Immune Transcriptional Response in Head Kidney Primary Cell Cultures Isolated from the Three Most Important Species in Chilean Salmonids Aquaculture
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Head Kidney Primary Cell Culture (HKPCC) Preparation
2.3. In Vitro Immunostimulation
2.4. Total RNA Extraction
2.5. qRT-PCR Analysis of Gene Expression
2.6. Statistical Analysis
3. Results
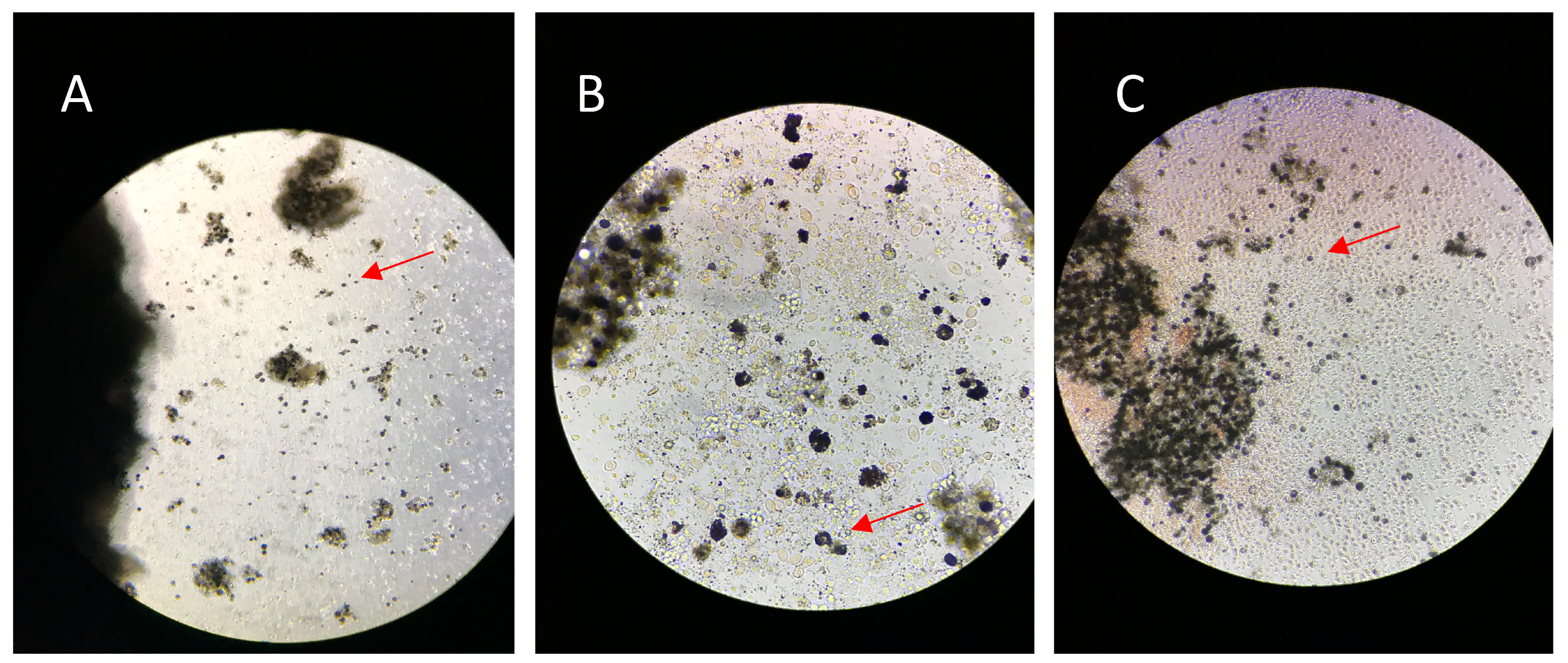
3.1. Morphology of Head Kidney Culture Cells
3.2. mRNA Gene Expression Changes
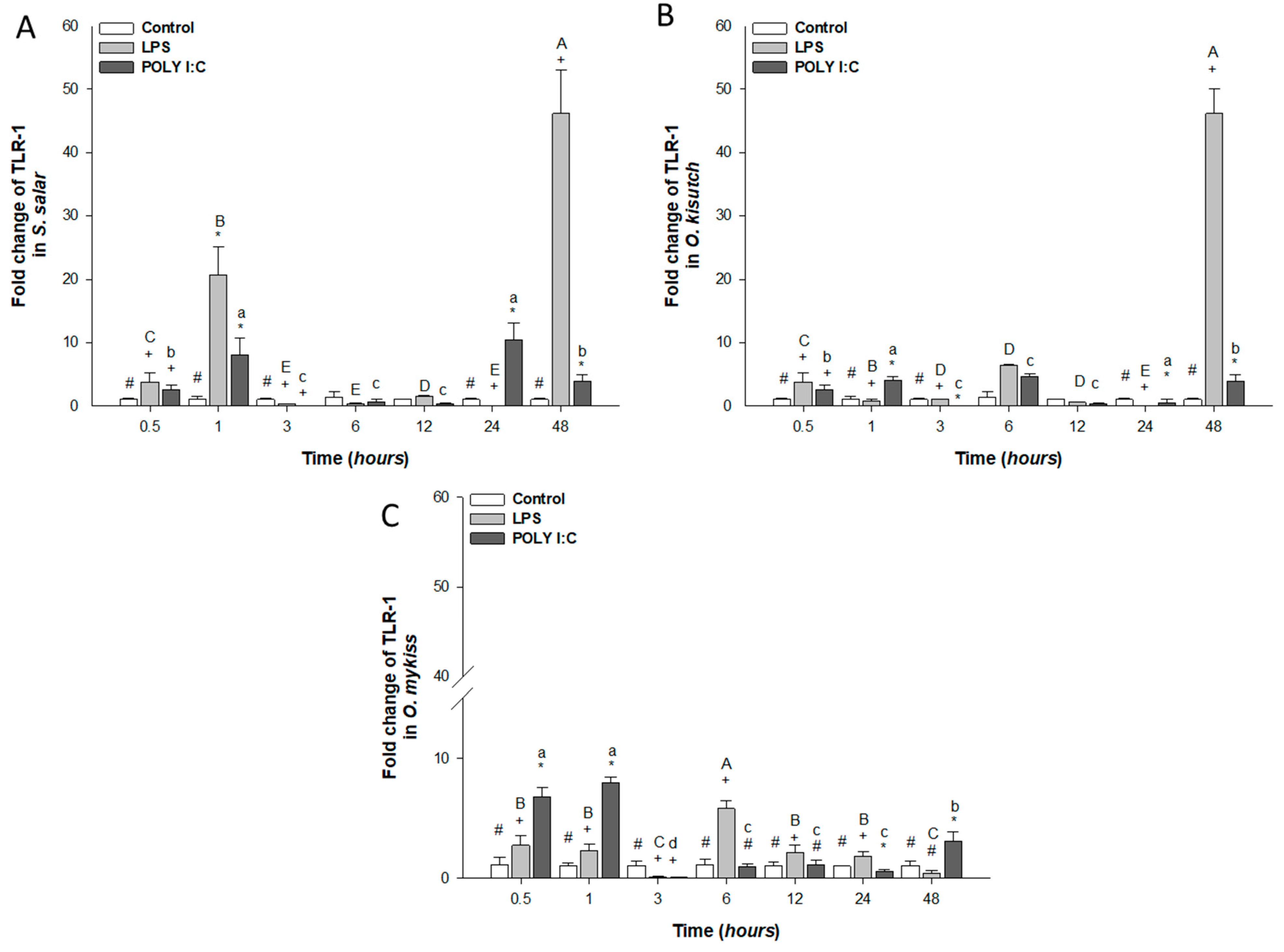
3.2.1. TLR-1 Expression
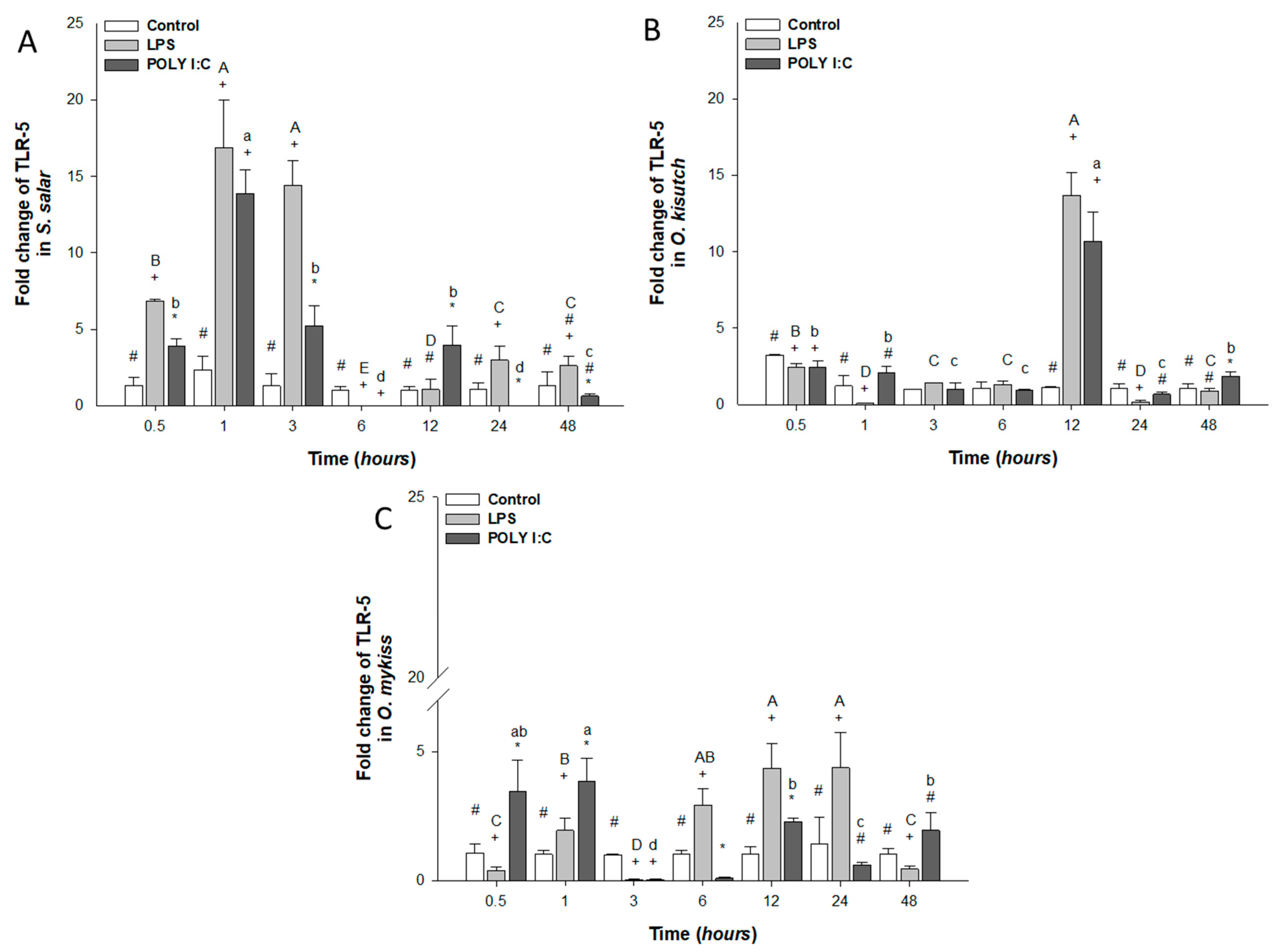
3.2.2. TLR-5 Expression
3.2.3. TLR-8 Expression
3.2.4. IgM Expression
3.2.5. MHC II Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. World Fisheries and Aquaculture the State of Sustainability in Action; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- FAO. El Estado Mundial de La Pesca y La Acuicultura 2020; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Figueroa, J.; Cárcamo, J.; Yañez, A.; Olavarria, V.; Ruiz, P.; Manríquez, R.; Muñoz, C.; Romero, A.; Avendaño-Herrera, R. Addressing Viral and Bacterial Threats to Salmon Farming in Chile: Historical Contexts and Perspectives for Management and Control. Rev. Aquac. 2019, 11, 299–324. [Google Scholar] [CrossRef]
- Navedo, J.G.; Vargas-Chacoff, L. Salmon Aquaculture Threatens Patagonia. Science 2021, 372, 695–696. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Hemmi, H. Recognition of Pathogen-Associated Molecular Patterns by TLR Family. Immunol. Lett. 2003, 85, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Janeway, C.A. Decoding the Patterns of Self and Nonself by the Innate Immune System. Science 2002, 296, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Stansberg, C.; Subramaniam, S.; Collet, B.; Secombes, C.J.; Cunningham, C. Cloning of the Atlantic Salmon (Salmo salar) IL-1 Receptor Associated Protein. Fish Shellfish Immunol. 2005, 19, 53–65. [Google Scholar] [CrossRef]
- Kigerl, K.A.; de Rivero Vaccari, J.P.; Dietrich, W.D.; Popovich, P.G.; Keane, R.W. Pattern Recognition Receptors and Central Nervous System Repair. Exp. Neurol. 2014, 258, 5–16. [Google Scholar] [CrossRef]
- Sahoo, B.R. Structure of Fish Toll-like Receptors (TLR) and NOD-like Receptors (NLR). Int. J. Biol. Macromol. 2020, 161, 1602–1617. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Duan, T.; Du, Y.; Xing, C.; Wang, H.Y.; Wang, R.F. Toll-Like Receptor Signaling and Its Role in Cell-Mediated Immunity. Front. Immunol. 2022, 13, 812774. [Google Scholar] [CrossRef]
- Kawasaki, T.; Kawai, T. Toll-like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef]
- Fierro-Castro, C.; Barrioluengo, L.; López-Fierro, P.; Razquin, B.E.; Villena, A.J. Fish Cell Cultures as Invitro Models of Inflammatory Responses Elicited by Immunostimulants. Expression of Regulatory Genes of the Innate Immune Response. Fish Shellfish Immunol. 2013, 35, 979–987. [Google Scholar] [CrossRef]
- Martínez-López, A.; Tyrkalska, S.D.; Alcaraz-Pérez, F.; Cabas, I.; Candel, S.; Martínez Morcillo, F.J.; Sepulcre, M.P.; García-Moreno, D.; Cayuela, M.L.; Mulero, V. Evolution of LPS Recognition and Signaling: The Bony Fish Perspective. Dev. Comp. Immunol. 2023, 145, 104710. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Toll-Like Receptors And Innate Immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Duffy, K.E.; San Mateo, L.R.; Amegadzie, B.Y.; Sarisky, R.T.; Mbow, M.L. A Pathway Analysis of Poly(I:C)-Induced Global Gene Expression Change in Human Peripheral Blood Mononuclear Cells. Physiol. Genom. 2006, 26, 125–133. [Google Scholar] [CrossRef]
- Hua, Z.; Hou, B. The Role of B Cell Antigen Presentation in the Initiation of CD4+ T Cell Response. Immunol. Rev. 2020, 296, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, C.; Chaves-Pozo, E. Antigen Presentation and Autophagy in Teleost Adaptive Immunity. Int. J. Mol. Sci. 2022, 23, 4899. [Google Scholar] [CrossRef]
- Magnadottir, B. Immunological Control of Fish Diseases. Mar. Biotechnol. 2010, 12, 361–379. [Google Scholar] [CrossRef]
- Mokhtar, D.M.; Zaccone, G.; Alesci, A.; Kuciel, M.; Hussein, M.T.; Sayed, R.K.A. Main Components of Fish Immunity: An Overview of the Fish Immune System. Fishes 2023, 8, 93. [Google Scholar] [CrossRef]
- Turley, S.J.; Inaba, K.; Garrett, W.S.; Ebersold, M.; Unternaehrer, J.; Steinman, R.M.; Mellman, I. Transport of Peptide–MHC Class II Complexes in Developing Dendritic Cells. Science 2000, 288, 522–527. [Google Scholar] [CrossRef]
- Salinas, I.; Zhang, Y.A.; Sunyer, J.O. Mucosal Immunoglobulins and B Cells of Teleost Fish. Dev. Comp. Immunol. 2011, 35, 1346–1365. [Google Scholar] [CrossRef]
- Wu, L.; Qin, Z.; Liu, H.; Lin, L.; Ye, J.; Li, J. Recent Advances on Phagocytic B Cells in Teleost Fish. Front. Immunol. 2020, 11, 824. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, L.V.G.; Xu, Z.; Takizawa, F.; Parra, D.; Go, D.; Lapatra, S.E.; Sunyer, J.O. Mucosal Immunoglobulins at Respiratory Surfaces Mark an Ancient Association That Predates the Emergence of Tetrapods. Nat. Commun. 2016, 7, 10728. [Google Scholar] [CrossRef]
- Flajnik, M.F.; Kasahara, M. Origin and Evolution of the Adaptive Immune System: Genetic Events and Selective Pressures. Nat. Rev. Genet. 2010, 11, 47–59. [Google Scholar] [CrossRef]
- Xing, J.G.; Lee, L.E.J.; Fan, L.; Collodi, P.; Holt, S.E.; Bols, N.C. Initiation of a Zebrafish Blastula Cell Line on Rainbow Trout Stromal Cells and Subsequent Development Under Feeder-Free Conditions into a Cell Line, ZEB2J. Zebrafish 2008, 5, 49–63. [Google Scholar] [CrossRef]
- Joerink, M.; Ribeiro, C.M.S.; Stet, R.J.M.; Hermsen, T.; Savelkoul, H.F.J.; Wiegertjes, G.F. Head Kidney-Derived Macrophages of Common Carp (Cyprinus carpio L.) Show Plasticity and Functional Polarization upon Differential Stimulation. J. Immunol. 2006, 177, 61–69. [Google Scholar] [CrossRef]
- Langan, L.M.; Harper, G.M.; Owen, S.F.; Purcell, W.M.; Jackson, S.K.; Jha, A.N. Application of the Rainbow Trout Derived Intestinal Cell Line (RTgutGC) for Ecotoxicological Studies: Molecular and Cellular Responses Following Exposure to Copper. Ecotoxicology 2017, 26, 1117. [Google Scholar] [CrossRef]
- Aarattuthodi, S.; Dharan, V.; Koshy, M.; Alimentos, L.A.S.D.E.L.O.S.; Goswami, M.; Yashwanth, B.S.; Trudeau, V.; Lakra, W.S. Role and Relevance of Fish Cell Lines in Advanced in Vitro Research. J. Aquac. Res. Dev. 2022, 49, 2393–2411. [Google Scholar] [CrossRef]
- Aarattuthodi, S.; Dharan, V.; Koshy, M. Fish Cell Cultures-Uses and Prospects. J. Aquac. Res. Dev. 2021, 13, 667. [Google Scholar] [CrossRef]
- Barlian, N.C.B.A.; Caldwell, S.J. Development of a Cell Line from Primary Cultures of Rainbow Trout, Oncorhynchus mykiss (Walbaum), Gills. J. Fish Dis. 1994, 17, 601–611. [Google Scholar] [CrossRef]
- Langan, L.M.; Owen, S.F.; Jha, A.N. Establishment and Long-Term Maintenance of Primary Intestinal Epithelial Cells Cultured from the Rainbow Trout, Oncorhynchus mykiss. Biol. Open 2018, 7, bio032870. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Chacoff, L.; Ortíz, E.; Oyarzún, R.; Martinez, D.; Saavedra, E.; Sá, R.; Olavarría, V.; Nualart, D.; Yáñez, A.; Bertrán, C.; et al. Stocking Density and Piscirickettsia salmonis Infection Effect on Patagonian Blennie (Eleginops maclovinus, Cuvier 1830) Skeletal Muscle Intermediate Metabolism. Fish Physiol. Biochem. 2014, 40, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Schnell, S.; Kawano, A.; Porte, C.; Lee, L.E.J.; Bols, N.C. Effects of Ibuprofen on the Viability and Proliferation of Rainbow Trout Liver Cell Lines and Potential Problems and Interactions in Effects Assessment. Environ. Toxicol. Int. J. 2008, 24, 157–165. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real- Time Quantitative PCR and the 2−ΔΔCTC Method. Methods 2001, 408, 402–408. [Google Scholar] [CrossRef]
- Martínez, D.; Díaz-ibarrola, D.; Vargas-lagos, C.; Oyarzún, R.; Pontigo, J.P. Fish and Shell Fi Sh Immunology Immunological Response of the Sub-Antarctic Notothenioid Fi Sh Eleginops maclovinus Injected with Two Strains of Piscirickettsia salmonis. Fish Shellfish Immunol. 2018, 75, 139–148. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Lekanne Deprez, R.H.; Moorman, A.F.M. Assumption-Free Analysis of Quantitative Real-Time Polymerase Chain Reaction (PCR) Data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Chettri, J.K.; Raida, M.K.; Holten-Andersen, L.; Kania, P.W.; Buchmann, K. PAMP Induced Expression of Immune Relevant Genes in Head Kidney Leukocytes of Rainbow Trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 2011, 35, 476–482. [Google Scholar] [CrossRef]
- Hart, K.M.; Murphy, A.J.; Barrett, K.T.; Wira, C.R.; Guyre, P.M.; Pioli, P.A. Functional Expression of Pattern Recognition Receptors in Tissues of the Human Female Reproductive Tract. J. Reprod. Immunol. 2009, 80, 33–40. [Google Scholar] [CrossRef]
- Silva, M.T. Efficacy of the Treatments Used for the Control of Caligus rogercresseyi Infecting Atlantic Salmon, Salmo salar L., in a New Fish-Farming Location in Region XI, Chile. J. Fish Dis. 2013, 36, 221–228. [Google Scholar] [CrossRef]
- Ghosh, M.; Shen, Z.; Fahey, J.V.; Crist, S.G.; Patel, M.; Smith, J.M.; Wira, C.R. Pathogen Recognition in the Human Female Reproductive Tract: Expression of Intracellular Cytosolic Sensors NOD1, NOD2, RIG-1, and MDA5 and Response to HIV-1 and Neisseria Gonorrhea. Am. J. Reprod. Immunol. 2013, 69, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.V.; Shen, Z.; Wira, C.R. Poly (I: C) and LPS Induce Distinct Immune Responses by Ovarian Stromal. J. Reprod. Immunol. 2018, 127, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; McFarland-Mancini, M.M.; Funk, H.M.; Husseinzadeh, N.; Mounajjed, T.; Drew, A.F. Toll-like Receptor Expression in Normal Ovary and Ovarian Tumors. Cancer Immunol. Immunother. 2009, 58, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Zhu, W.; Yu, L.; Li, N.; Zhang, X.; Liu, P.; Chen, Q.; Chen, Y.; Han, D. Toll-like Receptor 3 and RIG-I-like Receptor Activation Induces Innate Antiviral Responses in Mouse Ovarian Granulosa Cells. Mol. Cell. Endocrinol. 2013, 372, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Holen, E.; Lie, K.K.; Araujo, P.; Olsvik, P.A. Fish & Shell Fi Sh Immunology Pathogen Recognition and Mechanisms in Atlantic Cod (Gadus morhua) Head Kidney Cells Bacteria (LPS) and Virus (Poly I: C) Signals through Different Pathways and Affect Distinct Genes. Fish Shellfish Immunol. 2012, 33, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shimada, M.; Richards, J.S. The Involvement of the Toll-like Receptor Family in Ovulation. J. Assist. Reprod. Genet. 2008, 25, 223. [Google Scholar] [CrossRef]
- Xu, A.; Han, F.; Zhang, Y.; Zhou, T.; Gao, T. Comparative Transcriptomic Analyses Revealed the Effects of Poly (I:C) on the Liver and Spleen of Argyrosomus japonicus. Int. J. Mol. Sci. 2022, 23, 9801. [Google Scholar] [CrossRef]
- Falco, A.; Miest, J.J.; Pionnier, N.; Pietretti, D.; Forlenza, M.; Wiegertjes, G.F.; Hoole, D. β-Glucan-Supplemented Diets Increase Poly(I: C)-Induced Gene Expression of Mx, Possibly via Tlr3-Mediated Recognition Mechanism in Common Carp (Cyprinus carpio). Fish Shellfish Immunol. 2014, 36, 494–502. [Google Scholar] [CrossRef]
- Liu, S.; Jiang, C.; Duan, C.; Hu, L.; Zhang, S. Fish & Shell Fi Sh Immunology Expression of Virus-Responsive Genes and Their Response to Challenge with Poly (I: C) at Different Stages of the Annual Fi Sh Nothobranchius guentheri: Implications for an Asymmetric Decrease in Immunity. Fish Shellfish Immunol. 2015, 46, 493–500. [Google Scholar] [CrossRef]
- Pham, P.H.; Vo, N.T.K.; Tan, E.J.H.; Russell, S.; Jones, G.; Lumsden, J.S.; Bols, N.C. Development of an Atlantic Salmon Heart Endothelial Cell Line (ASHe) That Responds to Lysophosphatidic Acid (LPA). Vitr. Cell. Dev. Biol.-Anim. 2017, 53, 20–32. [Google Scholar] [CrossRef]
- Vargas-Chacoff, L.; Martínez, D.; Oyarzún, R.; Nualart, D.; Olavarría, V.; Yáñez, A.; Bertrán, C.; Ruiz-Jarabo, I.; Mancera, J.M. Combined Effects of High Stocking Density and Piscirickettsia salmonis Treatment on the Immune System, Metabolism and Osmoregulatory Responses of the Sub-Antarctic Notothenioid Fish Eleginops maclovinus. Fish Shellfish Immunol. 2014, 40, 424–434. [Google Scholar] [CrossRef]



| Primer | Nucleotide Sequences (5′→3′) | PCR Product Size | Efficiency HK (%) |
|---|---|---|---|
| MHCII Fw | CTACGAGTTCTACCCCAAACCCAT | 102 bp | 102.91 |
| MHCII Rv | CAGTCGCTGTCAGCCAGTTCTT | ||
| TLR1 Fw | CAACGCTATCTGATCCCCAAGCAA | 114 bp | 101.6 |
| TLR1 Rv | AAAGCCGACGCTCAGTGTTTGT | ||
| TLR5 Fw | TGGCTCACTACCAGCTGATGAA | 112 bp | 102.5 |
| TLR5 Rv | AGCCGCTCATAAAACCACTC | ||
| TLR8 Fw | TCCTGCAGAACTCTCACTTCCT | 122 bp | 101.9 |
| TLR8 Rv | TCTGACCACATTCCTCAGGTTT | ||
| IgMs Fw | TGAAAGACTTCTACCCGCATGAGG | 124 bp | 102.7 |
| IgMs Rv | AACTGGCCATAAGCGGAAAAGG | ||
| 18s het Fw | GTCCGGGAAACCAAAGTC | 116 bp | 103.1 |
| 18s het Rv | TTGAGTCAAATTAAGCCGCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nualart, D.P.; Dann, F.; Oyarzún-Salazar, R.; Morera, F.J.; Vargas-Chacoff, L. Immune Transcriptional Response in Head Kidney Primary Cell Cultures Isolated from the Three Most Important Species in Chilean Salmonids Aquaculture. Biology 2023, 12, 924. https://doi.org/10.3390/biology12070924
Nualart DP, Dann F, Oyarzún-Salazar R, Morera FJ, Vargas-Chacoff L. Immune Transcriptional Response in Head Kidney Primary Cell Cultures Isolated from the Three Most Important Species in Chilean Salmonids Aquaculture. Biology. 2023; 12(7):924. https://doi.org/10.3390/biology12070924
Chicago/Turabian StyleNualart, Daniela P., Francisco Dann, Ricardo Oyarzún-Salazar, Francisco J. Morera, and Luis Vargas-Chacoff. 2023. "Immune Transcriptional Response in Head Kidney Primary Cell Cultures Isolated from the Three Most Important Species in Chilean Salmonids Aquaculture" Biology 12, no. 7: 924. https://doi.org/10.3390/biology12070924
APA StyleNualart, D. P., Dann, F., Oyarzún-Salazar, R., Morera, F. J., & Vargas-Chacoff, L. (2023). Immune Transcriptional Response in Head Kidney Primary Cell Cultures Isolated from the Three Most Important Species in Chilean Salmonids Aquaculture. Biology, 12(7), 924. https://doi.org/10.3390/biology12070924







