Chromosome-Level Analysis of the Pelochelys cantorii Genome Provides Insights to Its Immunity, Growth and Longevity
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Evolutionary and Comparative Genomic Analyses
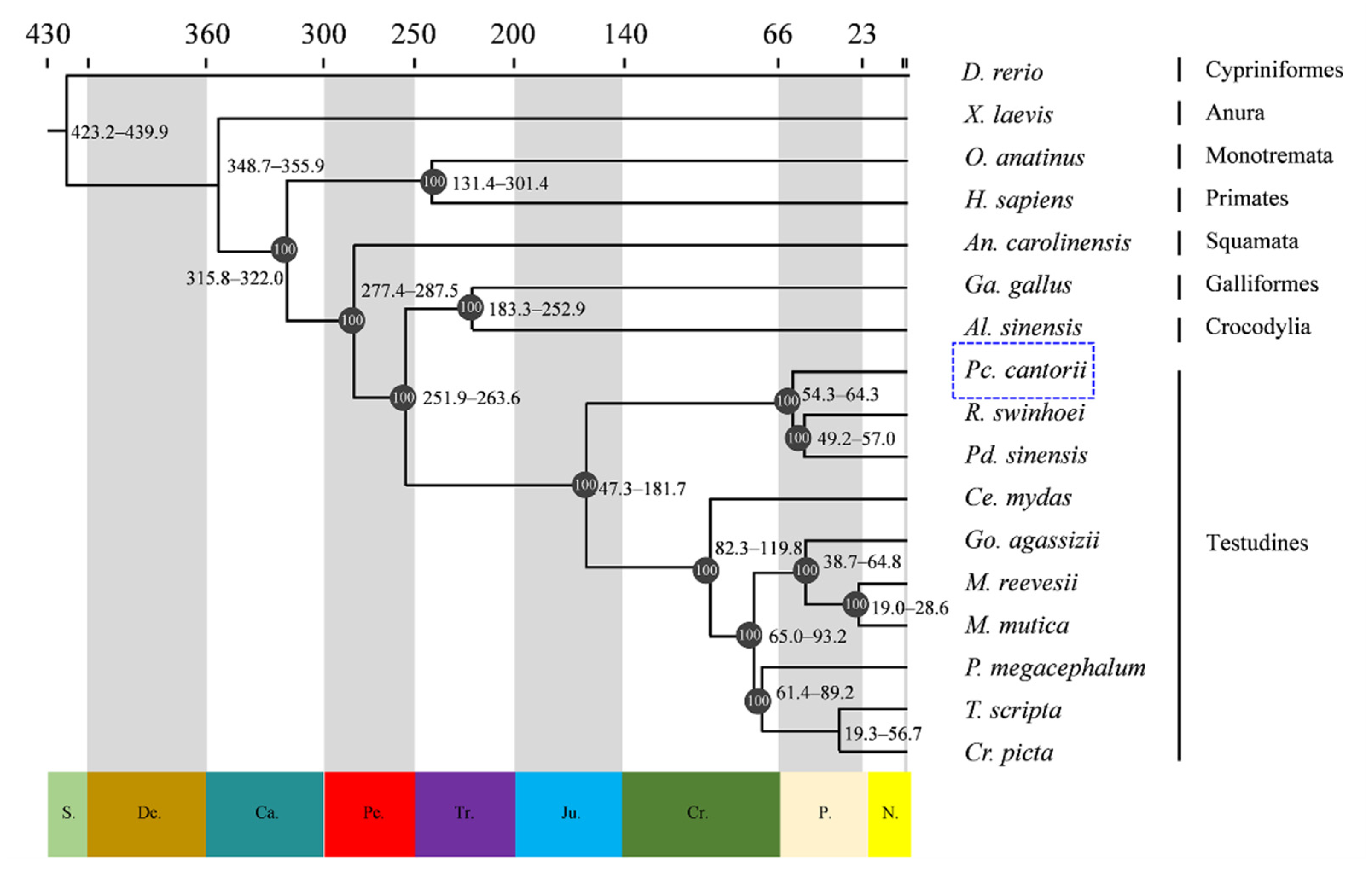
2.2. Estimation of Divergence Times
2.3. Collinearity Analysis
2.4. Duplicate Gene Analysis
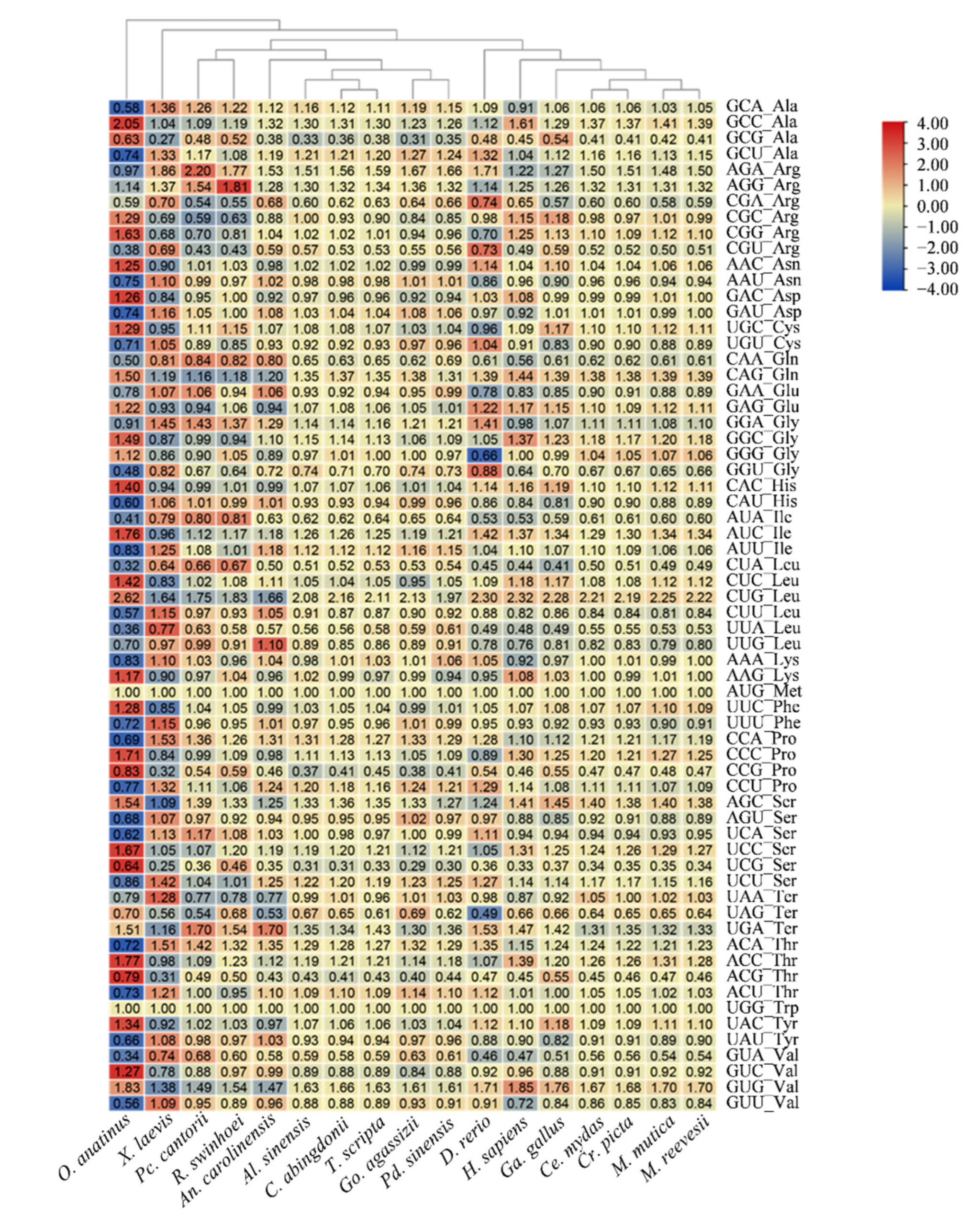
2.5. Relative Synonymous Codon Usage (RSCU) Analysis
3. Results
3.1. Genome Evolution
3.2. Collinearity Analysis
3.3. Relative Synonymous Codon Usage (RSCU) Analysis
3.4. Duplicate Genes in Turtles
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Das, I. Pelochelys cantorii Gray 1864-Asian Giant Softshell Turtle. In Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group; Rhodin, A.G.J., Pritchard, P.C.H., van Dijk, P.P., Saumure, R.A., Buhlmann, K.A., Iverson, J.B., Eds.; Chelonian Research Monographs; Chelonian Research Foundation: Arlington, VT, USA, 2008; pp. 1–6. [Google Scholar]
- Hong, X.; Cai, X.; Chen, C.; Liu, X.; Zhao, J.; Qiu, Q.; Zhu, X. Conservation Status of the Asian Giant Softshell Turtle (Pelochelys cantorii) in China. Chelonian Conserv. Biol. 2019, 18, 68–74. [Google Scholar]
- Xie, M.; Chen, C.; Wang, Y.; Li, W.; Yu, L.; Hong, X.; Zhu, X. Conservation Genetics of the Asian Giant Soft-Shelled Turtle (Pelochelys cantorii) with Novel Microsatellite Multiplexes. Animals 2022, 12, 3459. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Zhu, X.; Chen, C.; Zhao, J.; Ye, Z.; Qiu, Q. Reproduction traits of captive Asian giant soft-shelled turtles, Pelochelys cantorii. Acta Hydrobiol. Sin. 2018, 42, 794–799. [Google Scholar]
- Xie, M.; Hong, X.; Li, W.; Liu, X.; Chen, C.; Zhu, X. Skeletal system analysis of Asian giant soft-shelled turtles and comparison with Chinese soft-shell turtle. Acta Hydrobiol. Sin. 2022, 46, 654–663. [Google Scholar]
- van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. TIG 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. (Clifton NJ) 2009, 537, 39–64. [Google Scholar]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Yang, Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. CABIOS 1997, 13, 555–556. [Google Scholar] [CrossRef]
- Puttick, M.N. MCMCtreeR: Functions to prepare MCMCtree analyses and visualize posterior ages on trees. Bioinformatics 2019, 35, 5321–5322. [Google Scholar] [CrossRef] [PubMed]
- DeBarry, J.D.; Kissinger, J.C. Jumbled genomes: Missing Apicomplexan synteny. Mol. Biol. Evol. 2011, 28, 2855–2871. [Google Scholar] [CrossRef]
- Vishwanathan, N.; Bandyopadhyay, A.A.; Fu, H.Y.; Sharma, M.; Johnson, K.C.; Mudge, J.; Ramaraj, T.; Onsongo, G.; Silverstein, K.A.; Jacob, N.M.; et al. Augmenting Chinese hamster genome assembly by identifying regions of high confidence. Biotechnol. J. 2016, 11, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Evolution by gene duplication. Am. J. Hum. Genet. 1971, 23, 541. [Google Scholar]
- He, X.; Zhang, J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 2005, 169, 1157–1164. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar]
- Sharp, P.M.; Li, W.H. The codon Adaptation Index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, G.A.; Uddin, A.; Chakraborty, S. Preference of A/T ending codons in mitochondrial ATP6 gene under phylum Platyhelminthes: Codon usage of ATP6 gene in Platyhelminthes. Mol. Biochem. Parasitol. 2018, 225, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ding, H.; Kan, X. Codon usage patterns and evolution of HSP60 in birds. Int. J. Biol. Macromol. 2021, 183, 1002–1012. [Google Scholar] [CrossRef]
- Nie, L.; Cui, Y.; Wu, L.; Zhou, J.; Xu, Z.; Li, Y.; Li, X.; Wang, Y.; Yao, H. Gene losses and variations in chloroplast genome of parasitic plant macrosolen and phylogenetic relationships within santalales. Int. J. Mol. Sci. 2019, 20, 5812. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.; Al-Taher, A.; Al-Nazawi, M.; AI, A.-M.; Kandeel, M. Analysis of preferred codon usage in the coronavirus N genes and their implications for genome evolution and vaccine design. J. Virol. Methods 2020, 277, 113806. [Google Scholar] [CrossRef]
- Holland, P.W.; Marlétaz, F.; Maeso, I.; Dunwell, T.L.; Paps, J. New genes from old: Asymmetric divergence of gene duplicates and the evolution of development. Philos. Trans. R. Soc. Lond. Ser. B. Biol. Sci. 2017, 372, 20150480. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Wolf, U.; Atkin, N.B. Evolution from fish to mammals by gene duplication. Hereditas 1968, 59, 169–187. [Google Scholar] [CrossRef]
- Qian, W.; Zhang, J. Genomic evidence for adaptation by gene duplication. Genome Res. 2014, 24, 1356–1362. [Google Scholar] [CrossRef]
- Kurz, S.; Thieme, R.; Amberg, R.; Groth, M.; Jahnke, H.G.; Pieroh, P.; Horn, L.C.; Kolb, M.; Huse, K.; Platzer, M.; et al. The anti-tumorigenic activity of A2M-A lesson from the naked mole-rat. PLoS ONE 2017, 12, e0189514. [Google Scholar] [CrossRef]
- Lindner, I.; NY, H.; Buchold, M.; Huse, K.; Bigl, M.; Oerlecke, I.; Ricken, A.; Gaunitz, F.; Sack, U.; Naumann, A.; et al. Alpha2-macroglobulin inhibits the malignant properties of astrocytoma cells by impeding beta-catenin signaling. Cancer Res. 2010, 70, 277–287. [Google Scholar] [CrossRef]
- García, M.; Barrio, R.; García-Lavandeira, M.; Garcia-Rendueles, A.R.; Escudero, A.; Díaz-Rodríguez, E.; Gorbenko Del Blanco, D.; Fernández, A.; de Rijke, Y.B.; Vallespín, E.; et al. The syndrome of central hypothyroidism and macroorchidism: IGSF1 controls TRHR and FSHB expression by differential modulation of pituitary TGFβ and Activin pathways. Sci. Rep. 2017, 7, 42937. [Google Scholar] [CrossRef] [PubMed]
- Ong, J.; Woldhuis, R.R.; Boudewijn, I.M.; van den Berg, A.; Kluiver, J.; Kok, K.; Terpstra, M.; Guryev, V.; de Vries, M.; Vermeulen, C.; et al. Age-related gene and miRNA expression changes in airways of healthy individuals. Sci. Rep. 2019, 9, 3765. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Pascual-Anaya, J.; Zadissa, A.; Li, W.; Niimura, Y.; Huang, Z.; Li, C.; White, S.; Xiong, Z.; Fang, D.; et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat. Genet. 2013, 45, 701–706. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Yuan, J.; Liu, F.; Hong, X.; Yu, L.; Chen, C.; Li, W.; Ni, W.; Liu, H.; et al. Chromosome-level genome assembly of Asian yellow pond turtle (Mauremys mutica) with temperature-dependent sex determination system. Sci. Rep. 2022, 12, 7905. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhang, Q.; Yan, X.; Hou, D.; Huang, H.; Li, C.; Rao, D.; Li, Y. Genomic insights into the evolution of the critically endangered soft-shelled turtle Rafetus swinhoei. Moclecular Ecol. Resour. 2022, 22, 1972–1985. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Q.; Benton, M.J. The timing and pattern of biotic recovery following the end-Permian mass extinction. Nat. Geosci. 2012, 5, 375–383. [Google Scholar] [CrossRef]
- Scheyer, T.M.; Klein, N.; Evers, S.W.; Mautner, A.K.; Pabst, B. First evidence of Proganochelys quenstedtii (Testudinata) from the Plateosaurus bonebeds (Norian, Late Triassic) of Frick, Canton Aargau, Switzerland. Swiss. J. Palaeontol. 2022, 141, 17. [Google Scholar]
- Dong, S.; Ying, Z.; Yu, S.; Wang, Q.; Liao, G.; Ge, Y.; Cheng, R. Complete chloroplast genome of Stephania tetrandra (Menispermaceae) from Zhejiang Province: Insights into molecular structures, comparative genome analysis, mutational hotspots and phylogenetic relationships. BMC Genom. 2021, 22, 880. [Google Scholar] [CrossRef]
- Chaney, J.L.; Clark, P.L. Roles for Synonymous Codon Usage in Protein Biogenesis. Annu. Rev. Biophys. 2015, 44, 143–166. [Google Scholar] [CrossRef]
- Supek, F. The code of silence: Widespread associations between synonymous codon biases and gene function. J. Mol. Evol. 2016, 82, 65–73. [Google Scholar] [CrossRef]
- Romero, H.; Zavala, A.; Musto, H. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucleic Acids Res. 2000, 28, 2084–2090. [Google Scholar] [CrossRef] [PubMed]
- Gemmell, N.J.; Rutherford, K.; Prost, S.; Tollis, M.; Winter, D.; Macey, J.R.; Adelson, D.L.; Suh, A.; Bertozzi, T.; Grau, J.H.; et al. The tuatara genome reveals ancient features of amniote evolution. Nature 2020, 584, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.L. Evolution of Duplicate Gene Sequences, Expression Patterns, and Functions in the Brassicaceae and Other Rosids. Ph.D. Thesis, University of British Columbia, Vancouver, BC, Canada, 2011. [Google Scholar]
- Di, D.; Chen, L.; Guo, Y.; Wang, L.; Zhao, C.; Ju, J. BCSC-1 suppresses human breast cancer metastasis by inhibiting NF-κB signaling. Int. J. Oncol. 2018, 52, 1674–1684. [Google Scholar] [CrossRef] [PubMed]
- Silverton, L.; Dean, M.; Moitra, K. Variation and evolution of the ABC transporter genes ABCB1, ABCC1, ABCG2, ABCG5 and ABCG8: Implication for pharmacogenetics and disease. Drug. Metab. Drug. Interact. 2011, 26, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Zamani, M.; Mohammadi, M.; Zamani, H.; Tavasoli, A. Pharmacogenetic study on the impact of rivastigmine concerning genetic variants of A2M and IL-6 genes on Iranian Alzheimer’s patients. Mol. Neurobiol. 2016, 53, 4521–4528. [Google Scholar] [CrossRef]
- Guan, Y.; Wang, Y.; Bhandari, A.; Xia, E.; Wang, O. IGSF1: A novel oncogene regulates the thyroid cancer progression. Cell. Biochem. Funct. 2019, 37, 516–524. [Google Scholar] [CrossRef]
- Sun, Y.; Bak, B.; Schoenmakers, N.; van Trotsenburg, A.S.; Oostdijk, W.; Voshol, P.; Cambridge, E.; White, J.K.; le Tissier, P.; Gharavy, S.N.; et al. Loss-of-function mutations in IGSF1 cause an X-linked syndrome of central hypothyroidism and testicular enlargement. Nat. Genet. 2012, 44, 1375–1381. [Google Scholar] [CrossRef]
- Asakura, Y.; Abe, K.; Muroya, K.; Hanakawa, J.; Oto, Y.; Narumi, S.; Hasegawa, T.; Adachi, M. Combined growth hormone and thyroid-stimulating hormone deficiency in a Japanese patient with a novel frameshift mutation in IGSF1. Horm. Res. Paediatr. 2015, 84, 349–354. [Google Scholar] [CrossRef]
- Plassais, J.; Rimbault, M.; Williams, F.J.; Davis, B.W.; Schoenebeck, J.J.; Ostrander, E.A. Analysis of large versus small dogs reveals three genes on the canine X chromosome associated with body weight, muscling and back fat thickness. PLoS Genet. 2017, 13, e1006661. [Google Scholar] [CrossRef]
- Badwan, S.; Harper, J. Size Matters: Body size is correlated with longevity in Speckled Cockroaches (Nauphoeta cineria). Curr. Aging Sci. 2021, 14, 214–222. [Google Scholar] [CrossRef]
- Speakman, J.R. Body size, energy metabolism and lifespan. J. Exp. Biol. 2005, 208, 1717–1730. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Liu, H.; Wang, Y.; Li, M.; Ji, L.; Wang, K.; Wei, C.; Li, W.; Chen, C.; Yu, L.; et al. Chromosome-Level Analysis of the Pelochelys cantorii Genome Provides Insights to Its Immunity, Growth and Longevity. Biology 2023, 12, 939. https://doi.org/10.3390/biology12070939
Liu X, Liu H, Wang Y, Li M, Ji L, Wang K, Wei C, Li W, Chen C, Yu L, et al. Chromosome-Level Analysis of the Pelochelys cantorii Genome Provides Insights to Its Immunity, Growth and Longevity. Biology. 2023; 12(7):939. https://doi.org/10.3390/biology12070939
Chicago/Turabian StyleLiu, Xiaoli, Haiyang Liu, Yakun Wang, Mingzhi Li, Liqin Ji, Kaikuo Wang, Chengqing Wei, Wei Li, Chen Chen, Lingyun Yu, and et al. 2023. "Chromosome-Level Analysis of the Pelochelys cantorii Genome Provides Insights to Its Immunity, Growth and Longevity" Biology 12, no. 7: 939. https://doi.org/10.3390/biology12070939
APA StyleLiu, X., Liu, H., Wang, Y., Li, M., Ji, L., Wang, K., Wei, C., Li, W., Chen, C., Yu, L., Zhu, X., & Hong, X. (2023). Chromosome-Level Analysis of the Pelochelys cantorii Genome Provides Insights to Its Immunity, Growth and Longevity. Biology, 12(7), 939. https://doi.org/10.3390/biology12070939





