected with viruses, viral pathogens can persist and spread, resulting in a decline in flower quality, stunted growth, leaf yellowing, and plant death.
The study of plant viromes, encompassing the total viral population within a plant, is crucial for understanding the diversity and dynamics of plant-associated viruses [
12,
13]. Advanced molecular techniques, such as high-throughput sequencing (HTS), have revolutionized the study of plant viromes by enabling comprehensive analyses of viral populations present within host organisms [
14,
15]. In particular, RNA sequencing (RNA-seq) has emerged as a powerful tool for virome analysis, allowing for the identification and characterization of viral sequences in plant samples [
16,
17]. By leveraging RNA-seq data, researchers can gain insights into viral species, their genetic diversity, and their interactions with the narcissus host [
18]. Understanding viral infection diversity, prevalence, and the impact on narcissus is crucial for developing effective disease management strategies.
This study aims to investigate the viromes associated with Chinese narcissus flowers at different flowering stages using RNA-seq data. By capturing the viral genetic material in the flower samples, we can understand the viral diversity and dynamics throughout the flowering process. Additionally, by examining viral populations at different flowering stages, we can explore potential correlations between viral abundance and the developmental stages of the flowers.
3. Results
3.1. Identification of Viruses from Chinese Narcissus Transcriptomes
To investigate the viromes of narcissus flowers, we utilized Chinese narcissus transcriptomes comprising 20 distinct samples obtained from a previous study [
19]. Samples were collected at five different stages of flower development: the bud stage (S1), initial flowering stage (S2), full bloom stage (S3), full expansion stage (S4), and decay stage (S5), from two Chinese narcissus cultivars (
Table 1). Two independent biological replicates were used at each stage. To facilitate sample identification, we assigned the names W (white) and Y (yellow) to the plants based on the color of their tepals. For instance, W1R1 indicates White, Stage 1, Repetition 1. Each developmental stage is represented by numerical values ranging from 1 (S1) to 5 (S5). The biological replicates within each stage were classified as R1 (replicate 1) and R2 (replicate 2).
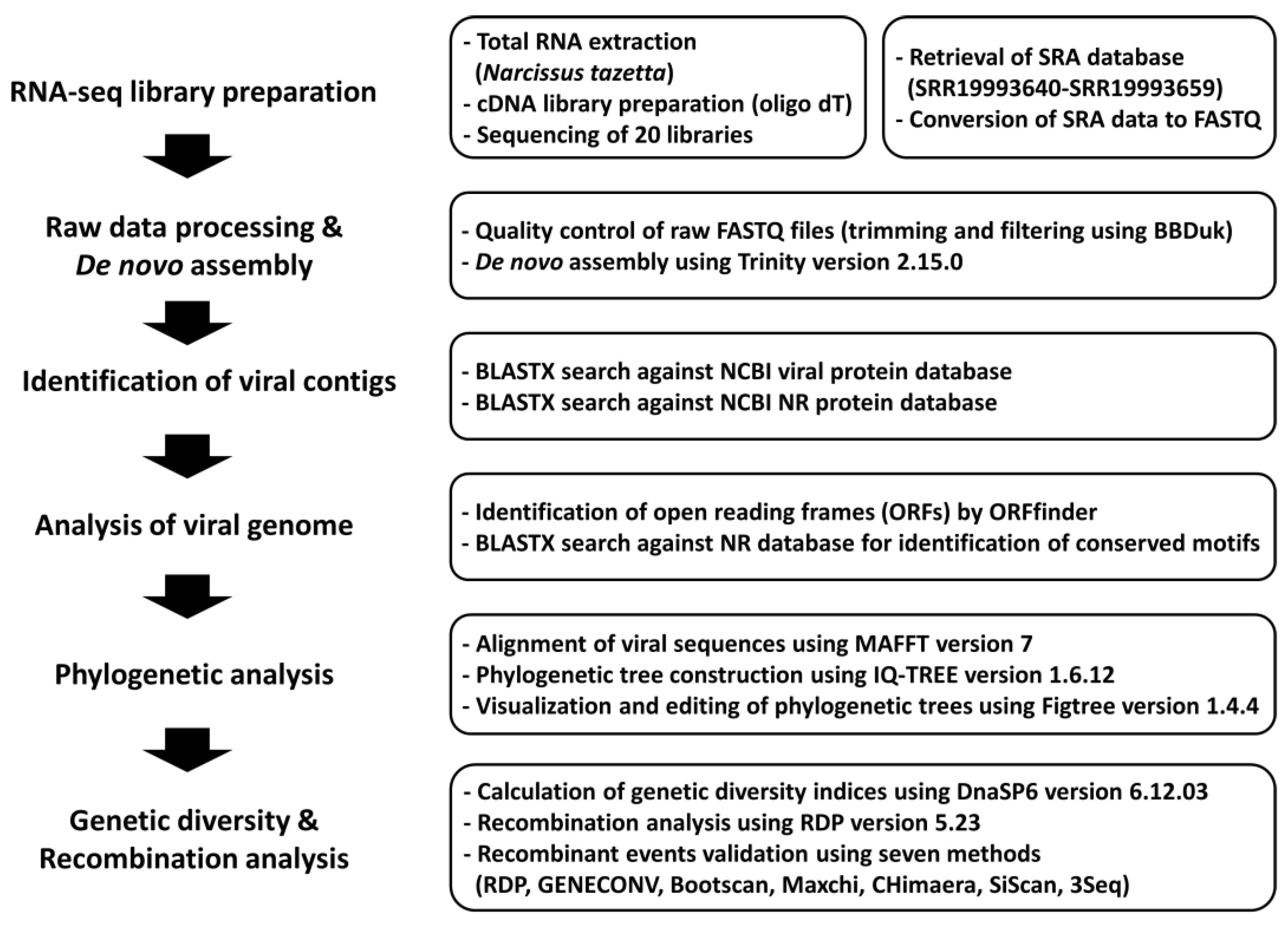
As depicted in
Figure 1, the viromes of narcissus were analyzed using data from 20 narcissus transcriptomes. Initially, raw data for each sample were retrieved from the NCBI SRA database to identify viruses present within the 20 Chinese narcissus flower transcriptomes. Subsequently, quality control measures were applied, and trimming was performed to ensure data integrity. The raw data within each library were subjected to
de novo assembly using Trinity assembler. The resulting assembled contigs were compared to a viral protein database using BLASTX. Contigs identified as contaminants originating from narcissus were removed from further analysis. Ultimately, we obtained 5893 viral contigs that matched seven distinct viral species.
Within the genus Carlavirus, we identified three viruses: NCLV (2913 contigs), narcissus mottling-associated virus (NMaV) (463 contigs), and NSV (30 contigs). In addition, six contigs were associated with the cucumber mosaic virus (CMV) genome, including RNA1 (three contigs), RNA2 (one contig), and RNA3 (two contigs). Furthermore, we identified three viruses within the genus Potyvirus: NDV (266 contigs), NLSYV (360 contigs), and NYSV (1855 contigs). Notably, NCLV (2913 contigs) demonstrated the highest abundance among the identified viruses, followed by NYSV (1855 contigs) and NMaV (463 contigs), as determined by the number of contigs.
Using viral sequences as references for the seven identified viruses, we performed read mapping of the raw data from each library to calculate the number of mapped reads and transcripts per million (TPM) for each virus (
Table 2). It should be noted that, for NMaV, only a partial sequence covering the viral replicase was available for analysis. Eighteen million, three hundred and seventy two thousand, three hundred and thirty one reads were successfully mapped to seven viral genomes (
Table 2) and based on the highest read count NYSV (40.6%) was followed by NDV (32.3%), NCLV (12.4%), and NMaV (10.7%).
3.2. Number of Identified Viruses and Proportion of Viral Reads in Each Sample
Six viruses were identified in the white tepal plants, whereas seven were detected in the yellow tepal plants. Specifically, the composition of the identified viruses in the white tepal samples remained consistent across all samples. In contrast, two samples, Y4R1 and Y4R2, were infected with all seven viruses, including CMV, whereas the remaining eight yellow tepal samples were infected with six viruses.
Subsequently, we assessed each sample’s proportion of viral reads (
Figure 2). Notably, in the white tepal samples, the proportion of viral reads gradually increased with the advancement of flower development (
Figure 2A). The viral proportions in W1R1 and W1R2 were 0.7% and 1.1%, respectively. However, the viral proportion increased significantly to 11.6% in W5R1 and 15.9% in W5R2. In contrast, the difference in the viral proportions among the yellow tepal samples was relatively small, ranging from 1.2% to 2.6% (
Figure 2B). When comparing the viral proportion at flower development stage 1, such as Y1R1 (1.5%) and Y1R2 (1.3%), with that at stage 5, such as Y5R1 (2.4%) and Y5R2 (2.6%), a slight increase in the viral proportion was observed (
Figure 2B).
3.3. Number of Identified Viruses and Proportion of Viral Reads in Each Sample
Examining the composition of the identified viruses in each sample can provide valuable insights into the dominant viruses in samples infected with multiple viruses. Among the ten white tepal samples, NDV emerged as the most abundant virus in nine samples, ranging from 56.7% (W1R2) to 99.4% (W2R2 and W4R1), except W1R1, where NSV (66.6%) was the dominant virus, followed by NDV (26.7%) (
Figure 3A). Furthermore, the proportion of NLSYV exceeded 5% in three samples: W1R1 (5.7%), W1R2 (9.9%), and W5R2 (15.6%) (
Figure 3A).
In the yellow tepal samples, NDV emerged as the dominant virus in eight samples, ranging from 39.5% (Y5R1) to 89% (Y4R1) (
Figure 3B). NYSV was also prominent in most of the yellow tepal samples, ranging from 5.6% (Y4R1) to 66.6% (Y1R2) (
Figure 3B).
Except for two samples, W1R1 and W1R2 contained several carlaviruses (
Figure 3C). In yellow tepal samples, potyviruses were dominant in eight samples, except for Y2R2 and Y5R1, which also harbored carlaviruses (
Figure 3D).
In this study, potyviruses appeared to be the major viruses infecting
Narcissus tazetta. Next, we analyzed the proportion of identified potyviruses in each sample to identify the dominant species. In the white tepal samples, NDV emerged as the dominant potyvirus in most samples, with NLSYV also being abundantly present in W1R1, W1R2, and W5R2 (
Figure 3E). In the yellow tepal samples, NDV was the most abundant potyvirus among the seven samples, followed by NYSV, which was prominently present in Y1R2 and Y2R2 (
Figure 3F). NLSYV was identified in all yellow tepal samples, but its proportion ranked third among the three potyviruses infecting Chinese narcissus (
Figure 3F).
3.4. Assembly and Annotation of Viral Genomes
Through
de novo transcriptome assembly, we obtained numerous contigs from five different viruses (
Table 3). The size of the acquired viral contigs ranged from 201 bp to 1000 bp. Specifically, most viral contigs associated with NMaV, for which the genome sequence was unavailable, were less than 1000 bp. Only six viral contigs associated with NMaV had sizes between 1001 bp and 2000 bp. The number of viral contigs linked to CMV and NSV was limited, and these contigs were shorter than those in the reference viral genomes.
The reference genome of NCLV (GenBank NC_008266.1) was 8539 nucleotides (nt). Among the 24 viral contigs associated with NCLV, more than 8001 base pairs (bp) were observed. The genome sizes of three potyviruses, NDV (GenBank NC_008824.1) with 9816 nt, NLSYV (GenBank NC_023628.1) with 9687 nt, and NYSV (GenBank NC_011541.1) with 9650 nt, exceeded 9000 nt. It is plausible to consider 41 viral contigs (NYSV), 5 viral contigs (NLSYV), and 27 viral contigs (NDV) as putative viral genome sequences using a cutoff of 9000 bp.
Candidate viral contigs for the identified viruses were further analyzed to identify open reading frames (ORFs). Only viral genomes encompassing complete open reading frames (ORFs) were selected. We obtained 18 NCLV, 27 NDV genomes, 2 NLSYV genomes, and 33 NYSV genomes (
Table S1).
3.5. Phylogenetic Analysis of Identified Viral Genomes
Phylogenetic trees were constructed for the five viruses based on the obtained viral genomes and known reference sequences from GenBank. For NCLV, NDV, NLSYV, and NYSV, only nucleotide sequences encompassing the complete open reading frames (ORFs) were used for tree construction.
Only one NCLV genome isolate, Zhangzhou, which belongs to the genus
Carlavirus and encodes six viral proteins, has been reported. Our study obtained 18 NCLV genomes from four yellow and 14 white tepal samples. The phylogenetic tree revealed two distinct genetic groups of NCLV: group A, comprising two isolates (Zhangzhou and Y2R2-1), and group B, consisting of three isolates from yellow tepal samples and 14 isolates from white tepal samples (
Figure 4A). Interestingly, different NCLV isolates, such as W3R2, W4R1, and W5R1, were identified in some samples. Specifically, 14 NCLV genomes from white tepal samples showed high similarity and low genetic diversity within the same clade. According to the phylogenetic tree, all 14 NCLV genomes from the white tepal samples originated from two NCLV isolates (Y3R1-1 and Y3R2-1) obtained from the yellow tepal samples.
The complete genome sequence of NMaV is currently unavailable, and only a partial sequence encoding the replicase domain has been reported. In the present study, we obtained many partial sequences encompassing the NMaV replicase domain. However, we did not identify any complete genome sequences or sequences covering other open reading frames (ORFs) of NMaV. From the obtained partial sequences, we selected 16 NMaV sequences with sizes exceeding 900 base pairs (
Table S1). A consensus sequence was also generated from all the NMaV partial sequences (
Table S1). Remarkably, all NMaV sequences were derived from yellow tepal samples, and multiple variants were identified within the same sample, such as five isolates from Y3R2. Phylogenetic analysis classified the NMaV sequences into three distinct groups: group A, comprising two isolates; group B, comprising six isolates; and group C, comprising ten isolates (
Figure 4B).
We obtained 27 NDV genome sequences, including 16 isolates from yellow tepal samples and 11 isolates from white tepal samples (
Table S1). Currently, only seven NDV genome sequences are available in public databases. Phylogenetic analysis revealed three distinct groups of NDV isolates: group A (2 isolates), group B (16 isolates), and group C (16 isolates) (
Figure 5A). It appears that the NDV isolates in group C originated from the NDV isolates in group B, derived from the NDV isolates in group A. NDV isolates within the same group exhibited low genetic variability. Interestingly, all 11 NDV isolates belonged to group C, including one isolate (NY-FK266) from
Narcissus jonquilla in Japan, three isolates from
Narcissus tazetta in Japan, and one isolate (Marijiniup2) from
Narcissus species in Australia. Group B included two known NDV isolates from Japan (NY-AC230) and China (Zhangzhou).
The 33 NLSYV isolates, including the two isolates from this study, were categorized into four groups: group A (eight isolates), group B (eight isolates), group C (nine isolates), and group D (eight isolates) (
Figure 5B). In this study, two isolates (Y2R2-1 and Y4R1-1) belonged to group C, including six isolates from Japan and one from China (Zhangzhou). Based on phylogenetic analysis, the NLSYV isolates in group A were the ancestors of the isolates in groups B, C, and D. Moreover, the seven NLSYV isolates from Japan and one isolate from China (Marijiniup8) in group B represented more recent evolutionary lineages, according to the phylogenetic tree.
Forty six NYSV isolates were obtained, comprising twenty one from white tepal samples and eleven from yellow tepal samples (
Figure 5C). The 46 NYSV isolates were broadly divided into three groups: A (11 isolates), B (18 isolates), and C (17 isolates). The 11 isolates in group A exhibited high sequence similarity. In contrast, 18 isolates in group B and 17 in group C showed high sequence diversity. Group A consisted primarily of seven NYSV isolates from yellow tepal samples and one isolate (W2R1-1) from a white tepal sample. Several variant genomes were identified, including two samples from Y3R2 and Y5R1 and three variants from Y2R1. The isolates in group B exhibited significant sequence diversity, and two isolates of narcissus virus 1 grouped with the NYSV reference genome, indicating their membership in the NYSV. Group B included seven isolates from white tepal samples and four from yellow tepal samples. Interestingly, group C was predominantly composed of white tepal samples (13 isolates) and showed high sequence divergence. It is worth noting that some variants displayed high sequence divergence despite being identified from the same sample, such as W2R1-1 in group A, W2R1-2 in group C, W2R2-4 and W2R2-5 in group B, and W2R2-2 and W2R2-3 in group D.
3.6. Genetic Diversity Analysis of NCLV, NDV, NLSYV, and NYSV Isolates
We conducted a comprehensive analysis of the genetic diversity of four viruses (
Table 4). The viral populations varied in size, with NYSV having the largest population (46 isolates) and NCLV having the smallest (19 isolates). Among the four viruses, NDV had the highest number of sites (9542 sites), whereas NCLV had the fewest (8513 sites). Notably, NYSV displayed the highest number of segregation sites (4520 sites), indicating greater genetic diversity, whereas NDV had the lowest number of segregation sites (1488 sites). NYSV also exhibited the highest effective number of alleles (6826), suggesting a larger gene pool, whereas NDV had the lowest (1548). The number of haplotypes ranged from 15 (NCLV) to 45 (NYSV), with NLSYV demonstrating the highest haplotype diversity (1) and NCLV the lowest (0.965). Nucleotide diversity was the highest in NYSV (0.20667) and lowest in NDV (0.03409). Additionally, NYSV displayed the highest θw value of 0.1682, indicating the highest estimated population mutation rate (0.1682 per site) among the four viruses. NLSYV exhibited the highest expected heterozygosity (33), indicating substantial genetic diversity. It also demonstrated the highest values for diversity (Hd), nucleotide diversity (Pi), and Watterson’s theta (θw) among the four viruses.
3.7. Recombination Analysis of NCLV, NDV, NLSYV, and NYSV
Next, we conducted recombination analyses of the four viruses (
Table S2). Our findings revealed a low number of recombination events for the NDV (four events), including five MDV recombinants: Y4R2-2, Y4R2-3, Y2R1-2, Y2R1-1, and Y3R2-2. For NLSYV, we identified four recombination events involving ten isolates: NY-CB1, Y4R1-1, Marijiniup9, NLSYV NY-F1, NY-OS1, NY-HG25, NY-HR39, NY-FK266, and Marijiniup9. For NCLV, eight recombination events were observed, including five recombinants: Y1R1-1, Y2R2-1, Zhangzhou, Y3R2-1, and Y3R1-1. The highest number of recombination events (25) was detected in NYSV, with 24 recombinants identified among the 21 isolates in our study.
Subsequently, we analyzed the recombination breakpoints to identify the specific genomic regions in which these events occurred. Of the four viruses, the NCLV genome exhibited the highest breakpoints (
Figure 6). Notably, Rho per bp analysis revealed three peaks in the RdRp region and a single highly elevated peak in the TGB1 region of NCLV, indicating potential recombination events (
Figure 6). In NYSV, multiple breakpoints were identified along the viral genome, including regions P1, P3, C1, NIa-VPg, NIb, and CP (
Figure 7). Statistical analysis strongly supported the presence of breakpoints in the P3 region. Among the various peaks observed in the Rho per bp graph, the peak in the P1 region exhibited the highest Rho per bp (
Figure 7).
The NDV genome displayed few breakpoints, with only the P3 region exhibiting a high breakpoint value (
Figure S1). Moreover, Rho per base pair analysis revealed no peaks, suggesting no recombination events occurred in the examined NDV genomes. For NLSYV, both the P1 and CP regions showed higher breakpoints (
Figure S2). Analysis of Rho per base pair revealed elevated peaks in the P1 and P3 regions of NLSYV, indicating potential recombination events.
4. Discussion
In this study, we investigated the viromes in Chinese narcissus flowers using transcriptome data from 20 samples collected at various stages of flower development. Through a series of data processing steps, we identified seven distinct virus species in the narcissus transcriptome: NCLV, NMaV, NSV, CMV, NDV, NLSYV, and NYSV. Similarly, previous studies have revealed coinfection with multiple viruses that infect
Narcissus species. For example, in China, three viruses (NCLV, NYSV, and NLSYV) have been identified in
Narcissus tazetta using reverse transcriptase-polymerase chain reaction (RT-PCR) with conserved carlavirus and potyvirus primers [
3]. In New Zealand, enzyme-linked immunosorbent assays (ELISA) and mechanical transmission tests have detected five viruses (CMV, NLV, narcissus mosaic virus [NMV], narcissus tip necrosis virus, and NYSV) from diverse
Narcissus species [
10]. In Australia, two viruses (NLSYV and vallota speciosa virus) were identified in domestic and wild
Narcissus species [
26]. Furthermore, in Japan, RT-PCR has detected two potyviruses (NLSYV and NDV) in the narcissus [
4]. Our study identified the highest number of viruses infecting Chinese narcissus plants using high-throughput sequencing (HTS) techniques and many plant samples. Based on previous results, plant samples and geographical regions are important factors in determining virus populations.
HTS and bioinformatics analyses enabled us to quantify the viral load in each sample. In particular, the analysis of viral reads and transcripts per million (TPM) provides insights into the abundance of each virus in samples [
12,
13]. NYSV was most abundant among the identified viruses, followed by NDV, NCLV, and NMaV. Interestingly, the proportion of viral reads varied between the white and yellow tepal plants. White tepal plants showed an increasing proportion of viral reads with the flower development stage, whereas the difference in viral proportion among yellow tepal plants was relatively small. Based on these results, we hypothesized that the viral load increases as the plant grows; however, this phenomenon highly depends on the plant cultivar or species. Further examination of the composition of the identified viruses in each sample revealed that NDV was the most abundant virus in most of the white and yellow tepal samples. NYSV was also prominent in yellow tepal samples. Potyviruses, including NDV, NLSYV, and NYSV, were the major viruses infecting Chinese narcissus in this study. This is the first study to report viral populations in the diverse flower tissues of higher plants. Therefore, our results provide information about the dominant virus in each sample, which cannot be readily determined using other virus detection techniques, such as RT-PCR and ELISA.
Moreover, our study demonstrated that flower tissue is suitable for virus detection in narcissus plants because of the high viral load, regardless of the developmental stage. The viral abundance in narcissus flower tissues during the examined developmental stages was significantly higher than in other plant species, such as sweet potato [
27] and lily [
28]. Selecting the appropriate plant tissue is crucial for virus detection, as extracting high-quality total RNA from plant leaves enriched with polysaccharides and polyphenols can be challenging [
29]. Therefore, we strongly suggest that flower tissue from
Narcissus spp. is an excellent material for virus detection.
De novo transcriptome assembly enabled us to obtain the viral contigs for further analysis. The obtained contigs varied in size, with some contigs associated with NMaV being less than 1000 bp. We identified the putative viral genome sequences for NYSV, NLSYV, and NDV by applying a size cutoff. Obtaining a large number of viral genomes in a virome study facilitates studies related to viral genome-associated aspects, such as the phylogenetics, diversity, and evolutionary rates of the identified viruses [
4,
11,
30]. Remarkably, 80 viral genome sequences encompassing open reading frames (ORFs) were obtained within a single study. Interestingly, we found that carlaviruses, such as NCLV and the three potyviruses, had poly(A) tails that could be detected by RNA sequencing generated from cDNA using oligo-d(T) [
31]. Moreover, based on our experience, longer sequence lengths achieved through high-throughput sequencing (HTS) (here, 150 bp was used instead of 100 bp) are crucial for obtaining complete viral genome sequences. For NMaV, only partial sequences were obtained, indicating that an alternative approach should be considered for obtaining the complete NMaV genome soon.
Phylogenetic analyses of the identified viral genomes provided insights into their genetic relationships. The NCLV, NMaV, NDV, NLSYV, and NYSV have distinct genetic groups. Multiple NCLV isolates were identified in the same sample, indicating the presence of genetic variants. The NMaV sequences were classified into three distinct groups, with multiple variants identified within the samples. The NDV isolates exhibited three distinct groups, where group C isolates originated from group B and were derived from group A. NLSYV isolates were categorized into four groups, with group A isolates being the ancestors of groups B, C, and D. NYSV isolates were broadly divided into three groups, with groups B and C exhibiting high sequence diversity.
One of the most intriguing findings of this study was the phylogenetic analysis of both NDV and NCLV, which showed that the two viruses found in yellow tepal plants were ancestors of the viruses present in white tepal plants. Previous studies suggested that yellow tepal plants mutated from the white tepal plant known as the ‘Jinzhan Yintai’ cultivar [
19]; however, no biological evidence supported this claim. Based on our results, it is likely that white tepal plants originated from yellow tepal plants. Furthermore, many viruses identified in yellow tepal plants exhibited more significant divergence than those in white tepal plants. This suggests that yellow tepal plants may represent wild populations in diverse regions, whereas white tepal plants are domesticated narcissus plants clonally propagated on a large scale and commonly found in many places. However, further experiments are required to confirm this hypothesis.
Analysis of genetic diversity among the NCLV, NDV, NLSYV, and NYSV isolates provided insights into the variation within these viral populations. Variants were identified even within samples, indicating the presence of diverse viral genomes within the same host. These results suggest that the genetic diversity of the identified viruses was significantly higher in yellow tepal plants than in white tepal plants, indicating host-dependent viral mutations and evolution [
32].
Overall, this study revealed the presence of multiple viruses in Chinese narcissus flowers and shed light on their abundance, composition, genetic diversity, and relationships. These findings enhance our understanding of the narcissus flower virome and emphasize the prevalence of potyviruses in this plant species. Further research is warranted to investigate the impact of these viruses on the health and productivity of narcissus.
5. Conclusions
In conclusion, this study explored the viromes in Chinese narcissus flowers using transcriptome data from 20 samples collected at various stages of flower development. Seven distinct viral species have been identified: NCLV, NMaV, NSV, CMV, NDV, NLSYV, and NYSV. This study identified the highest number of viruses infecting Narcissus tazetta plants to date, using high-throughput sequencing techniques and a large number of plant samples. The results revealed that the viral loads varied between white and yellow tepal plants, with white tepal plants showing an increasing proportion of viral reads during flower development. NDV was the most abundant virus in most samples, followed by NYSV, NCLV, and NMaV. Potyviruses, including NDV, NLSYV, and NYSV, are the major viruses infecting Chinese daffodils.
Furthermore, this study demonstrated that flower tissue is suitable for virus detection in narcissus plants because of its high viral load, regardless of the developmental stage. Flower tissues exhibited significantly higher viral abundance than other plant species. The use of high-throughput sequencing and bioinformatic analysis has allowed the identification of viral contigs and putative viral genome sequences. Phylogenetic analysis provided insights into the genetic relationships between the identified viruses, revealing the distinct genetic groups of NCLV, NMaV, NDV, NLSYV, and NYSV. Notably, phylogenetic analyses suggested that the viruses found in yellow tepal plants were the ancestors of white tepal plants, challenging previous assumptions regarding the origin of different narcissus cultivars.
This study contributes to our understanding of the virome in Chinese narcissus flowers by highlighting the prevalence of potyviruses and providing information on the dominant viruses in each sample. These findings also emphasize the importance of selecting appropriate plant tissues for virus detection and the impact of host-dependent viral mutations and evolution on genetic diversity. Further research is needed to investigate the implications of these viruses on the health and productivity of narcissus.