Biodiversity of Endophytic Microbes in Diverse Tea Chrysanthemum Cultivars and Their Potential Promoting Effects on Plant Growth and Quality
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and PCR Amplification
2.3. Illumina Sequencing
2.4. Data Analysis
2.5. Isolation and Identification of Endophytic Bacteria and Fungi
2.6. Determination of IAA Content in the Fermentation Broth of Endophytic Bacteria
2.7. Field Experiment Design and Inoculation of IAA-Producing Endophytic Stains
2.8. Measurement of Plant Growth and Quality
3. Results
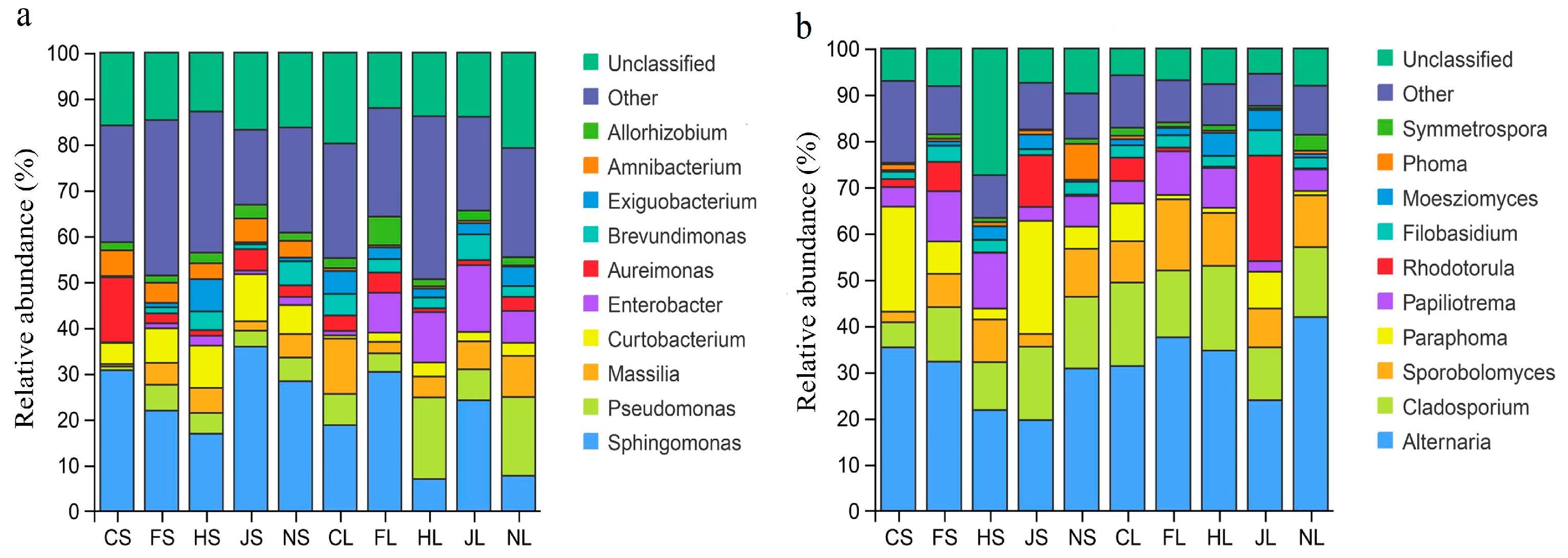
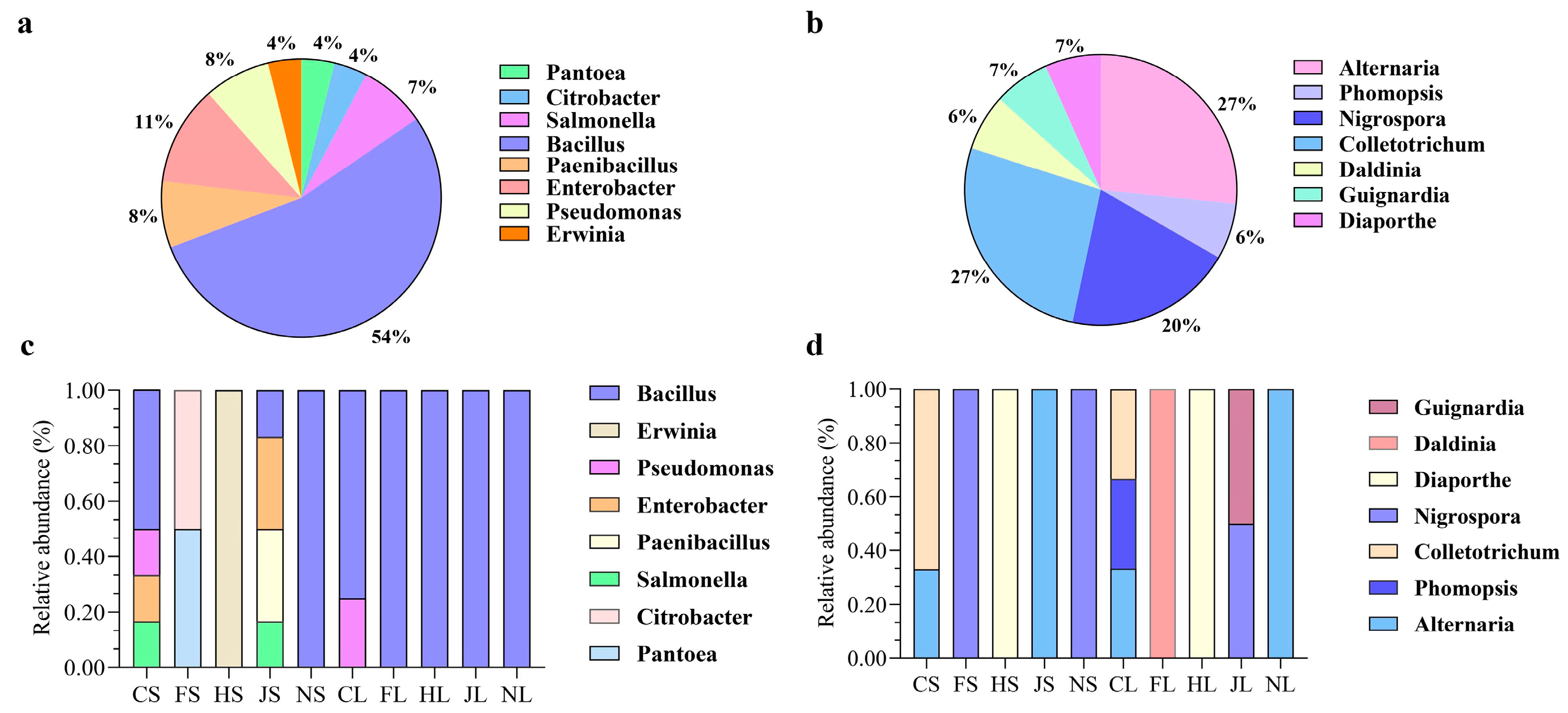
3.1. Composition of Endophytic Microbiomes
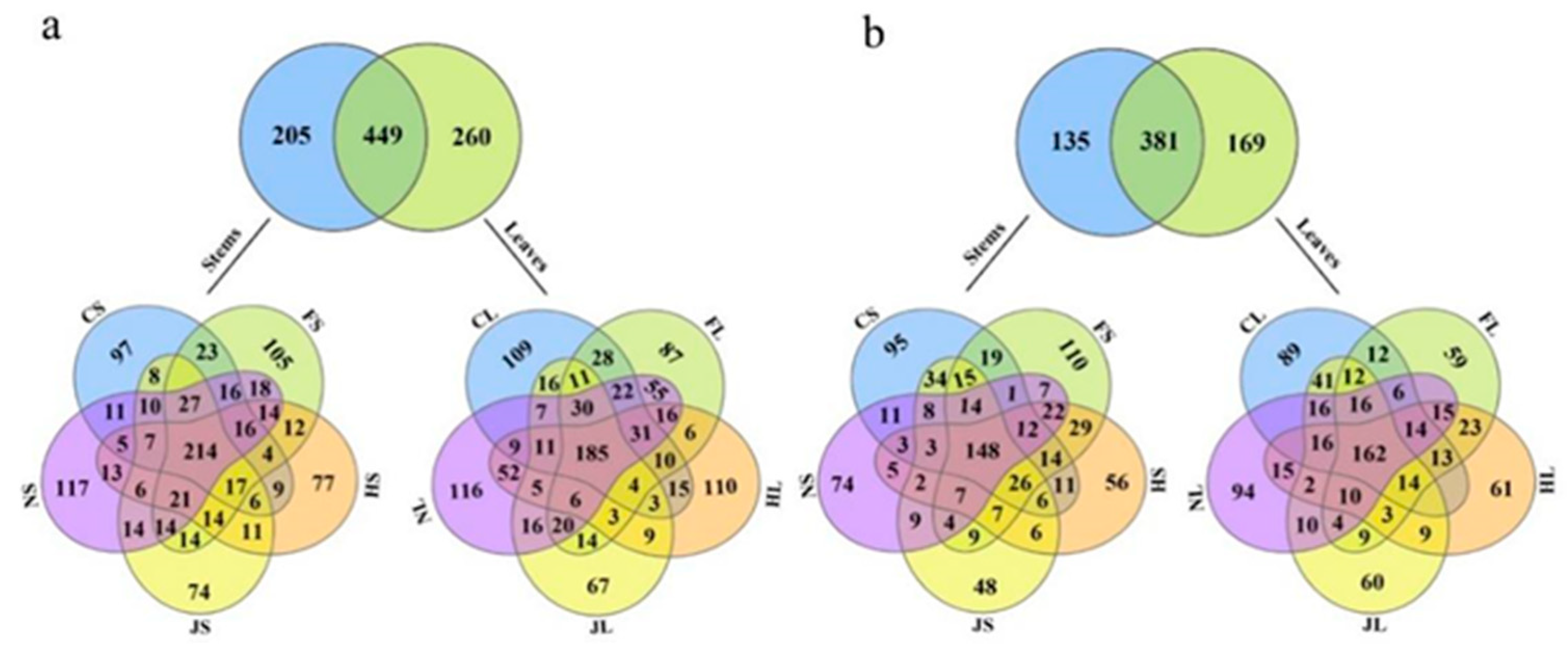
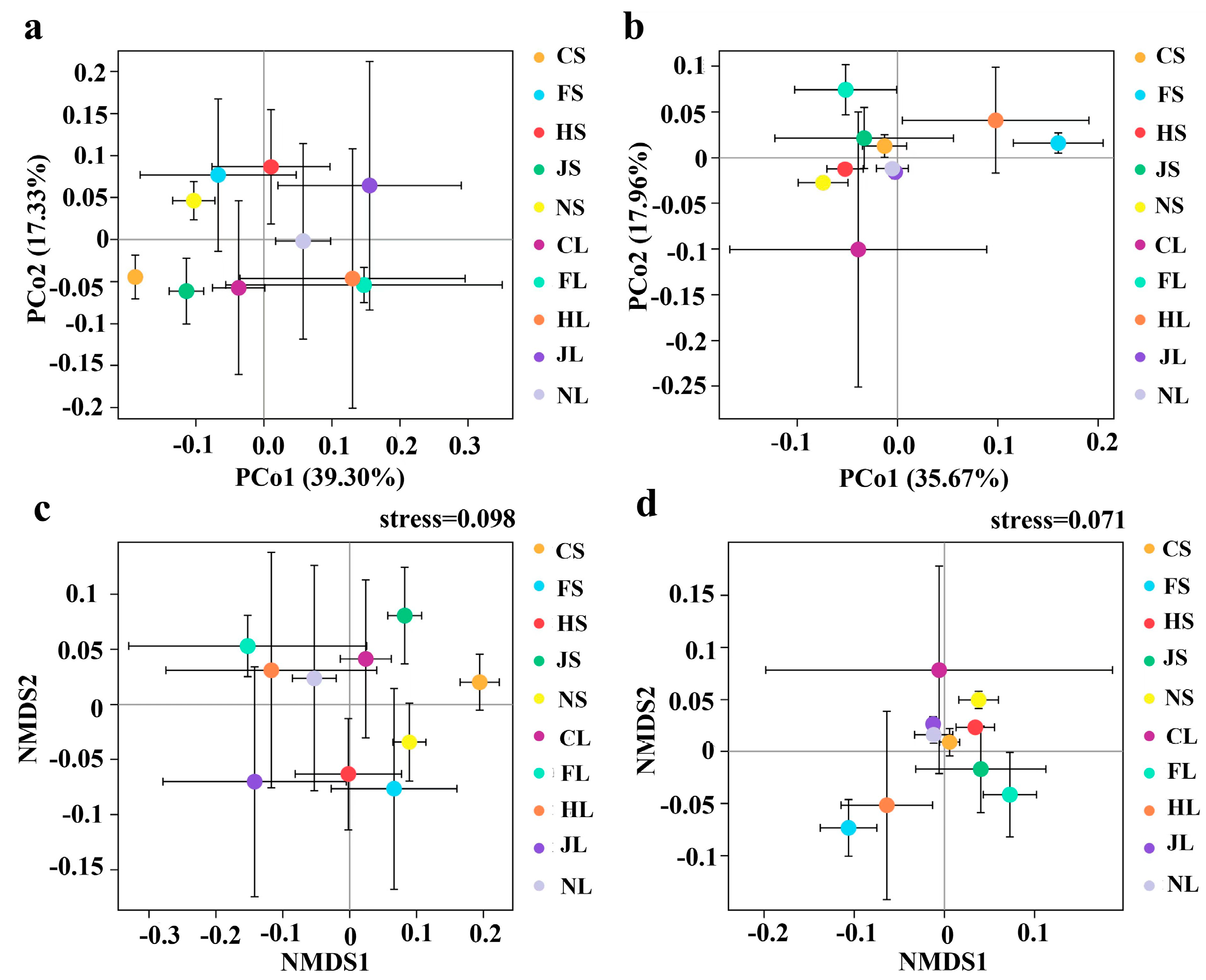
3.2. Diversity of Endophytic Microbiome
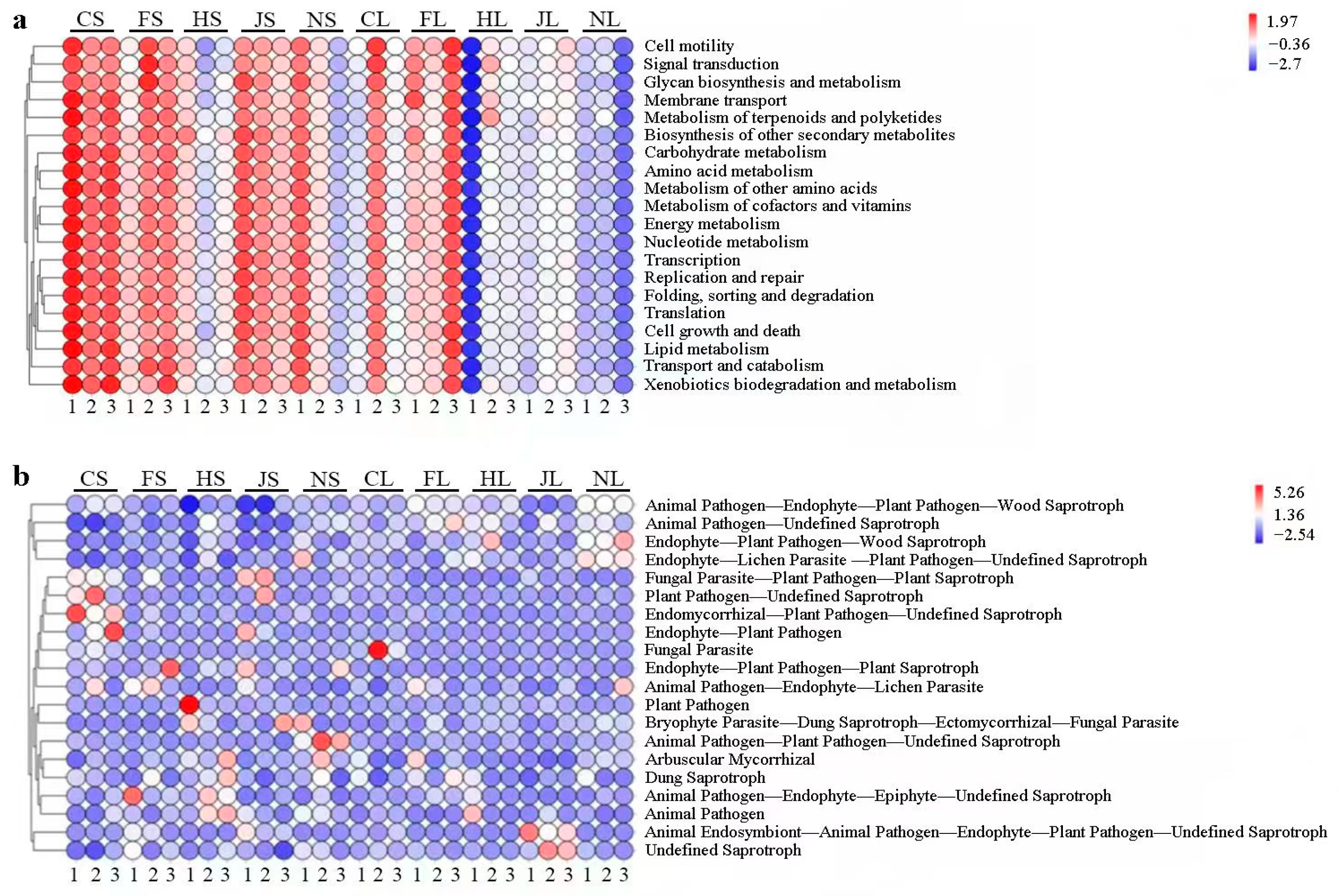
3.3. Gene-Oredicted Functional Profiles of Endophytic Bacterial and Fungi
3.4. Composition of Endophytic Bacterial and Fungal Communities Based on the Culture-Dependent Method
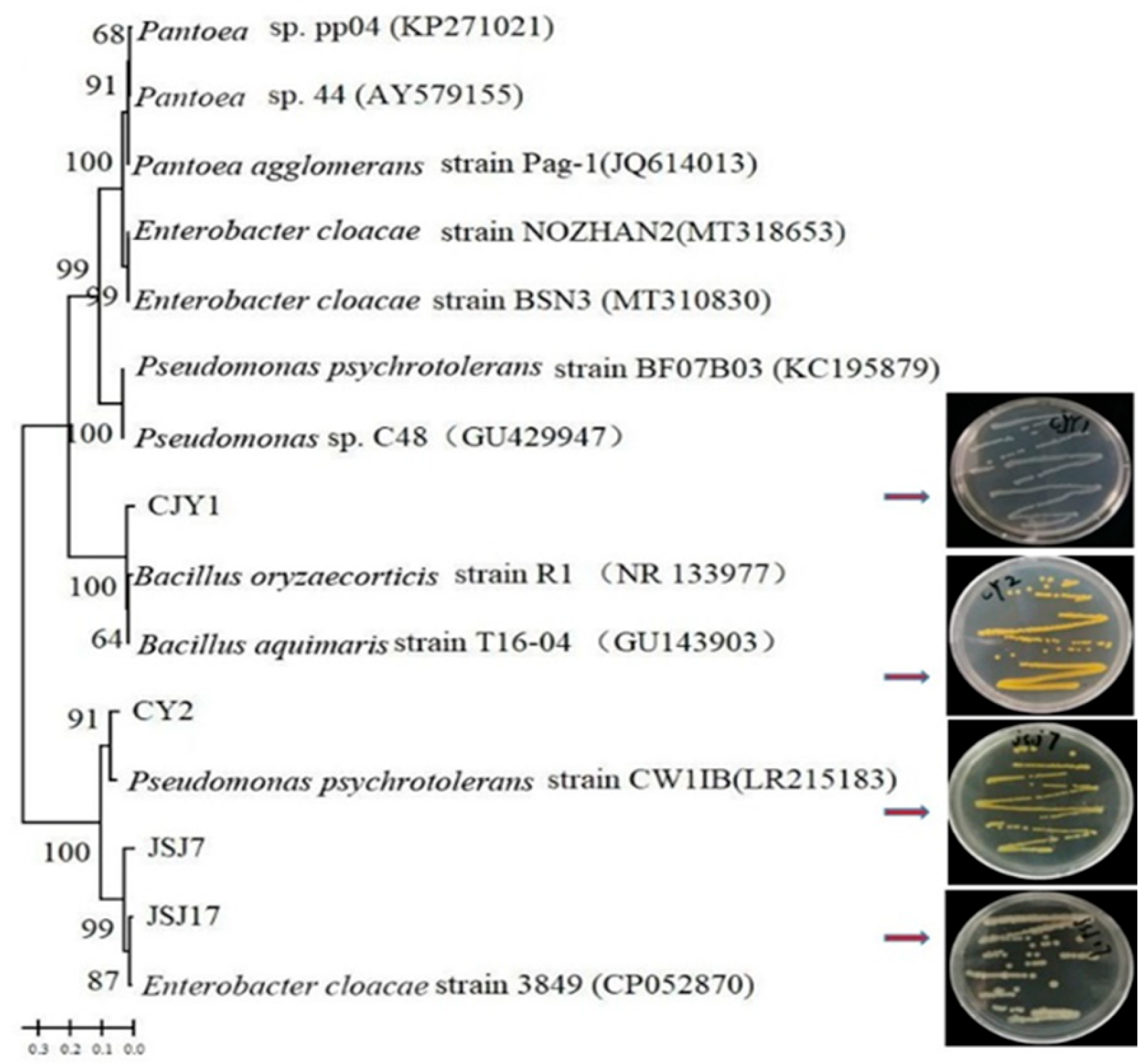
3.5. Selection and Identification of IAA-Producing Endophytic Strains
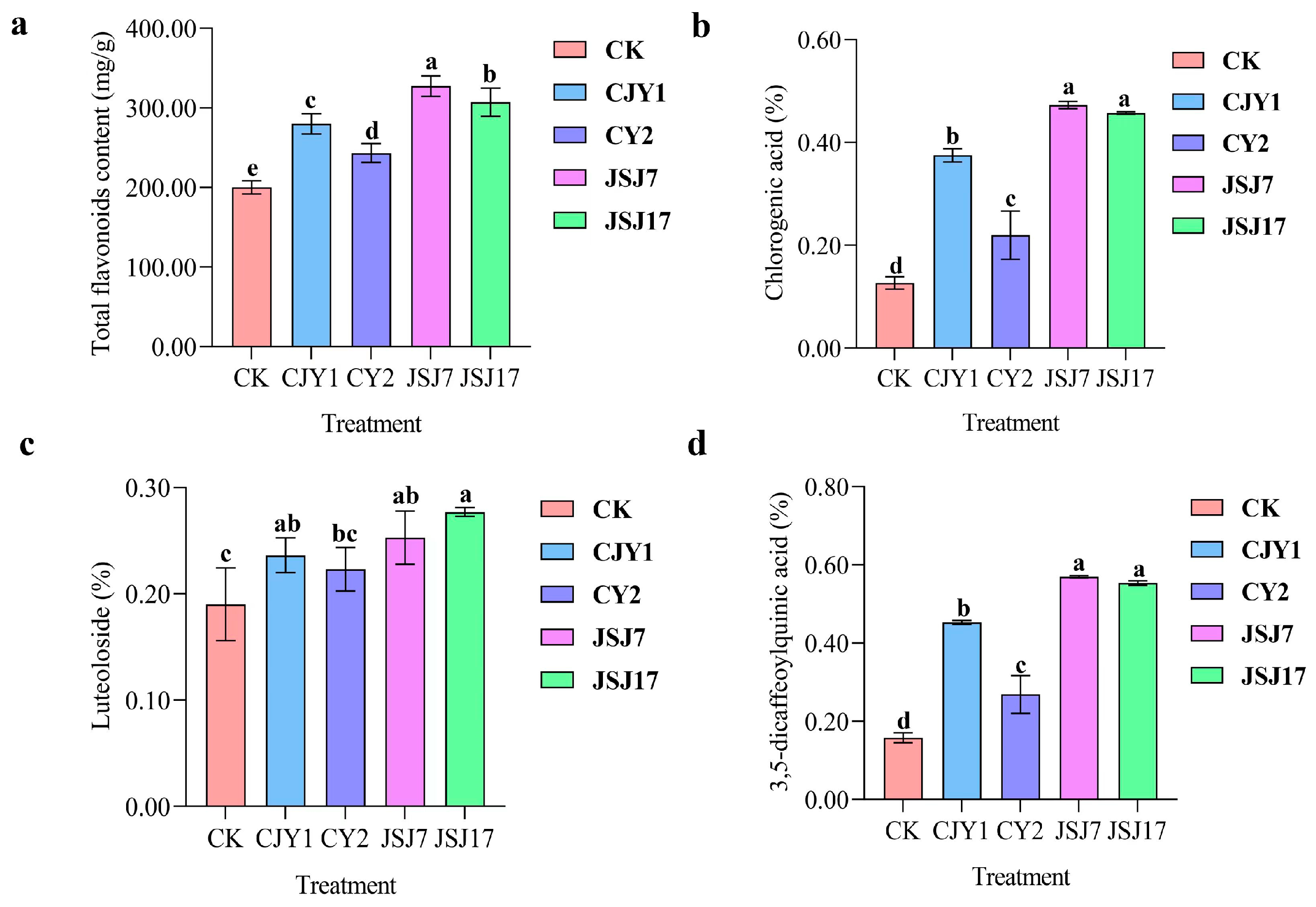
3.6. Effect of IAA-Producing Endophytic Bacterial Inoculation on Plant Growth and Quality
3.7. Effect of IAA-Producing Endophytic Bacterial Inoculation on Plant Yield and Quality
4. Discussion
4.1. Endophytic Community Composition and Diversity in Tea Chrysanthemum Cultivars
4.2. Selection and Evaluation of Plant Growth-Promoting Endophytic Microbes
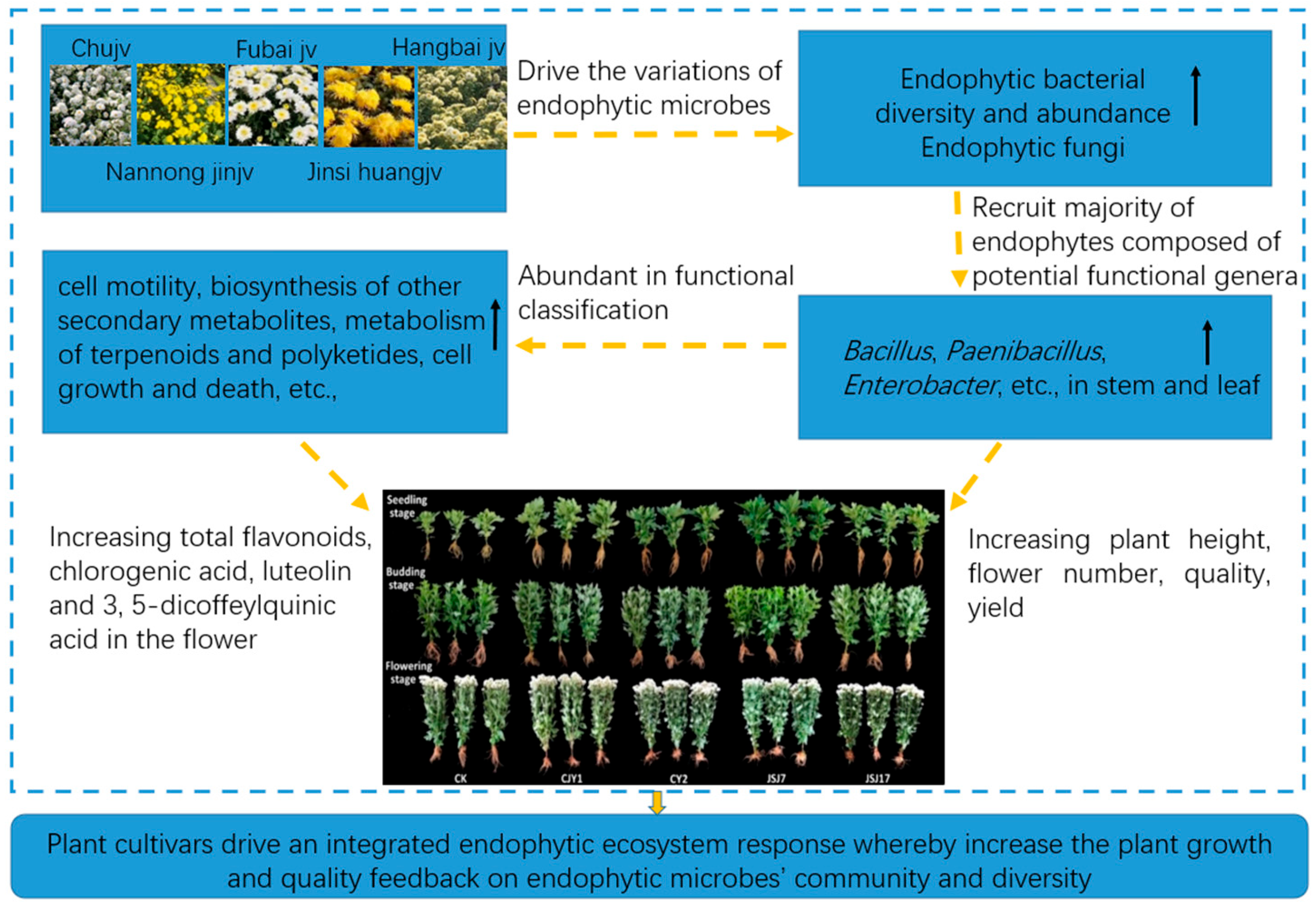
4.3. Interactions of Endophytic Microbes with Plant Cultivars and Their Promoting Effects on Plant Growth and Quality
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hadizadeh, H.; Samiei, L.; Shakeri, A. Chrysanthemum, an ornamental genus with considerable medicinal value: A comprehensive review. S. Afr. J. Bot. 2022, 144, 23–43. [Google Scholar] [CrossRef]
- Chinese Pharmacopoeia Commission. Chinese Pharmacopoeia; China Medical Science and Technology Press: Beijing, China, 2015; pp. 310–311.
- Miao, W.H.; Xiao, X.Y.; Wang, Y.A.; Ge, L.J.; Yang, Y.Y.; Liu, Y.; Liao, Y.; Guan, Z.Y.; Chen, S.M.; Fang, W.M.; et al. CmWRKY6-1–CmWRKY15-like transcriptional cascade negatively regulates the resistance to Fusarium oxysporum infection in Chrysanthemum morifolium. Hortic. Res. 2023, 10, uhad101. [Google Scholar] [CrossRef]
- Chen, H.J.; Zhao, S.; Zhao, J.M.; Zhang, K.K.; Jiang, J.; Guan, Z.Y.; Chen, S.M.; Chen, F.D.; Fang, W.M. Deep tillage combined with bio-fertilizer following soil fumigation improved chrysanthemum growth by regulating the soil microbiome. Microbiology 2020, 9, e1045. [Google Scholar]
- Yang, C.B.; Yan, K.R.; Ma, C.N.A.; Xie, L.; Wang, W.; Chen, W.L.; Mao, B.Z. Insight into the root growth, soil quality, and assembly of the root-associated microbiome in the virus-free chrysanthemum morifolium. Ind. Crops Prod. 2022, 176, 114362. [Google Scholar] [CrossRef]
- Minuto, A.; Gullino, M.L.; Lamberti, F.; D’Addabbo, T.; Tescari, E.; Ajwa, H.; Garibaldi, A. Application of an emulsifiable mixture of 1,3-dichloropropene and chloropicrin against root knot nematodes and soilborne fungi for greenhouse tomatoes in Italy. Crop Prot. 2006, 25, 1244–1252. [Google Scholar] [CrossRef]
- Omar, I.; O’Neill, T.M.; Rossall, S. Biological control of fusarium crown and root rot of tomato with antagonistic bacteria and integrated control when combined with the fungicide carbendazim. Plant Pathol. 2006, 55, 92–99. [Google Scholar] [CrossRef]
- Yang, X.M.; Chen, L.H.; Yong, X.Y.; Shen, Q.R. Formulations can affect rhizosphere colonization and biocontrol efficiency of Trichoderma harzianum SQR-T037 against Fusarium wilt of cucumbers. Biol. Fert. Soils 2011, 47, 239–248. [Google Scholar] [CrossRef]
- Ryan, R.P.; Germaine, K.; Franks, A.; Ryan, D.J.; Dowling, D.N. Bacterial endophytes: Recent developments and applications. FEMS Microbiol. Lett. 2008, 278, 1–9. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Hardoim, C.C.P.; van Overbeek, L.S.; van Elsas, J.D. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE 2012, 7, e30438. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellin, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Hori, K.; Matsumoto, S. Bacterial adhesion: From mechanism to control. Biochem. Eng. J. 2010, 48, 424–434. [Google Scholar] [CrossRef]
- Friesen, M.L.; Porter, S.S.; Stark, S.C.; von Wettberg, E.J.; Sachs, J.L.; Martinez-Romero, E. Microbially mediated plant functional traits. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 23–46. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; van Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; Orozco-Mosqueda, M.D.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Khare, E.; Mishra, J.; Arora, N.K. Multifaceted interactions between endophytes and plant: Developments and prospects. Front. Microbiol. 2018, 9, 2732. [Google Scholar] [CrossRef] [PubMed]
- Prisana, W.; Shin-Ichi, I.; Anurag, S. Volatile organic compounds emitted from endophytic fungus Trichoderma asperellum T1 mediate antifungal activity, defense response and promote plant growth in lettuce (Lactuca sativa). Fungal Ecol. 2020, 43, 100867. [Google Scholar]
- Mashiane, A.R.; Adeleke, R.A.; Bezuidenhout, C.C.; Chirima, G.J. Community composition and functions of endophytic bacteria of Bt maize. S. Afr. J. Sci. 2018, 114, 88–97. [Google Scholar]
- Del Barrio-Duque, A.; Ley, J.; Samad, A.; Antonielli, L.; Sessitsch, A.; Compant, S. Beneficial endophytic bacteria-serendipita indica interaction for crop enhancement and resistance to phytopathogens. Front. Microbiol. 2019, 10, 2888. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef]
- Liu, D.F.; Sun, H.J.; Ma, H.W. Deciphering Microbiome related to rusty roots of Panax ginseng and evaluation of antagonists against pathogenic Ilyonectria. Front. Microbiol. 2019, 10, 1350. [Google Scholar] [CrossRef] [PubMed]
- Haro, C.; Anguita-Maeso, M.; Metsis, M.; Juan ANavas-Cortés Landa, B.B. Evaluation of established methods for DNA extraction and primer pairs targeting 16S rRNA gene for bacterial microbiome profiling of olive xylem sap. Front. Plant Sci. 2021, 12, 296. [Google Scholar] [CrossRef] [PubMed]
- Vancov, T.; Keen, B. Amplification of soil fungal community DNA using the ITS86F and ITS4 primers. FEMS Microbio. Lett. 2009, 296, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.J.; Zhao, J.M.; Jiang, J.; Zhao, Z.G.; Guan, Z.Y.; Chen, S.M.; Chen, F.D.; Fang, W.M.; Zhao, S. Effects of inorganic, organic and bio-organic fertilizer on growth, rhizosphere soil microflora and soil function sustainability in chrysanthemum monoculture. Agriculture 2021, 11, 1214. [Google Scholar] [CrossRef]
- Liu, H.; Li, X.; Wang, X.S. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: Application in study of rice-bacterium interaction. Plant Methods 2012, 8, 2. [Google Scholar] [CrossRef]
- Longley, R.; Noel, Z.A.; Benucci, G.M.N.; Chilvers, M.I.; Trail, F.; Bonito, G. Crop management impacts the soybean (Glycine max) microbiome. Front. Microbiol. 2020, 11, 1116. [Google Scholar] [CrossRef]
- Fan, Z.J.; Xiao, S.M.; Hu, H.Y.; Zhang, P.F.; Chao, J.; Guo, S.; Hou, D.Y.; Xu, J. Endophytic bacterial and fungal community compositions in different organs of ginseng (Panax ginseng). Arch. Microbiol. 2022, 204, 208. [Google Scholar] [CrossRef]
- Wang, S.S.; Liu, J.M.; Sun, J.; Sun, Y.F.; Liu, J.N.; Jia, N.; Fan, B.; Dai, X.F. Diversity of culture-independent bacteria and antimicrobial activity of culturable endophytic bacteria isolated from different Dendrobium stems. Sci. Rep. 2019, 9, 10389. [Google Scholar] [CrossRef]
- Cregger, M.A.; Veach, A.M.; Yang, Z.K.; Crouch, M.J.; Vilgalys, R.; Tuskan, G.A.; Schadt, C.W. The Populus holobiont: Dissecting the effects of plant niches and genotype on the microbiome. Microbiome 2018, 6, 31. [Google Scholar] [CrossRef]
- Wang, W.F.; Zhai, Y.Y.; Cao, L.X.; Tan, H.M.; Zhang, R.D. Endophytic bacterial and fungal microbiota in sprouts, roots and stems of rice (Oryza sativa L.). Microbiol. Res. 2016, 188, 1–8. [Google Scholar] [CrossRef]
- Sturz, A.V.; Nowak, J. Endophytic communities of rhizobacteria and the strategies required to create yield enhancing associations with crops. Appl. Soil. Ecol. 2000, 15, 183–190. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, C.C. Endophytes: A potential resource for biosynthesis, biotransformation, and biodegradation. Ann. Microbiol. 2011, 61, 207–215. [Google Scholar] [CrossRef]
- De Almeida Lopes, K.B.; Carpentieri-Pipolo, V.; Oro, T.H.; Stefani Pagliosa, E.; Degrassi, G. Culturable endophytic bacterial communities associated with field-grown soybean. J. Appl. Microbiol. 2016, 120, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.S.; Patra, J.K. Endophytes: A treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 2016, 7, 1538. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.E.; Kim, J.U.; Lee, J.W.; Lee, S.W.; Jo, I.H. Diversity of bacterial endophytes in Panax ginseng and their protective effects against pathogens. 3 Biotech 2018, 8, 397. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Babalola, O.O. The endosphere microbial communities, a great promise in agriculture. Int. Microbiol. 2021, 24, 1–17. [Google Scholar] [CrossRef]
- Singha, K.M.; Singh, B.; Pandey, P. Host specific endophytic microbiome diversity and associated functions in three varieties of scented black rice are dependent on growth stage. Sci. Rep. 2021, 11, 12259. [Google Scholar] [CrossRef]
- Muller, H.; Berg, C.; Landa, B.B.; Auerbach, A.; Moissl-Eichinger, C.; Berg, G. Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize mediterranean olive trees. Front. Microbiol. 2015, 6, 138. [Google Scholar] [CrossRef]
- Bashir, S.; Iqbal, A.; Hasnain, S. Comparative analysis of endophytic bacterial diversity between two varieties of sunflower Helianthus annuus with their PGP evaluation. Saudi J. Biol. Sci. 2020, 27, 720–726. [Google Scholar] [CrossRef]
- Alawiye, T.T.; Babalola, O.O. Bacterial diversity and community structure in typical plant rhizosphere. Diversity 2019, 11, 179. [Google Scholar] [CrossRef]
- Verma, S.C.; Ladha, J.K.; Tripathi, A.K. Evaluation of plant growth promoting and colonization ability of endophytic diazotrophs from deep water rice. J. Biotechnol. 2001, 91, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Kosaka, Y.; Tsuge, S.; Kubo, Y.; Horino, O. Evaluation of the endophyte Enterobacter cloacae SM10 isolated from spinach roots for biological control against Fusarium wilt of spinach. J. Gen. Plant Pathol. 2001, 67, 78–84. [Google Scholar] [CrossRef]
- Montanez, A.; Abreu, C.; Gill, P.R.; Hardarson, G.; Sicardi, M. Biological nitrogen fixation in maize (Zea mays L.) by 15N isotope-dilution and identification of associated culturable diazotrophs. Biol. Fert. Soils 2009, 45, 253–263. [Google Scholar] [CrossRef]
- Lodewyckx, C.; Vangronsveld, J.; Porteous, F.; Moore, E.R.B.; Taghavi, S.; Mezgeay, M.; van der Lelie, D. Endophytic bacteria and their potential applications. Crit. Rev. Plant Sci. 2002, 21, 583–606. [Google Scholar] [CrossRef]
- Ahmad, T.; Farooq, S.; Mirza, D.N.; Kumar, A.; Mir, R.A.; Riyaz-Ul-Hassan, S. Insights into the endophytic bacterial microbiome of Crocus sativus: Functional characterization leads to potential agents that enhance the plant growth, productivity, and key metabolite content. Microb. Ecol. 2022, 83, 669–688. [Google Scholar] [CrossRef]
- Wu, T.; Li, X.B.; Xu, J.; Liu, L.X.; Ren, L.L.; Dong, B.; Li, W.; Xie, W.J.; Yao, Z.G.; Chen, Q.F.; et al. Diversity and functional characteristics of endophytic bacteria from two grass species growing on an oil-contaminated site in the Yellow River Delta, China. Sci. Total Environ. 2021, 767, 144340. [Google Scholar] [CrossRef]
- Huang, K.; Chen, C.; Zhang, J.; Tang, Z.; Shen, Q.R.; Rosen, B.P.; Zhao, F.J. Efficient arsenic methylation and volatilization mediated by a novel bacterium from an arsenic-contaminated paddy soil. Environ. Sci. Technol. 2016, 50, 6389–6396. [Google Scholar] [CrossRef]
- Glick, B.R.; Bashan, Y. Genetic manipulation of plant growth-promoting bacteria to enhance biocontrol of phytopathogens. Biotechnol. Adv. 1997, 15, 353–378. [Google Scholar] [CrossRef]
| Samples | Alpha Diversity Indices of Endophytic Bacteria | Alpha Diversity Indices of Endophytic Fungi | ||||||
|---|---|---|---|---|---|---|---|---|
| Chao 1 | Ace | Simpson | Shannon | Chao 1 | Ace | Shannon | Simpson | |
| 619.86 ± 45.23 a | 620.85 ± 35.24 a | 5.51 ± 0.29 a | 0.94 ± 0.004 a | 426.26 ± 28.78 c | 416.89 ± 19.68 c | 3.34 ± 0.32 c | 0.86 ± 0.004 c | |
| FS | 549.32 ± 25.12 c | 544.05 ± 27.12 c | 4.98 ± 0.15 c | 0.92 ± 0.007 c | 421.24 ± 32.56 c | 418.57 ± 21.58 c | 3.44 ± 0.25 c | 0.87 ± 0.004 c |
| HS | 496.90 ± 29.35 d | 498.47 ± 47.21 d | 4.24 ± 0.36 d | 0.90 ± 0.002 d | 457.10 ± 31.26 a | 443.66 ± 15.74 a | 4.86 ± 0.25 a | 0.80 ± 0.005 a |
| JS | 569.84 ± 15.28 b | 578.02 ± 38.12 b | 5.24 ± 0.35 b | 0.93 ± 0.005 b | 439.64 ± 19.56 b | 431.84 ± 15.2 b | 3.77 ± 0.18 b | 0.83 ± 0.003 b |
| NS | 502.56 ± 47.26 d | 510.24 ± 42.59 d | 4.51 ± 0.43 d | 0.90 ± 0.007 d | 424.14 ± 24.36 c | 420.84 ± 11.26 c | 3.39 ± 0.24 c | 0.86 ± 0.004 c |
| CL | 682.85 ± 51.23 a | 679.16 ± 58.42 a | 6.75 ± 0.31 a | 0.94 ± 0.006 a | 421.54 ± 17.56 b | 399.45 ± 34.57 b | 3.54 ± 0.17 b | 0.83 ± 0.003 b |
| FL | 516.82 ± 38.69 c | 514.21 ± 47.58 c | 5.29 ± 0.25 c | 0.91 ± 0.002 c | 418.42 ± 35.64 b | 415.83 ± 23.69 b | 3.49 ± 0.24 b | 0.84 ± 0.002 a |
| HL | 507.52 ± 34.89 c | 508.83 ± 41.29 c | 5.14 ± 0.27 c | 0.91 ± 0.005 c | 415.03 ± 23.56 b | 407.82 ± 31.56 b | 3.61 ± 0.29 b | 0.83 ± 0.003 b |
| JL | 575.55 ± 38.21 b | 545.07 ± 35.21 b | 5.98 ± 0.38 b | 0.93 ± 0.007 b | 453.50 ± 22.25 a | 449.65 ± 21.35 a | 3.93 ± 0.32 a | 0.80 ± 0.001 a |
| NL | 510.90 ± 59.32 c | 522.43 ± 54.67 c | 5.26 ± 0.47 c | 0.90 ± 0.007 a | 419.93 ± 21.25 b | 412.57 ± 20.45 b | 3.59 ± 0.33 b | 0.83 ± 0.001 b |
| Shoot | Stem | Leave | Root | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Growth Stages | Treatment | Height (cm) | Crown Diameter (cm) | Diameter (mm) | Fresh wt (g) | Dry wt (g) | Fresh wt (g) | Dry wt (g) | Dry wt (g) |
| Seedling stage (30 d) | CK | 16.08 ± 1.08 c | 7.41 ± 13.4 b | 4.19 ± 0.35 b | 4.22 ± 1.09 c | 0.66 ± 0.11 c | 12.45 ± 2.92 c | 1.45 ± 0.28 c | 0.33 ± 0.63 b |
| CJY1 | 21.05 ± 1.12 ab | 9.57 ± 5.22 a | 5.05 ± 0.27 a | 8.04 ± 1.25 ab | 1.21 ± 0.18 ab | 21.08 ± 2.85 ab | 2.45 ± 0.32 ab | 0.38 ± 0.10 b | |
| CY2 | 20.05 ± 1.10 b | 9.82 ± 8.52 a | 4.86 ± 0.48 a | 6.64 ± 0.95 b | 1.03 ± 0.15 b | 18.08 ± 1.47 b | 2.19 ± 0.23 b | 0.52 ± 0.12 a | |
| JSJ7 | 21.90 ± 1.03 a | 9.99 ± 4.36 a | 4.96 ± 0.29 a | 8.03 ± 1.49 a | 1.34 ± 0.20 a | 22.28 ± 2.09 a | 2.67 ± 0.23 a | 0.58 ± 0.06 a | |
| JSJ17 | 20.47 ± 0.84 b | 8.17 ± 8.61 b | 4.40 ± 0.30 b | 6.94 ± 1.06 b | 1.09 ± 0.13 b | 19.61 ± 1.68 ab | 2.36 ± 0.29 ab | 0.58 ± 0.08 a | |
| Budding stage (60 d) | CK | 40.46 ± 1.93 b | 13.78 ± 29.7 c | 5.49 ± 0.78 b | 25.85 ± 5.54 c | 4.41 ± 0.68 c | 53.52 ± 5.54 d | 6.38 ± 0.60 c | 1.02 ± 0.19 b |
| CJY1 | 45.38 ± 1.47 a | 15.25 ± 17.3 bc | 6.42 ± 0.71 a | 35.24 ± 3.93 b | 6.60 ± 0.82 b | 69.20 ± 0.19 c | 8.63 ± 0.68 b | 1.01 ± 0.95 b | |
| CY2 | 45.31 ± 1.81 a | 17.20 ± 5.87 ab | 6.77 ± 0.32 a | 37.26 ± 2.83 ab | 7.55 ± 0.68 ab | 73.85 ± 3.61 bc | 9.84 ± 0.76 ab | 1.61 ± 0.20 a | |
| JSJ7 | 46.80 ± 2.20 a | 19.60 ± 0.18 a | 6.89 ± 0.16 a | 43.80 ± 5.58 a | 8.19 ± 0.89 a | 82.78 ± 8.08 a | 10.19 ± 0.70 a | 1.56 ± 0.14 a | |
| JSJ17 | 46.32 ± 1.02 a | 18.63 ± 12.04 a | 6.89 ± 0.34 a | 42.09 ± 4.91 a | 7.80 ± 0.99 ab | 80.40 ± 2.44 ab | 10.06 ± 0.55 ab | 1.28 ± 0.17 b | |
| Flowering stage (90 d) | CK | 47.98 ± 0.8 d | 25.09 ± 22.67 b | 8.17 ± 0.88 ab | 54.52 ± 9.91 b | 13.83 ± 2.45 b | 60.03 ± 8.25 b | 7.24 ± 1.06 c | 2.39 ± 0.62 b |
| CJY1 | 66.23 ± 2.45 b | 28.23 ± 11.53 b | 8.07 ± 0.41 b | 80.52 ± 12.44 a | 20.43 ± 2.23 a | 89.61 ± 14.39 a | 10.62 ± 1.78 b | 2.43 ± 0.62 ab | |
| CY2 | 61.93 ± 1.29 c | 29.15 ± 17.99 b | 8.58 ± 0.53 ab | 81.45 ± 9.97 a | 21.71 ± 2.39 a | 85.02 ± 10.67 a | 11.59 ± 1.27 ab | 3.31 ± 0.79 a | |
| JSJ7 | 68.98 ± 1.71 a | 33.62 ± 35.67 a | 8.35 ± 0.44 ab | 85.77 ± 6.70 a | 24.06 ± 2.11 a | 96.93 ± 6.69 a | 12.86 ± 1.11 a | 3.17 ± 0.54 ab | |
| JSJ17 | 62.70 ± 2.91 c | 27.33 ± 23.96 b | 8.91 ± 0.15 b | 91.83 ± 14.06 a | 24.16 ± 3.84 a | 102.01 ± 17.21 a | 12.60 ± 2.20 ab | 3.26 ± 0.61 ab | |
| Flower | ||||||
|---|---|---|---|---|---|---|
| Treatment | Diameter (mm) | Inflorescence Number | Fresh wt (g) | Fresh wt Single Flower/g | Fresh wt Per Plant/g | Estimated Yield Per mu/kg |
| CK | 44.84 ± 1.20 b | 49.17 ± 4.14 c | 1.32 ± 0.09 c | 0.18 ± 0.01 b | 94.90 ± 5.46 b | 758.20 ± 43.72 b |
| CJY1 | 49.77 ± 1.40 a | 79.67 ± 10.84 ab | 2.00 ± 0.12 b | 0.26 ± 0.02 a | 159.33 ± 21.68 a | 1274.68 ± 173.48 a |
| CY2 | 50.40 ± 0.73 a | 73.67 ± 4.96 b | 1.94 ± 0.11 b | 0.26 ± 0.01 a | 142.91 ± 9.61 a | 1143.32 ± 76.91 a |
| JSJ7 | 50.36 ± 0.84 a | 85.17 ± 5.02 a | 1.91 ± 0.05 b | 0.26 ± 0.01 a | 165.92 ± 6.19 a | 1325.50 ± 77.14 a |
| JSJ17 | 51.11 ± 0.79 a | 74.33 ± 11.00 ab | 2.14 ± 0.10 a | 0.27 ± 0.01 a | 171.07 ± 17.71 a | 1366.84 ± 203.93 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, T.; Yang, Y.; Duan, K.; Liao, Y.; Zhang, Z.; Guan, Z.; Chen, S.; Fang, W.; Chen, F.; Zhao, S. Biodiversity of Endophytic Microbes in Diverse Tea Chrysanthemum Cultivars and Their Potential Promoting Effects on Plant Growth and Quality. Biology 2023, 12, 986. https://doi.org/10.3390/biology12070986
Sun T, Yang Y, Duan K, Liao Y, Zhang Z, Guan Z, Chen S, Fang W, Chen F, Zhao S. Biodiversity of Endophytic Microbes in Diverse Tea Chrysanthemum Cultivars and Their Potential Promoting Effects on Plant Growth and Quality. Biology. 2023; 12(7):986. https://doi.org/10.3390/biology12070986
Chicago/Turabian StyleSun, Tong, Yanrong Yang, Kuolin Duan, Yuan Liao, Zhi Zhang, Zhiyong Guan, Sumei Chen, Weimin Fang, Fadi Chen, and Shuang Zhao. 2023. "Biodiversity of Endophytic Microbes in Diverse Tea Chrysanthemum Cultivars and Their Potential Promoting Effects on Plant Growth and Quality" Biology 12, no. 7: 986. https://doi.org/10.3390/biology12070986
APA StyleSun, T., Yang, Y., Duan, K., Liao, Y., Zhang, Z., Guan, Z., Chen, S., Fang, W., Chen, F., & Zhao, S. (2023). Biodiversity of Endophytic Microbes in Diverse Tea Chrysanthemum Cultivars and Their Potential Promoting Effects on Plant Growth and Quality. Biology, 12(7), 986. https://doi.org/10.3390/biology12070986








