Identification and Expressional Analysis of Putative PRDI-BF1 and RIZ Homology Domain-Containing Transcription Factors in Mulinia lateralis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of M. lateralis PRDMs
2.2. PRDMs Sequence Analysis
2.3. Phylogenetic Analysis
2.4. Gene Expression Analysis of M. lateralis prdms
2.5. Collection of Embryos and Larvae
2.6. Quantitative RT-PCR Analysis of Ml-prdm1 and Ml-prdm14
2.7. Whole-Mount In Situ Hybridization of Ml-prdm1 and Ml-prdm14
3. Results
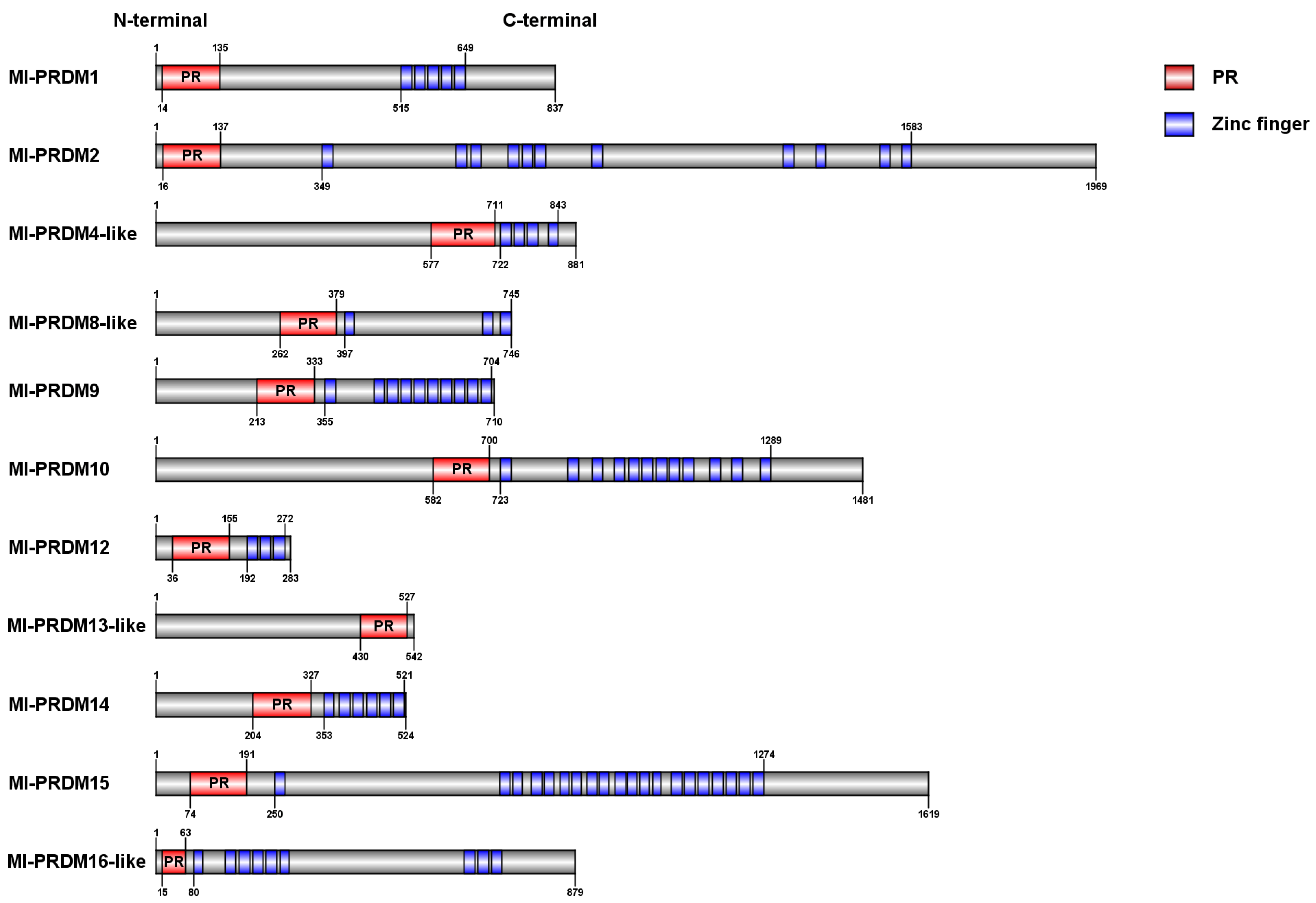
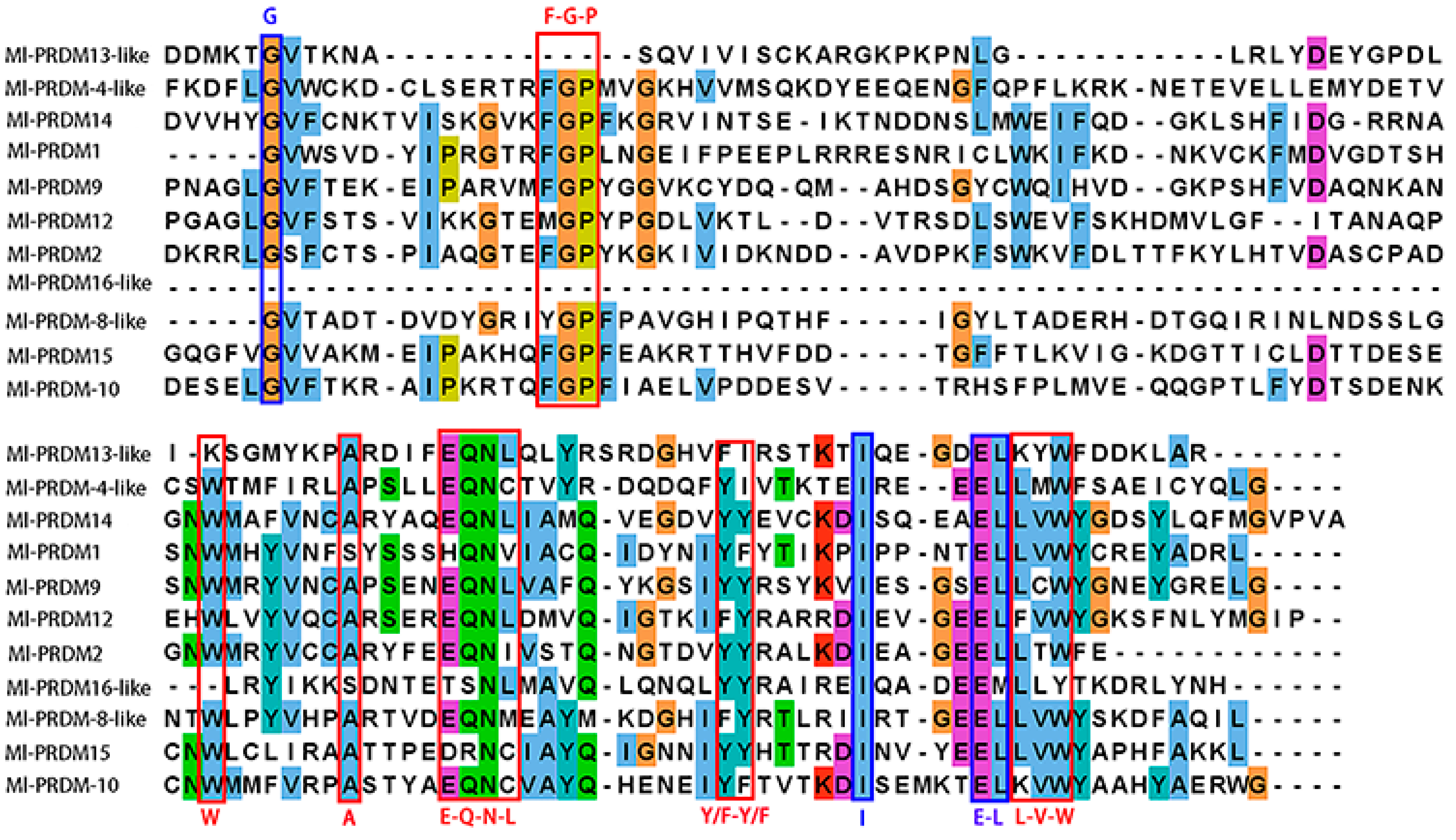
3.1. Identification and Sequence Analysis of M. lateralis Putative PRDMs
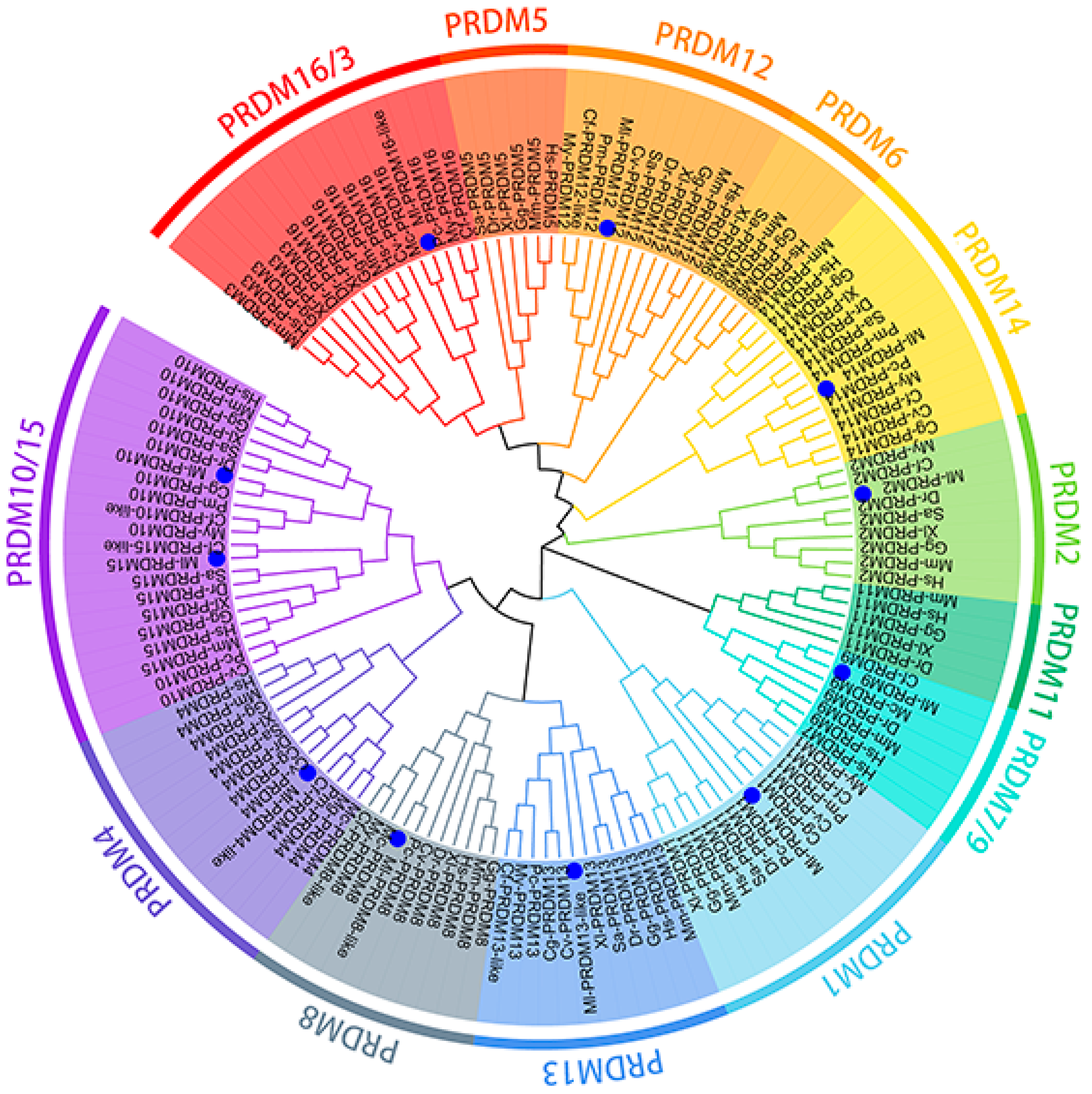
3.2. Phylogenetic Analysis of Putative M. lateralis PRDMs
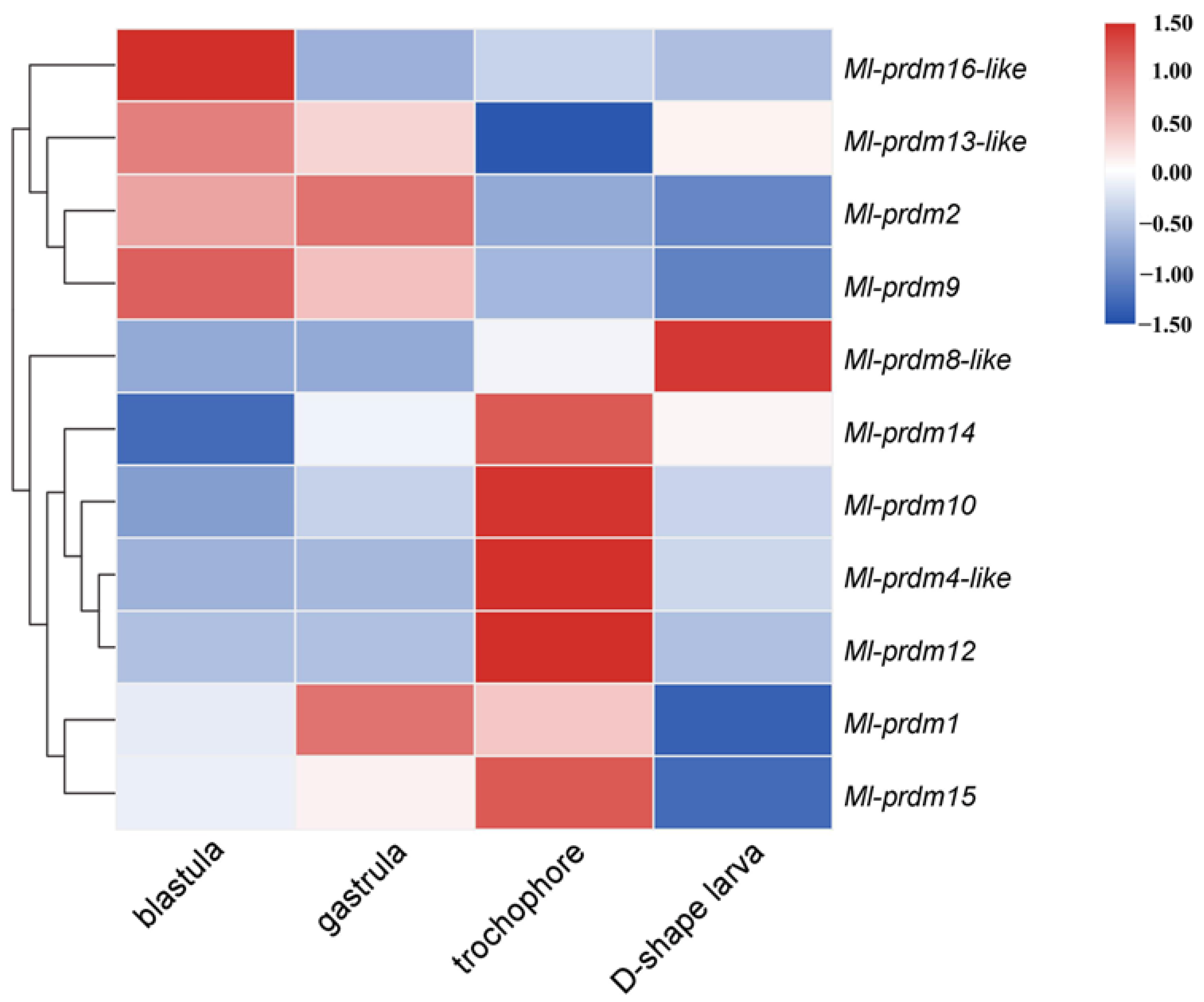
3.3. Temporal Expression Pattern of Putative M. lateralis prdms during Early Development Stages
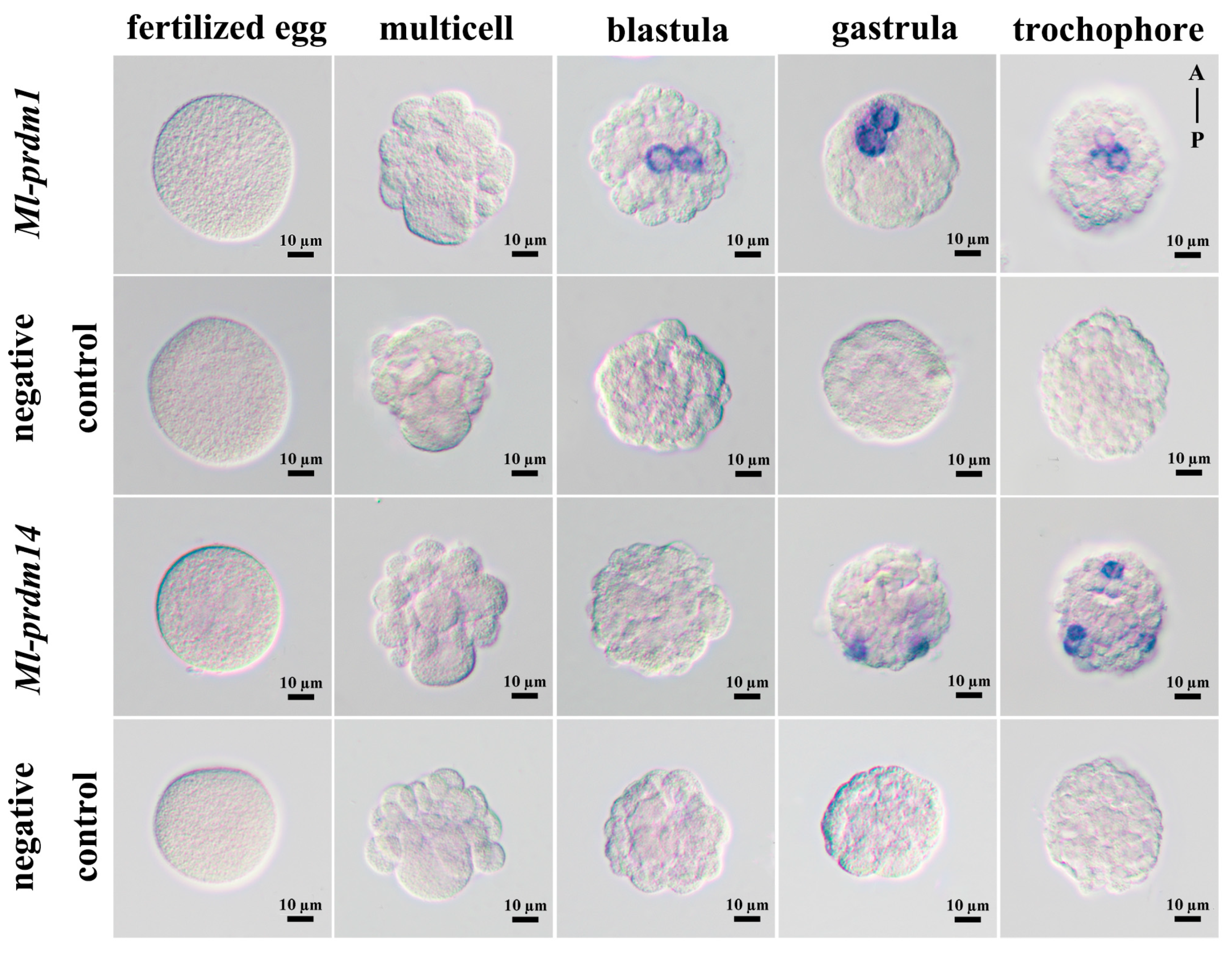
3.4. Spatial Expression Pattern of M. lateralis prdm1 and prdm14 in Embryo and Larva
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Magnúsdóttir, E.; Surani, M.A. How to make a primordial germ cell. Development 2014, 141, 245–252. [Google Scholar] [CrossRef]
- Zeng, F.; Huang, F.; Guo, J.; Hu, X.; Liu, C.; Wang, H. Emerging methods to generate artificial germ cells from stem cells. Biol. Reprod. 2015, 92, 89. [Google Scholar] [CrossRef] [PubMed]
- Extavour, C.G.; Akam, M. Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development 2003, 130, 5869–5884. [Google Scholar] [CrossRef]
- Juliano, C.E.; Swartz, S.Z.; Wessel, G.M. A conserved germline multipotency program. Development 2010, 137, 4113–4126. [Google Scholar] [CrossRef] [PubMed]
- Rebscher, N.; Zelada-González, F.; Banisch, T.U.; Raible, F.; Arendt, D. Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev. Biol. 2007, 306, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Extavour, C.G. Evolution of the bilaterian germ line: Lineage origin and modulation of specification mechanisms. Integr. Comp. Biol. 2007, 47, 770–785. [Google Scholar] [CrossRef]
- Fabioux, C.; Huvet, A.; Lelong, C.; Robert, R.; Pouvreau, S.; Daniel, J.Y.; Minguant, C.; Le Pennec, M. Oyster vasa-like gene as a marker of the germline cell development in Crassostrea gigas. Biochem. Biophys. Res. Commun. 2004, 320, 592–598. [Google Scholar] [CrossRef]
- Kranz, A.M.; Tollenaere, A.; Norris, B.J.; Degnan, B.M.; Degnan, S.M. Identifying the germline in an equally cleaving mollusc: Vasa and Nanos expression during embryonic and larval development of the vetigastropod Haliotis asinina. J. Exp. Zool. B Mol. Dev. Evol. 2010, 314, 267–279. [Google Scholar] [CrossRef]
- Liu, L.; Liu, T.; Wu, S.; Li, Y.; Wei, H.; Zhang, L.; Shu, Y.; Yang, Y.; Xing, Q.; Wang, S.; et al. Discovery of nanos1 and nanos2/3 as germ cell markers during scallop gonadal development. Mar. Biotechnol. 2022, 24, 408–416. [Google Scholar] [CrossRef]
- Xu, R.; Li, Q.; Yu, H.; Kong, L. Oocyte maturation and origin of the germline as revealed by the expression of Nanos-like in the Pacific oyster Crassostrea gigas. Gene 2018, 663, 41–50. [Google Scholar] [CrossRef]
- Huang, S.; Shao, G.; Liu, L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J. Biol. Chem. 1998, 273, 15933–15939. [Google Scholar] [CrossRef] [PubMed]
- Fumasoni, I.; Meani, N.; Rambaldi, D.; Scafetta, G.; Alcalay, M.; Ciccarelli, F.D. Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol. Biol. 2007, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Ohinata, Y.; Payer, B.; O’Carroll, D.; Ancelin, K.; Ono, Y.; Sano, M.; Barton, S.C.; Obukhanych, T.; Nussenzweig, M.; Tarakhovsky, A.; et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005, 436, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Kurimoto, K.; Yamaji, M.; Seki, Y.; Saitou, M. Specification of the germ cell lineage in mice: A process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle 2008, 7, 3514–3518. [Google Scholar] [CrossRef] [PubMed]
- Vincent, S.D.; Dunn, N.R.; Sciammas, R.; Shapiro-Shalef, M.; Davis, M.M.; Calame, K.; Bikoff, E.K.; Robertson, E.J. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of PGCs in the mouse. Development 2005, 132, 1315–1325. [Google Scholar] [CrossRef]
- Yamaji, M.; Seki, Y.; Kurimoto, K.; Yabuta, Y.; Yuasa, M.; Shigeta, M.; Yamanaka, K.; Ohinata, Y.; Saitou, M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008, 40, 1016–1022. [Google Scholar] [CrossRef]
- Nakaki, F.; Hayashi, K.; Ohta, H.; Kurimoto, K.; Yabuta, Y.; Saitou, M. Induction of mouse germ-cell fate by transcription factors In Vitro. Nature 2013, 501, 222–226. [Google Scholar] [CrossRef]
- Irie, N.; Weinberger, L.; Tang, W.W.; Kobayashi, T.; Viukov, S.; Manor, Y.S.; Dietmann, S.; Hanna, J.H.; Surani, M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell 2015, 160, 253–268. [Google Scholar] [CrossRef]
- Sybirna, A.; Tang, W.W.C.; Pierson Smela, M.; Dietmann, S.; Gruhn, W.H.; Brosh, R.; Surani, M.A. A critical role of PRDM14 in human primordial germ cell fate revealed by inducible degrons. Nat. Commun. 2020, 11, 1282. [Google Scholar] [CrossRef]
- Okuzaki, Y.; Kaneoka, H.; Suzuki, T.; Hagihara, Y.; Nakayama, Y.; Murakami, S.; Murase, Y.; Kuroiwa, A.; Iijima, S.; Nishijima, K.I. PRDM14 and BLIMP1 control the development of chicken PGCs. Dev. Biol. 2019, 455, 32–41. [Google Scholar] [CrossRef]
- Nakamura, T.; Extavour, C.G. The transcriptional repressor Blimp-1 acts downstream of BMP signaling to generate PGCs in the cricket Gryllus bimaculatus. Development 2016, 143, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Di Zazzo, E.; Porcile, C.; Bartollino, S.; Moncharmont, B. Critical function of PRDM2 in the neoplastic growth of testicular germ cell tumors. Biology 2016, 5, 54. [Google Scholar] [CrossRef]
- Eom, G.H.; Kim, K.; Kim, S.M.; Kee, H.J.; Kim, J.Y.; Jin, H.M.; Kim, J.R.; Kim, J.H.; Choe, N.; Kim, K.B.; et al. Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis. Biochem. Biophys. Res. Commun. 2009, 388, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Baudat, F.; Buard, J.; Grey, C.; Fledel-Alon, A.; Ober, C.; Przeworski, M.; Coop, G.; de Massy, B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 2010, 327, 836–840. [Google Scholar] [CrossRef] [PubMed]
- Parvanov, E.D.; Petkov, P.M.; Paigen, K. Prdm9 controls activation of mammalian recombination hotspots. Science 2010, 327, 835. [Google Scholar] [CrossRef] [PubMed]
- Cheedipudi, S.; Puri, D.; Saleh, A.; Gala, H.P.; Rumman, M.; Pillai, M.S.; Sreenivas, P.; Arora, R.; Sellathurai, J.; Schrøder, H.D.; et al. A fine balance: Epigenetic control of cellular quiescence by the tumor suppressor PRDM2/RIZ at a bivalent domain in the cyclin a gene. Nucleic Acids Res. 2015, 43, 6236–6256. [Google Scholar] [CrossRef]
- Mzoughi, S.; Zhang, J.; Hequet, D.; Teo, S.X.; Fang, H.; Xing, Q.R.; Bezzi, M.; Seah, M.K.Y.; Ong, S.L.M.; Shin, E.M.; et al. PRDM15 safeguards naive pluripotency by transcriptionally regulating WNT and MAPK-ERK signaling. Nat. Genet. 2017, 49, 1354–1363. [Google Scholar] [CrossRef]
- Bogani, D.; Morgan, M.A.; Nelson, A.C.; Costello, I.; McGouran, J.F.; Kessler, B.M.; Robertson, E.J.; Bikoff, E.K. The PR/SET domain zinc finger protein Prdm4 regulates gene expression in embryonic stem cells but plays a nonessential role in the developing mouse embryo. Mol. Cell. Biol. 2013, 33, 3936–3950. [Google Scholar] [CrossRef][Green Version]
- Galli, G.G.; Honnens de Lichtenberg, K.; Carrara, M.; Hans, W.; Wuelling, M.; Mentz, B.; Multhaupt, H.A.; Fog, C.K.; Jensen, K.T.; Rappsilber, J.; et al. Prdm5 regulates collagen gene transcription by association with RNA polymerase II in developing bone. PLoS Genet. 2012, 8, e1002711. [Google Scholar] [CrossRef]
- Davis, C.A.; Haberland, M.; Arnold, M.A.; Sutherland, L.B.; McDonald, O.G.; Richardson, J.A.; Childs, G.; Harris, S.; Owens, G.K.; Olson, E.N. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol. Cell. Biol. 2006, 26, 2626–2636. [Google Scholar] [CrossRef]
- Wu, Y.; Ferguson, J.E.; Wang, H.; Kelley, R.; Ren, R.; McDonough, H.; Meeker, J.; Charles, P.C.; Wang, H.; Patterson, C. PRDM6 is enriched in vascular precursors during development and inhibits endothelial cell proliferation, survival, and differentiation. J. Mol. Cell. Cardiol. 2008, 44, 47–58. [Google Scholar] [CrossRef]
- Zhang, Y.; Stehling-Sun, S.; Lezon-Geyda, K.; Juneja, S.C.; Coillard, L.; Chatterjee, G.; Wuertzer, C.A.; Camargo, F.; Perkins, A.S. PR-domain-containing Mds1-Evi1 is critical for long-term hematopoietic stem cell function. Blood 2011, 118, 3853–3861. [Google Scholar] [CrossRef] [PubMed]
- Konantz, M.; Alghisi, E.; Müller, J.S.; Lenard, A.; Esain, V.; Carroll, K.J.; Kanz, L.; North, T.E.; Lengerke, C. Evi1 regulates Notch activation to induce zebrafish hematopoietic stem cell emergence. EMBO J. 2016, 35, 2315–2331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, X.; Xue, H.; Liu, X.; Dai, A.; Song, Y.; Ke, K.; Cao, M. Upregulation of PRDM5 is associated with astrocyte proliferation and neuronal apoptosis caused by lipopolysaccharide. J. Mol. Neurosci. 2016, 59, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, R.; Yoshigai, E.; Koga, T.; Kuhara, S.; Tashiro, K. Spatiotemporal expression of Prdm genes during Xenopus development. Cytotechnology 2015, 67, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Auer-Grumbach, M.; Matsukawa, S.; Zitzelsberger, M.; Themistocleous, A.C.; Strom, T.M.; Samara, C.; Moore, A.W.; Cho, L.T.; Young, G.T.; et al. Transcriptional regulator PRDM12 is essential for human pain perception. Nat. Genet. 2015, 47, 803–808. [Google Scholar] [CrossRef]
- Mona, B.; Uruena, A.; Kollipara, R.K.; Ma, Z.; Borromeo, M.D.; Chang, J.C.; Johnson, J.E. Repression by PRDM13 is critical for generating precision in neuronal identity. Elife 2017, 6, e25787. [Google Scholar] [CrossRef]
- Shimada, I.S.; Acar, M.; Burgess, R.J.; Zhao, Z.; Morrison, S.J. Prdm16 is required for the maintenance of neural stem cells in the postnatal forebrain and their differentiation into ependymal cells. Genes Dev. 2017, 31, 1134–1146. [Google Scholar] [CrossRef]
- Su, L.; Lei, X.; Ma, H.; Feng, C.; Jiang, J.; Jiao, J. PRDM16 orchestrates angiogenesis via neural differentiation in the developing brain. Cell Death Differ. 2020, 27, 2313–2329. [Google Scholar] [CrossRef]
- Lannan, J.E.; Robinson, A.; Breese, W.P. Broodstock management of Crassostrea gigas: II. Broodstock conditioning to maximize larval survival. Aquaculture 1980, 21, 337–345. [Google Scholar] [CrossRef]
- de Siqueira-Silva, D.H.; Saito, T.; Dos Santos-Silva, A.P.; da Silva Costa, R.; Psenicka, M.; Yasui, G.S. Biotechnology applied to fish reproduction: Tools for conservation. Fish. Physiol. Biochem. 2018, 44, 1469–1485. [Google Scholar] [CrossRef] [PubMed]
- Rivers, N.; Daly, J.; Temple-Smith, P. New directions in assisted breeding techniques for fish conservation. Reprod. Fertil. Dev. 2020, 32, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Kakoi, S.; Kin, K.; Miyazaki, K.; Wada, H. Early development of the Japanese spiny oyster (Saccostrea kegaki): Characterization of some genetic markers. Zool. Sci. 2008, 25, 455–464. [Google Scholar] [CrossRef][Green Version]
- Rui, X.; Qi, L.; Hong, Y. Expression pattern of Piwi-like gene implies the potential role in germline development in the Pacific oyster Crossosrea gigas. Aquac. Rep. 2020, 18, 100486. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009, 23, 205–211. [Google Scholar] [CrossRef]
- Li, Y.; Sun, X.; Hu, X.; Xun, X.; Zhang, J.; Guo, X.; Jiao, W.; Zhang, L.; Liu, W.; Wang, J.; et al. Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins. Nat. Commun. 2017, 8, 1721. [Google Scholar] [CrossRef]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, X.; Zhao, F.; Liu, D.; Yang, Z.; Li, M.; Li, Y.; Wei, H.; Wang, H.; Qin, Z.; et al. Full-length transcriptome analysis provides insights into larval shell formation in Mulinia lateralis. Front. Mar. Sci. 2023, 9, 1111241. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, J.; Jiao, W.; Li, J.; Xun, X.; Sun, Y.; Guo, X.; Huan, P.; Dong, B.; Zhang, L.; et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 2017, 1, 120. [Google Scholar] [CrossRef]
- Vervoort, M.; Meulemeester, D.; Béhague, J.; Kerner, P. Evolution of Prdm genes in animals: Insights from comparative genomics. Mol. Biol. Evol. 2016, 33, 679–696. [Google Scholar] [CrossRef]
- Huang, S. Histone methyltransferases, diet nutrients and tumour suppressors. Nat. Rev. Cancer. 2002, 2, 469–476. [Google Scholar] [CrossRef]
- Hohenauer, T.; Moore, A.W. The Prdm family: Expanding roles in stem cells and development. Development 2012, 139, 2267–2282. [Google Scholar] [CrossRef]
- Derunes, C.; Briknarová, K.; Geng, L.; Li, S.; Gessner, C.R.; Hewitt, K.; Wu, S.; Huang, S.; Woods, V.I., Jr.; Ely, K.R. Characterization of the PR domain of RIZ1 histone methyltransferase. Biochem. Biophys. Res. Commun. 2005, 333, 925–934. [Google Scholar] [CrossRef]
- Pinheiro, I.; Margueron, R.; Shukeir, N.; Eisold, M.; Fritzsch, C.; Richter, F.M.; Mittler, G.; Genoud, C.; Goyama, S.; Kurokawa, M.; et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 2012, 150, 948–960. [Google Scholar] [CrossRef]
- Gewies, A.; Castineiras-Vilarino, M.; Ferch, U.; Jährling, N.; Heinrich, K.; Hoeckendorf, U.; Przemeck, G.K.; Munding, M.; Groß, O.; Schroeder, T.; et al. Prdm6 is essential for cardiovascular development in vivo. PLoS ONE 2013, 8, e81833. [Google Scholar] [CrossRef]
- Koh-Stenta, X.; Joy, J.; Poulsen, A.; Li, R.; Tan, Y.; Shim, Y.; Min, J.H.; Wu, L.; Ngo, A.; Peng, J.; et al. Characterization of the histone methyltransferase PRDM9 using biochemical, biophysical and chemical biology techniques. Biochem. J. 2014, 461, 323–334. [Google Scholar] [CrossRef]
- Gyory, I.; Wu, J.; Fejér, G.; Seto, E.; Wright, K.L. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 2004, 5, 299–308. [Google Scholar] [CrossRef]
- Duan, Z.; Person, R.E.; Lee, H.H.; Huang, S.; Donadieu, J.; Badolato, R.; Grimes, H.L.; Papayannopoulou, T.; Horwitz, M.S. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Mol. Cell Biol. 2007, 27, 6889–6902. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Angelin-Duclos, C.; Greenwood, J.; Liao, J.; Calame, K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 2000, 20, 2592–2603. [Google Scholar] [CrossRef]
- Fog, C.K.; Galli, G.G.; Lund, A.H. PRDM proteins: Important players in differentiation and disease. Bioessays 2012, 34, 50–60. [Google Scholar] [CrossRef]
- Voronezhskaya, E.E.; Tyurin, S.A.; Nezlin, L.P. Neuronal development in larval chiton Ischnochiton hakodadensis (Mollusca: Polyplacophora). J. Comp. Neurol. 2002, 444, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Dyachuk, V.; Odintsova, N. Development of the larval muscle system in the mussel Mytilus trossulus (Mollusca, Bivalvia). Dev. Growth Differ. 2009, 51, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Pavlicek, A.; Schwaha, T.; Wanninger, A. Towards a ground pattern reconstruction of bivalve nervous systems: Neurogenesis in the zebra mussel Dreissena polymorpha. Org. Divers. Evol. 2018, 18, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, O.V.; Skiteva, O.I.; Voronezhskaya, E.E.; Dyachuk, V.A. Nervous system development in the Pacific oyster, Crassostrea gigas (Mollusca: Bivalvia). Front. Zool. 2018, 15, 10. [Google Scholar] [CrossRef]
- Liu, C.; Ma, W.; Su, W.; Zhang, J. Prdm14 acts upstream of islet2 transcription to regulate axon growth of primary motoneurons in zebrafish. Development 2012, 139, 4591–4600. [Google Scholar] [CrossRef]
- Chittka, A.; Nitarska, J.; Grazini, U.; Richardson, W.D. Transcription factor positive regulatory domain 4 (PRDM4) recruits protein arginine methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J. Biol. Chem. 2012, 287, 42995–43006. [Google Scholar] [CrossRef] [PubMed]
- Kinameri, E.; Inoue, T.; Aruga, J.; Imayoshi, I.; Kageyama, R.; Shimogori, T.; Moore, A.W. Prdm proto-oncogene transcription factor family expression and interaction with the Notch-Hes pathway in mouse neurogenesis. PLoS ONE 2008, 3, e3859. [Google Scholar] [CrossRef]
- Woods, F.H. History of the germ cells in Sphaerium striatinum (Lam.). Int. J. Morphol. 1931, 51, 545–595. [Google Scholar] [CrossRef]
- Lyons, D.C.; Perry, K.J.; Lesoway, M.P.; Henry, J.Q. Cleavage pattern and fate map of the mesentoblast, 4d, in the gastropod Crepidula: A hallmark of spiralian development. Evodevo 2012, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Akagi, K.; Ueda, H. Regulatory mechanisms of ecdysone-inducible Blimp-1 encoding a transcriptional repressor that is important for the prepupal development in Drosophila. Dev. Growth Differ. 2011, 53, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, N.; Tong, L.; Bronner, M.E. Ancestral network module regulating prdm1 expression in the lamprey neural plate border. Dev. Dyn. 2011, 240, 2265–2271. [Google Scholar] [CrossRef] [PubMed]
- Livi, C.B.; Davidson, E.H. Expression and function of blimp1/krox, an alternatively transcribed regulatory gene of the sea urchin endomesoderm network. Dev. Biol. 2006, 293, 513–525. [Google Scholar] [CrossRef]
- Wilm, T.P.; Solnica-Krezel, L. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development 2005, 132, 393–404. [Google Scholar] [CrossRef]
- de Souza, F.S.; Gawantka, V.; Gómez, A.P.; Delius, H.; Ang, S.L.; Niehrs, C. The zinc finger gene Xblimp1 controls anterior endomesodermal cell fate in Spemann’s organizer. EMBO J. 1999, 18, 6062–6072. [Google Scholar] [CrossRef]
- Chatfield, J.; O’Reilly, M.A.; Bachvarova, R.F.; Ferjentsik, Z.; Redwood, C.; Walmsley, M.; Patient, R.; Loose, M.; Johnson, A.D. Stochastic specification of PGCs from mesoderm precursors in axolotl embryos. Development 2014, 141, 2429–2440. [Google Scholar] [CrossRef]
- Wan, Z.; Rui, L.; Li, Z. Expression patterns of prdm1 during chicken embryonic and germline development. Cell Tissue Res. 2014, 356, 341–356. [Google Scholar] [CrossRef]
- Saitou, M.; Yamaji, M. Primordial germ cells in mice. Cold Spring Harb. Perspect. Biol. 2012, 4, a008375. [Google Scholar] [CrossRef] [PubMed]
- Saitou, M.; Yamaji, M. Germ cell specification in mice: Signaling, transcription regulation, and epigenetic consequences. Reproduction 2010, 139, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Magnúsdóttir, E.; Dietmann, S.; Murakami, K.; Günesdogan, U.; Tang, F.; Bao, S.; Diamanti, E.; Lao, K.; Gottgens, B.; Azim Surani, M. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 2013, 15, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Yabuta, Y.; Kurimoto, K.; Ohinata, Y.; Seki, Y.; Saitou, M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol. Reprod. 2006, 75, 705–716. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Ml-prdm1 | AGGAGAGGAAATGTTACAAGCA | GGCAGGAGTAGGTGGAGTCTTA |
| Ml-prdm14 | ACTGACGCCTCGGTTTTATC | GTCGGACTCGGTGTTTCTGA |
| Ml-ef1b | GGGCATTACTTCACTCTAAAT | TGTGCTATCTGAGGGTCTACT |
| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Ml-prdm1 | ATTTAGGTGACACTATAGAAGCG 1 CATTTAAGAGCCCGAAGT | TAATACGACTCACTATAGGGAGACA 2 CATAGGAGGGTAAGGTGG |
| Ml-prdm14 | ATTTAGGTGACACTATAGAAGCG 1 GATTGGTTTCCCATTTC | TAATACGACTCACTATAGGGAGACA 2 TCGTGCTCCTCTGACTT |
| Protein | Protein Length | Domain Number | PR Domain Length | pI | Mw (Da) | |
|---|---|---|---|---|---|---|
| PR | Zinc Finger | |||||
| Ml-PRDM1 | 837 | 1 | 5 | 122 | 6.96 | 95,462.42 |
| Ml-PRDM2 | 1969 | 1 | 11 | 122 | 6.04 | 221,527.46 |
| Ml-PRDM4-like | 881 | 1 | 4 | 135 | 5.74 | 98,698.34 |
| Ml-PRDM8-like | 746 | 1 | 3 | 118 | 7.96 | 84,010.47 |
| Ml-PRDM9 | 710 | 1 | 10 | 121 | 8.54 | 81,667.44 |
| Ml-PRDM10 | 1481 | 1 | 12 | 119 | 6.25 | 165,430.32 |
| Ml-PRDM12 | 283 | 1 | 3 | 120 | 9.2 | 32,469.99 |
| Ml-PRDM13-like | 542 | 1 | 0 | 98 | 8.54 | 60,496.27 |
| Ml-PRDM14 | 524 | 1 | 6 | 124 | 8.91 | 59,197.42 |
| Ml-PRDM15 | 1619 | 1 | 20 | 118 | 5.96 | 181,519.82 |
| Ml-PRDM16-like | 879 | 1 | 9 | 49 | 6.76 | 98,759.51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, F.; Guo, X.; Li, X.; Liu, F.; Fu, Y.; Sun, X.; Yang, Z.; Zhang, Z.; Qin, Z. Identification and Expressional Analysis of Putative PRDI-BF1 and RIZ Homology Domain-Containing Transcription Factors in Mulinia lateralis. Biology 2023, 12, 1059. https://doi.org/10.3390/biology12081059
Zhao F, Guo X, Li X, Liu F, Fu Y, Sun X, Yang Z, Zhang Z, Qin Z. Identification and Expressional Analysis of Putative PRDI-BF1 and RIZ Homology Domain-Containing Transcription Factors in Mulinia lateralis. Biology. 2023; 12(8):1059. https://doi.org/10.3390/biology12081059
Chicago/Turabian StyleZhao, Feng, Xiaolin Guo, Xixi Li, Fang Liu, Yifan Fu, Xiaohan Sun, Zujing Yang, Zhifeng Zhang, and Zhenkui Qin. 2023. "Identification and Expressional Analysis of Putative PRDI-BF1 and RIZ Homology Domain-Containing Transcription Factors in Mulinia lateralis" Biology 12, no. 8: 1059. https://doi.org/10.3390/biology12081059
APA StyleZhao, F., Guo, X., Li, X., Liu, F., Fu, Y., Sun, X., Yang, Z., Zhang, Z., & Qin, Z. (2023). Identification and Expressional Analysis of Putative PRDI-BF1 and RIZ Homology Domain-Containing Transcription Factors in Mulinia lateralis. Biology, 12(8), 1059. https://doi.org/10.3390/biology12081059






