Diversity, Genomics and Symbiotic Characteristics of Sinorhizobia That Nodulate Desmanthus spp. in Northwest Argentina
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation, Cultivation, and Preservation of Rhizobia from Northwest Argentina with the Ability to Nodulate Desmanthus spp.
2.2. Evaluation of the Bacterial Tolerance to Abiotic Stresses That Are Frequently Present in Soils Populated with D. virgatus and D. paspalaceus
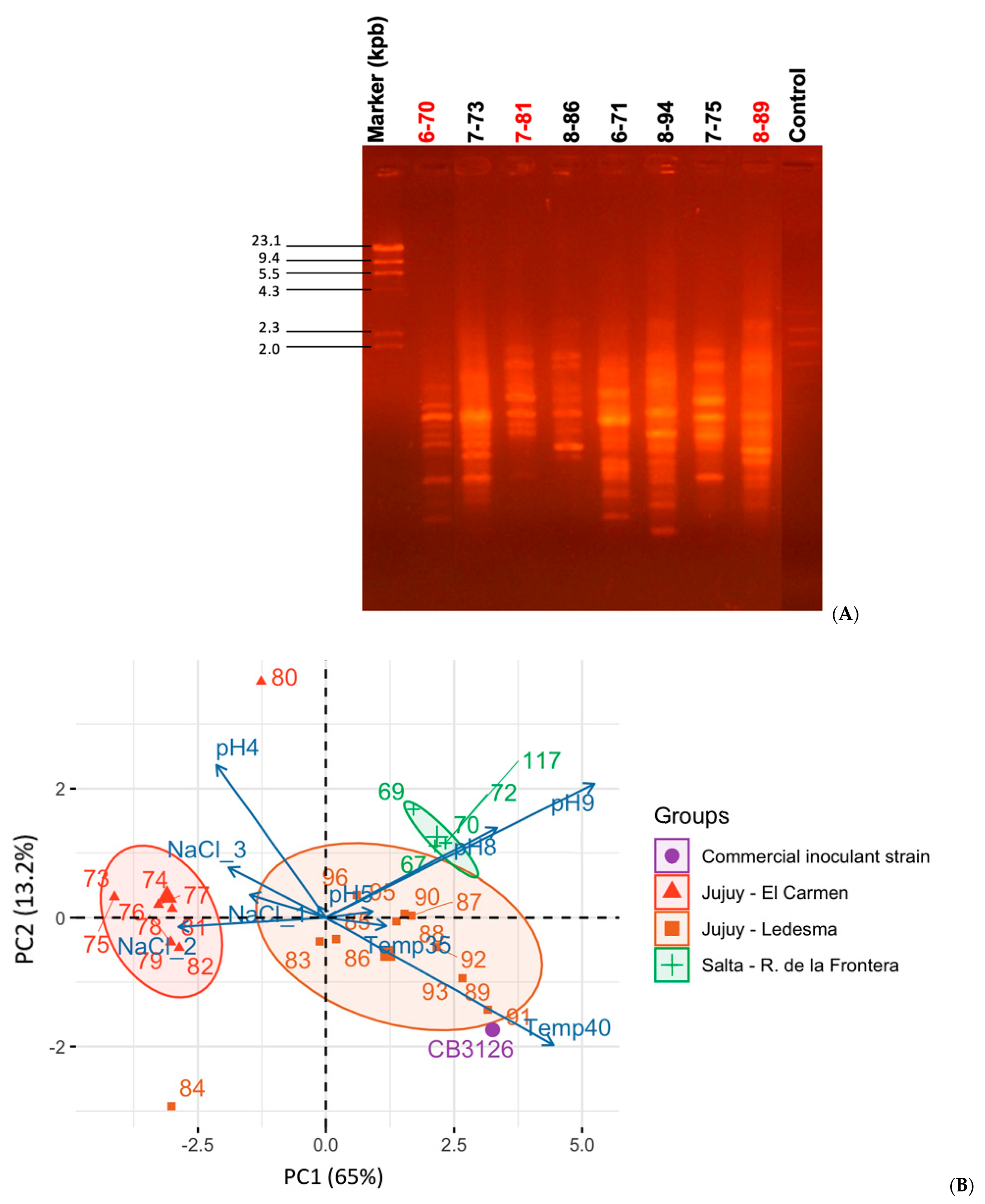
2.3. Typing and Analysis of Diversity in the Collection of Desmanthus-Nodulating Isolates
2.3.1. Biotyping by Matrix-Assisted Laser Desorption-Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF)
2.3.2. Evaluation of Genomic Diversity by BOXA1R PCR-Fingerprint Analysis
2.3.3. Genomic Sequencing and Annotation of Selected Root-Nodule Bacteria
2.4. Phylogenetic Analyses of Selected Root-Nodule Bacteria
2.4.1. Amplification of a Partial Sequence of the 16S rDNA
2.4.2. Analysis of Average Nucleotide Identity (ANIb) and Whole Genome Distance-Based Phylogenetic Tree
2.4.3. Phylogeny of Nod Symbiotic Markers
2.5. Nodulation Tests to Characterize the Symbiotic Performances of Selected Sinorhizobial Isolates from Jujuy and Salta
2.6. Statistical Calculations
3. Results
3.1. Sinorhizobium spp., the Most Abundant Desmanthus-Nodulating Rhizobia Isolated from Soils of the Provinces of Salta and Jujuy, Argentina: Diversity and Phenotypic Characteristics
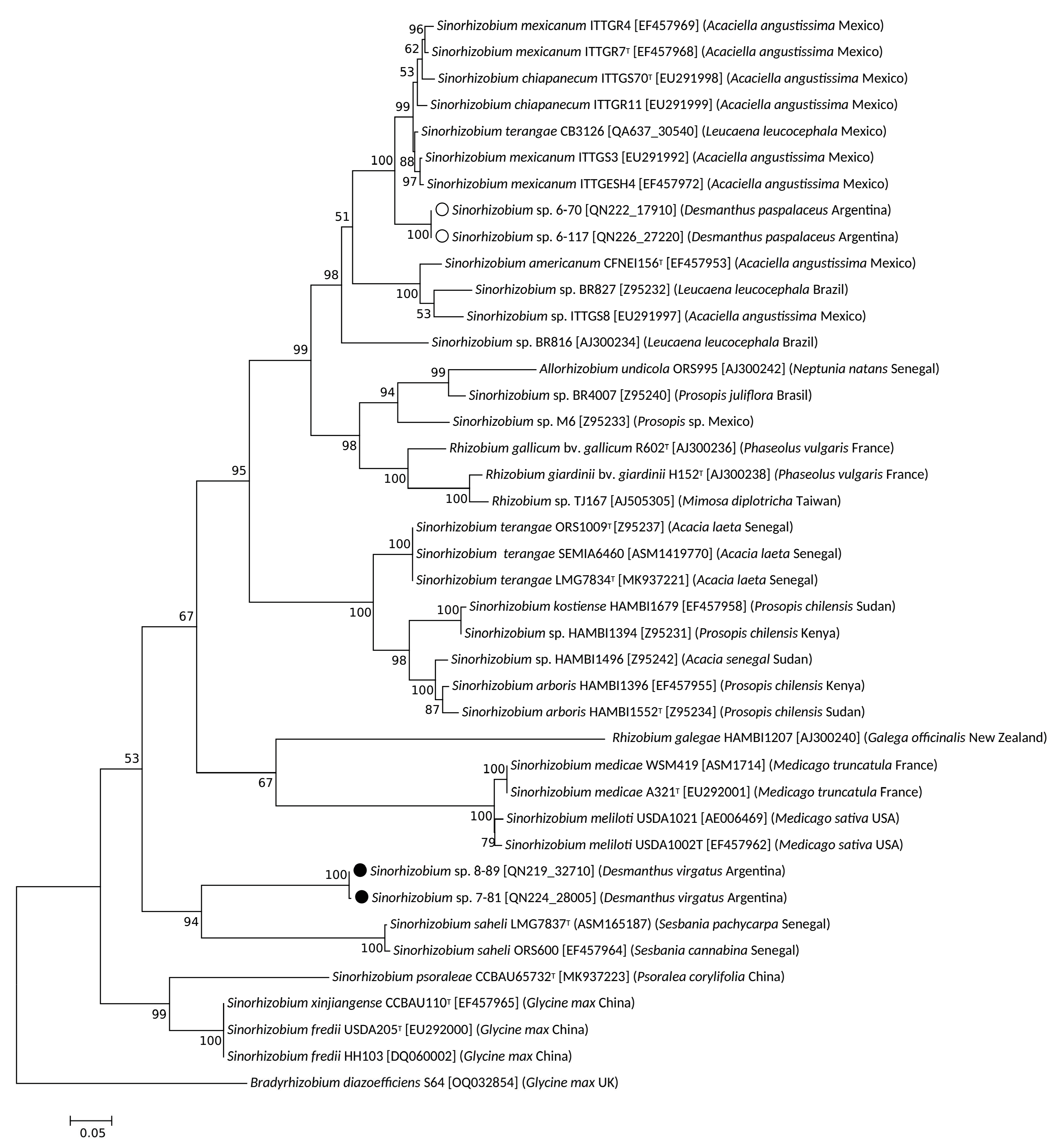
3.2. Taxonomic Position of Selected Desmanthus-Nodulating Sinorhizobial Isolates
3.3. Symbiotic Genes in Desmanthus-Nodulating Sinorhizobia and Nod-Gene Phylogenies
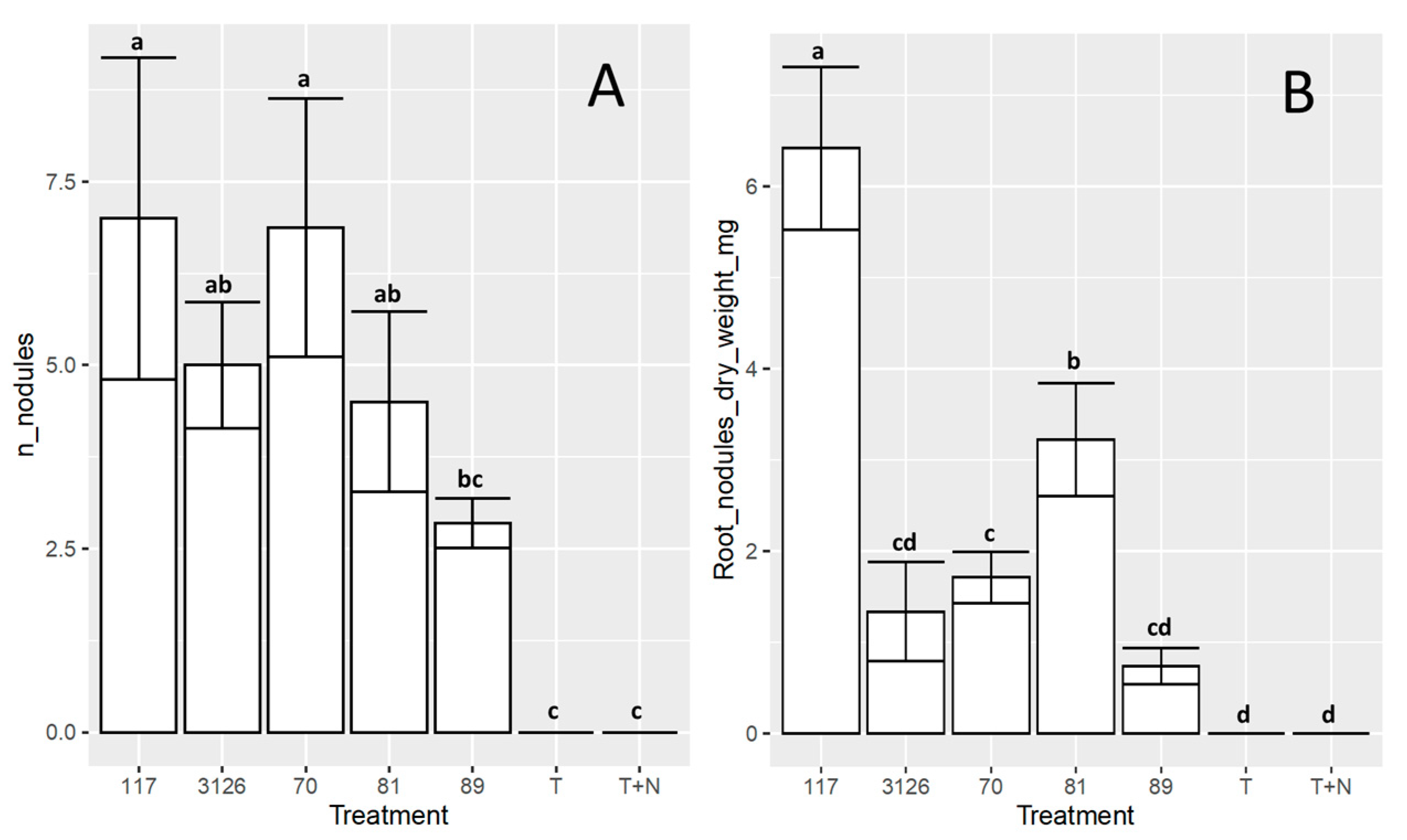
3.4. Symbiosis between the Sinorhizobial Isolates 6-70, 6-117, 7-81, or 8-89 and D. virgatus under Laboratory Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lewis, G.P. (Ed.) Legumes of the World; Royal Botanic Gardens, Kew: Richmond, UK, 2005; ISBN 978-1-900347-80-8. [Google Scholar]
- The Legume Phylogeny Working Group; Bruneau, A.; Doyle, J.J.; Herendeen, P.; Hughes, C.; Kenicer, G.; Lewis, G.; Mackinder, B.; Pennington, R.T.; Sanderson, M.J.; et al. Legume Phylogeny and Classification in the 21st Century: Progress, Prospects and Lessons for Other Species-Rich Clades. Taxon 2013, 62, 217–248. [Google Scholar] [CrossRef]
- Rao, I.M.; Peters, M.; Castro, A.; Schultze-Kraft, R.; White, D.; Fisher, M.; Miles, J.W.; Lascano Aguilar, C.E.; Blümmel, M.; Bungenstab, D.J.; et al. LivestockPlus—The Sustainable Intensification of Forage-Based Agricultural Systems to Improve Livelihoods and Ecosystem Services in the Tropics. Trop. Grassl. 2015, 3, 59–82. [Google Scholar] [CrossRef]
- Schultze-Kraft, R.; Rao, I.M.; Peters, M.; Clements, R.J.; Bai, C.; Liu, G. Tropical Forage Legumes for Environmental Benefits: An Overview. Trop. Grassl. Trop. 2018, 6, 1–14. [Google Scholar] [CrossRef]
- Dewhurst, R.; Delaby, L.; Moloney, A.; Boland, T.; Lewis, E. Nutritive Value of Forage Legumes Used for Grazing and Silage. Ir. J. Agric. Food Res. 2009, 48, 167–187. [Google Scholar]
- Calado, T.B.; Cunha, M.V.D.; Teixeira, V.I.; Santos, M.V.F.D.; Cavalcanti, H.S.; Lira, C.C. Morphology And Productivity Of “Jureminha” Genotypes (Desmanthus spp.) Under Different Cutting Intensities. Rev. Caatinga 2016, 29, 742–752. [Google Scholar] [CrossRef]
- Fischbach, J.A.; Peterson, P.R.; Ehlke, N.J.; Wyse, D.L.; Sheaffer, C.C. Illinois Bundleflower Forage Potential in the Upper Midwestern USA: II. Forage Quality. Agron. J. 2005, 97, 895–903. [Google Scholar] [CrossRef]
- Fontenele, A.C.F.; Aragão, W.M.; Rangel, J.H.d.A.; Almeida, S.A. Leguminosas Tropicais: Desmanthus virgatus (L.) Willd. Uma Forrageira Promissora. Curr. Agric. Sci. Technol. 2009, 15, 121–123. [Google Scholar]
- Pensiero, J.F.; Zabala, J.M. Recursos Fitogenéticos Forrajeros Nativos y Naturalizados Para Los Bajos Submeridionales: Prospección y Priorización de Especies Para Planes de Introducción a Cultivo. Fave. Sección Cienc. Agrar. 2017, 16, 67–98. [Google Scholar] [CrossRef]
- Gardiner, C.; Kempe, N.; Hannah, I.; McDonald, J. Progardes: A Legume for Tropical/Subtropical Semi Arid Tropical Clay Soils. Trop. Grassl. 2013, 1, 78–80. [Google Scholar] [CrossRef]
- Jones, R.; Clem, R. The Role of Genetic Resources in Developing Improved Pastures in Semi-Arid and Subhumid Northern Australia. Trop. Grassl. 1997, 31, 315–319. [Google Scholar]
- Luckow, M. Monograph of Desmanthus (Leguminosae-Mimosoideae). Syst. Bot. Monogr. 1993, 38, 1–166. [Google Scholar] [CrossRef]
- Zuloaga, F.O.; Morrone, O. Catálogo de Las Plantas Vasculares de La República Argentina II; Missouri Botanical Garden Press: St. Louis, MO, USA, 1999. [Google Scholar]
- Clem, R.; Hall, T. Persistence and Productivity of Tropical Pasture Legumes on Three Cracking Clay Soils (Vertisols) in North-Eastern Queensland. Aust. J. Exp. Agric. 1994, 34, 161–171. [Google Scholar] [CrossRef]
- Jones, R.; Brandon, N. Persistence and Productivity of Eight Accessions of Desmanthus virgatus under a Range of Grazing Pressures in Subtropical Queensland. Trop. Grassl. 1998, 32, 145–162. [Google Scholar]
- Zabala, J.; Pensiero, J.; Tomas, P.A.; Giavedoni, J. Morphological Characterisation of Populations of Desmanthus virgatus Complex from Argentina. Trop. Grassl. 2008, 42, 229–236. [Google Scholar]
- Skerman, P.J.; Cameron, D.G.; Riveros, F. Tropical Forage Legumes; FAO Plant Production and Protection Series; FAO: Rome, Italy, 1988. [Google Scholar]
- Burrows, D.M.; Porter, J.F. Regeneration and Survival of Desmanthus virgatus 78382 in Grazed and Ungrazed Pastures. Trop. Grassl. Aust. 1993, 27, 100–107. [Google Scholar]
- Bahnisch, G.A.; Date, R.A.; Brandon, N.J.; Pittaway, P. Growth Responses of Desmanthus virgatus to Inoculation with Rhizobium Strain CB3126. I. A Pot Trial with 8 Clay Soils from Central and Southern Queensland. Trop. Grassl. 1998, 32, 13–19. [Google Scholar]
- Date, R.A. Nitrogen Fixation in Desmanthus: Strain Specificity of Rhizobium and Responses to Inoculation in Acidic and Alkaline Soil. Trop. Grassl. Aust. 1991, 25, 47–55. [Google Scholar]
- Brandon, N.J.; Date, R.A.; Clem, R.L.; Robertson, B.A.; Graham, T.W.G. Growth Responses of Desmanthus virgatus to Inoculation with Rhizobium Strain CB3126. II. A Field Trial at 4 Sites in South-East Queensland. Trop. Grassl. 1998, 32, 20–27. [Google Scholar]
- Fornasero, L.V.; Del Papa, M.F.; López, J.L.; Albicoro, F.J.; Zabala, J.M.; Toniutti, M.A.; Pensiero, J.F.; Lagares, A. Phenotypic, Molecular and Symbiotic Characterization of the Rhizobial Symbionts of Desmanthus paspalaceus (Lindm.) Burkart That Grow in the Province of Santa Fe, Argentina. PLoS ONE 2014, 9, e104636. [Google Scholar] [CrossRef]
- Zamora-Natera, J.; Garcia, P.; Rodríguez, M.; Ruiz, M. Rendimiento y Composición Química Del Forraje de Huizachillo (Desmanthus virgatus L. Var. Depressus Willd) Bajo Condiciones de Cultivo. Rev. Fitotec. Mex. 2002, 25, 317–320. [Google Scholar] [CrossRef]
- Beyhaut, E.; DeHaan, L.; Byun, J.; Sheaffer, C.; Graham, P. Response to Inoculation in Illinois Bundleflower. Can. J. Plant Sci. 2006, 86, 919–926. [Google Scholar] [CrossRef]
- Beyhaut, E.; Tlusty, B.; van Berkum, P.; Graham, P.H. Rhizobium giardinii Is the Microsymbiont of Illinois Bundleflower (Desmanthus illinoensis (Michx.) Macmillan) in Midwestern Prairies. Can. J. Microbiol 2006, 52, 903–907. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.A.; Willems, A.; Nesme, X.; de Lajudie, P.; Lindström, K. Revised Phylogeny of Rhizobiaceae: Proposal of the Delineation of Pararhizobium Gen. Nov., and 13 New Species Combinations. Syst. Appl. Microbiol. 2015, 38, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Del Papa, M.F.; Pistorio, M.; Draghi, W.O.; Lozano, M.J.; Giusti, M.A.; Medina, C.; van Dillewijn, P.; Martínez-Abarca, F.; Flores, B.M.; Ruiz-Sainz, J.E.; et al. Identification and Characterization of a NodH Ortholog from the Alfalfa-Nodulating Or191-Like Rhizobia. Mol. Plant Microbe Interact. 2007, 20, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Fåhraeus, G. The Infection of Clover Root Hairs by Nodule Bacteria Studied by a Simple Glass Slide Technique. Microbiology 1957, 16, 374–381. [Google Scholar] [CrossRef]
- Vincent, J.M. A Manual for the Practical Study of the Root-Nodule Bacteria. Available online: https://www.cabdirect.org/cabdirect/abstract/19710700726 (accessed on 6 May 2023).
- Beringer, J.E. R Factor Transfer in Rhizobium leguminosarum. Microbiology 1974, 84, 188–198. [Google Scholar] [CrossRef]
- Rong Juan Zhang, R.Z.J. Identification and Classification of Rhizobia by Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight Mass Spectrometry. J. Proteom. Bioinform. 2015, 8, 98–107. [Google Scholar] [CrossRef]
- Ferreira, L.; Sánchez-Juanes, F.; García-Fraile, P.; Rivas, R.; Mateos, P.F.; Martínez-Molina, E.; González-Buitrago, J.M.; Velázquez, E. MALDI-TOF Mass Spectrometry Is a Fast and Reliable Platform for Identification and Ecological Studies of Species from Family Rhizobiaceae. PLoS ONE 2011, 6, e20223. [Google Scholar] [CrossRef]
- Sánchez-Juanes, F.; Ferreira, L.; Alonso de la Vega, P.; Valverde, A.; Barrios, M.L.; Rivas, R.; Mateos, P.F.; Martínez-Molina, E.; González-Buitrago, J.M.; Trujillo, M.E.; et al. MALDI-TOF Mass Spectrometry as a Tool for Differentiation of Bradyrhizobium Species: Application to the Identification of Lupinus Nodulating Strains. Syst. Appl. Microbiol. 2013, 36, 565–571. [Google Scholar] [CrossRef]
- Toniutti, M.A.; Fornasero, L.V.; Albicoro, F.J.; Martini, M.C.; Draghi, W.; Alvarez, F.; Lagares, A.; Pensiero, J.F.; Del Papa, M.F. Nitrogen-Fixing Rhizobial Strains Isolated from Desmodium incanum DC in Argentina: Phylogeny, Biodiversity and Symbiotic Ability. Syst. Appl. Microbiol. 2017, 40, 297–307. [Google Scholar] [CrossRef]
- Sogawa, K.; Watanabe, M.; Sato, K.; Segawa, S.; Ishii, C.; Miyabe, A.; Murata, S.; Saito, T.; Nomura, F. Use of the MALDI BioTyper System with MALDI–TOF Mass Spectrometry for Rapid Identification of Microorganisms. Anal. Bioanal. Chem. 2011, 400, 1905–1911. [Google Scholar] [CrossRef] [PubMed]
- Meade, H.M.; Long, S.R.; Ruvkun, G.B.; Brown, S.E.; Ausubel, F.M. Physical and Genetic Characterization of Symbiotic and Auxotrophic Mutants of Rhizobium meliloti Induced by Transposon Tn5 Mutagenesis. J. Bacteriol. 1982, 149, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J.; Schneider, M.; De Bruijn, F.; Lupski, J. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 1994, 5, 25–40. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Ciufo, S.; Li, W. Prokaryotic Genome Annotation Pipeline. In The NCBI Handbook, 2nd ed.; National Center for Biotechnology Information: Bethesda, MD, USA, 2013. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A Taxonomically United Database of 16S RNA Gene Sequences and Whole-Genome Assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal, W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for MacOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A Web Server for Prokaryotic Species Circumscription Based on Pairwise Genome Comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Rodriguez, R.L.M.; Konstantinidis, K.T. The Enveomics Collection: A Toolbox for Specialized Analyses of Microbial Genomes and Metagenomes. PeerJ 2016, 4, e1900v1. [Google Scholar]
- Hopkinson, J.M.; English, B.H. Germination and Hardseededness in Desmanthus. Trop. Grassl. 2004, 38, 1–16. [Google Scholar]
- Somasegaran, P. Handbook for Rhizobia: Methods in Legume Rhizobium Technology; Springer: New York, NY, USA, 2012; ISBN 978-1-4613-8375-8. [Google Scholar]
- Jensen, H.L. Nitrogen Fixation in Leguminous Plants. I. General Characters of Root-Nodule Bacteria Isolated from Species of Medicago and Trifolium in Australia. Proc. Linn. Soc. N. S. W. 1942, 67, 98–108. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio: Boston, MA, USA, 2020. [Google Scholar]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 3rd ed.; Sage Publications: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- de Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research; R Package Version 1.3-1; RStudio: Boston, MA, USA, 2020. [Google Scholar]
- Wickham, H. Data Analysis. In ggplot2: Elegant Graphics for Data Analysis; Wickham, H., Ed.; Use R! Springer International Publishing: Cham, Switzerland, 2016; pp. 189–201. ISBN 978-3-319-24277-4. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; ISBN 3-900051-07-0. [Google Scholar]
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. Posit Software Dplyr: A Grammar of Data Manipulation; 2023. [Google Scholar]
- Bache, S.; Wickham, H. Magrittr: A Forward-Pipe Operator for R. 2022. Available online: https://magrittr.tidyverse.org, https://github.com/tidyverse/magrittr (accessed on 26 June 2023).
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline Reach with Protein Family Model Curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef]
- López, J.L.; Alvarez, F.; Príncipe, A.; Salas, M.E.; Lozano, M.J.; Draghi, W.O.; Jofré, E.; Lagares, A. Isolation, Taxonomic Analysis, and Phenotypic Characterization of Bacterial Endophytes Present in Alfalfa (Medicago sativa) Seeds. J. Biotechnol. 2018, 267, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Lloret, L.; Ormeño-Orrillo, E.; Rincón, R.; Martínez-Romero, J.; Rogel-Hernández, M.A.; Martínez-Romero, E. Ensifer mexicanum sp. Nov. a New Species Nodulating Acacia angustissima (Mill.) Kuntze in Mexico. Syst. Appl. Microbiol. 2007, 30, 280–290. [Google Scholar] [CrossRef]
- Rincón-Rosales, R.; Lloret, L.; Ponce, E.; Martínez-Romero, E. Rhizobia with Different Symbiotic Efficiencies Nodulate Acaciella angustissima in Mexico, Including Sinorhizobium chiapanecum sp. Nov. Which Has Common Symbiotic Genes with Sinorhizobium mexicanum. FEMS Microbiol. Ecol. 2009, 67, 103–117. [Google Scholar] [CrossRef]
- Boivin, C.; Ndoye, I.; Lortet, G.; Ndiaye, A.; De Lajudie, P.; Dreyfus, B. The Sesbania Root Symbionts Sinorhizobium saheli and S. terangae bv. sesbaniae Can Form Stem Nodules on Sesbania rostrata, Although They Are Less Adapted to Stem Nodulation than Azorhizobium caulinodans. Appl. Environ. Microbiol. 1997, 63, 1040–1047. [Google Scholar] [CrossRef]
- Amarger, N. Rhizobia in the Field. Adv. Agron. 2001, 73, 109–168. [Google Scholar] [CrossRef]
- Andrews, M.; De Meyer, S.; James, E.; Stępkowski, T.; Hodge, S.; Simon, M.; Young, J. Horizontal Transfer of Symbiosis Genes within and Between Rhizobial Genera: Occurrence and Importance. Genes 2018, 9, 321. [Google Scholar] [CrossRef]
- Lagares, A.; Sanjuán, J.; Pistorio, M. The Plasmid Mobilome of the Model Plant-Symbiont Sinorhizobium meliloti: Coming up with New Questions and Answers. Microbiol. Spectr. 2014, 2, 1128. [Google Scholar] [CrossRef]
- Pistorio, M.; Giusti, M.A.; Del Papa, M.F.; Draghi, W.O.; Lozano, M.J.; Torres Tejerizo, G.; Lagares, A. Conjugal Properties of the Sinorhizobium meliloti Plasmid Mobilome. FEMS Microbiol. Ecol. 2008, 65, 372–382. [Google Scholar] [CrossRef] [PubMed]




| Isolate ID 1 | Place of Origin | Natural Host | Origen of the Root Nodules | MALDI-TOF 2 Identification Group |
|---|---|---|---|---|
| 6-67 | Salta | D. paspalaceus | Trapping plant in the laboratory | β |
| 6-69 | Salta | D. paspalaceus | Trapping plant in the laboratory | β |
| 6-70 | Salta | D. paspalaceus | Trapping plant in the laboratory | α |
| 6-71 | Salta | D. paspalaceus | Trapping plant in the laboratory | α |
| 6-72 | Salta | D. paspalaceus | Trapping plant in the laboratory | β |
| 6-117 | Salta | D. paspalaceus | Trapping plant in the laboratory | α |
| 7-73 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 7-74 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 7-75 | Jujuy | D. vitgatus | Trapping plant in the laboratory | α |
| 7-76 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 7-77 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 7-78 | Jujuy | D. vitgatus | Trapping plant in the laboratory | α |
| 7-79 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 7-80 | Jujuy | D. virgatus | Plant collected in the field | α |
| 7-81 | Jujuy | D. vitgatus | Plant collected in the field | α |
| 7-82 | Jujuy | D. virgatus | Plant collected in the field | α |
| 8-83 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 8-84 | Jujuy | D. vitgatus | Trapping plant in the laboratory | α |
| 8-85 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 8-86 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 8-87 | Jujuy | D. vitgatus | Trapping plant in the laboratory | α |
| 8-88 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 8-89 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 8-90 | Jujuy | D. vitgatus | Trapping plant in the laboratory | α |
| 8-91 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 8-92 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 8-93 | Jujuy | D. vitgatus | Trapping plant in the laboratory | α |
| 8-94 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 8-95 | Jujuy | D. virgatus | Trapping plant in the laboratory | α |
| 8-96 | Jujuy | D. vitgatus | Trapping plant in the laboratory | α |
| Average Nucleotide Identity (ANIb) (%) | ||||
|---|---|---|---|---|
| Sinorhizobial Strains 1 | Sinorhizobia from Northwest Argentina Which Nodulate Desmanthus spp. | |||
| 6-70 2 | 6-117 2 | 7-81 3 | 8-89 3 | |
| Sinorhizobium alkalisoli YIC4027T [GCA_008932245.1] | 79.64 | 79.66 | 79.92 | 79.89 |
| Sinorhizobium aridi LMR001T [GCA_002078505.1] | 80.84 | 81.55 | 81.57 | 80.84 |
| Sinorhizobium mexicanum ITTG R7T [GCA_013488225.1] | 92.90 4 | 92.90 4 | 85.97 | 85.95 |
| Sinorhizobium psoraleae CCBAU 65732T [GCA_013283645.1] | 86.06 | 86.07 | 96.29 4 | 92.90 4 |
| Ensifer sesbaniae CCBAU 65729T [GCA_013283665.1] | 78.77 | 78.77 | 78.83 | 78.77 |
| Sinorhizobium americanum CFNEI 156T [GCA_001651855.1] | 80.48 | 80.51 | 80.32 | 80.66 |
| Sinorhizobium arboris LMG 14919T [GCA_000427465.1] | 79.70 | 79.79 | 80.35 | 80.14 |
| Sinorhizobium saheli LMG 7837T [GCA_001651875.1] | 81.16 | 81.18 | 81.42 | 81.40 |
| Sinorhizobium terangae SEMIA 6460T [GCA_014197705.1] | 88.98 | 88.98 | 86.51 | 86.48 |
| Sinorhizobium terangae CB3126 [GCA_029714365.1] | 89.46 | 89.45 | 86.82 | 86.71 |
| Sinorhizobium sp. 6-70 2 [GCA_030124375.1] | - | 99.99 | 86.66 | 86.64 |
| Sinorhizobium sp. 6-117 2 [GCA_030124365.1] | 99.95 | - | 86.70 | 86.71 |
| Sinorhizobium sp. 7-81 3 [GCA_030124405.1] | 86.47 | 86.48 | - | 95.24 |
| Sinorhizobium sp. 8-89 3 [GCA_030124325.1] | 86.54 | 86.55 | 94.15 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuber, N.E.; Fornasero, L.V.; Erdozain Bagolín, S.A.; Lozano, M.J.; Sanjuán, J.; Del Papa, M.F.; Lagares, A. Diversity, Genomics and Symbiotic Characteristics of Sinorhizobia That Nodulate Desmanthus spp. in Northwest Argentina. Biology 2023, 12, 958. https://doi.org/10.3390/biology12070958
Zuber NE, Fornasero LV, Erdozain Bagolín SA, Lozano MJ, Sanjuán J, Del Papa MF, Lagares A. Diversity, Genomics and Symbiotic Characteristics of Sinorhizobia That Nodulate Desmanthus spp. in Northwest Argentina. Biology. 2023; 12(7):958. https://doi.org/10.3390/biology12070958
Chicago/Turabian StyleZuber, Nicolás Emilio, Laura Viviana Fornasero, Sofía Agostina Erdozain Bagolín, Mauricio Javier Lozano, Juan Sanjuán, María Florencia Del Papa, and Antonio Lagares. 2023. "Diversity, Genomics and Symbiotic Characteristics of Sinorhizobia That Nodulate Desmanthus spp. in Northwest Argentina" Biology 12, no. 7: 958. https://doi.org/10.3390/biology12070958
APA StyleZuber, N. E., Fornasero, L. V., Erdozain Bagolín, S. A., Lozano, M. J., Sanjuán, J., Del Papa, M. F., & Lagares, A. (2023). Diversity, Genomics and Symbiotic Characteristics of Sinorhizobia That Nodulate Desmanthus spp. in Northwest Argentina. Biology, 12(7), 958. https://doi.org/10.3390/biology12070958







