Symbiont Identity Impacts the Microbiome and Volatilome of a Model Cnidarian-Dinoflagellate Symbiosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Experimental Organisms
2.2. Microbe Sampling and Microbiome Analysis
2.3. BVOC Sampling and Volatilome Characterisation
2.4. Symbiont Cell Density and Protein Determination
2.5. Data Analysis
3. Results
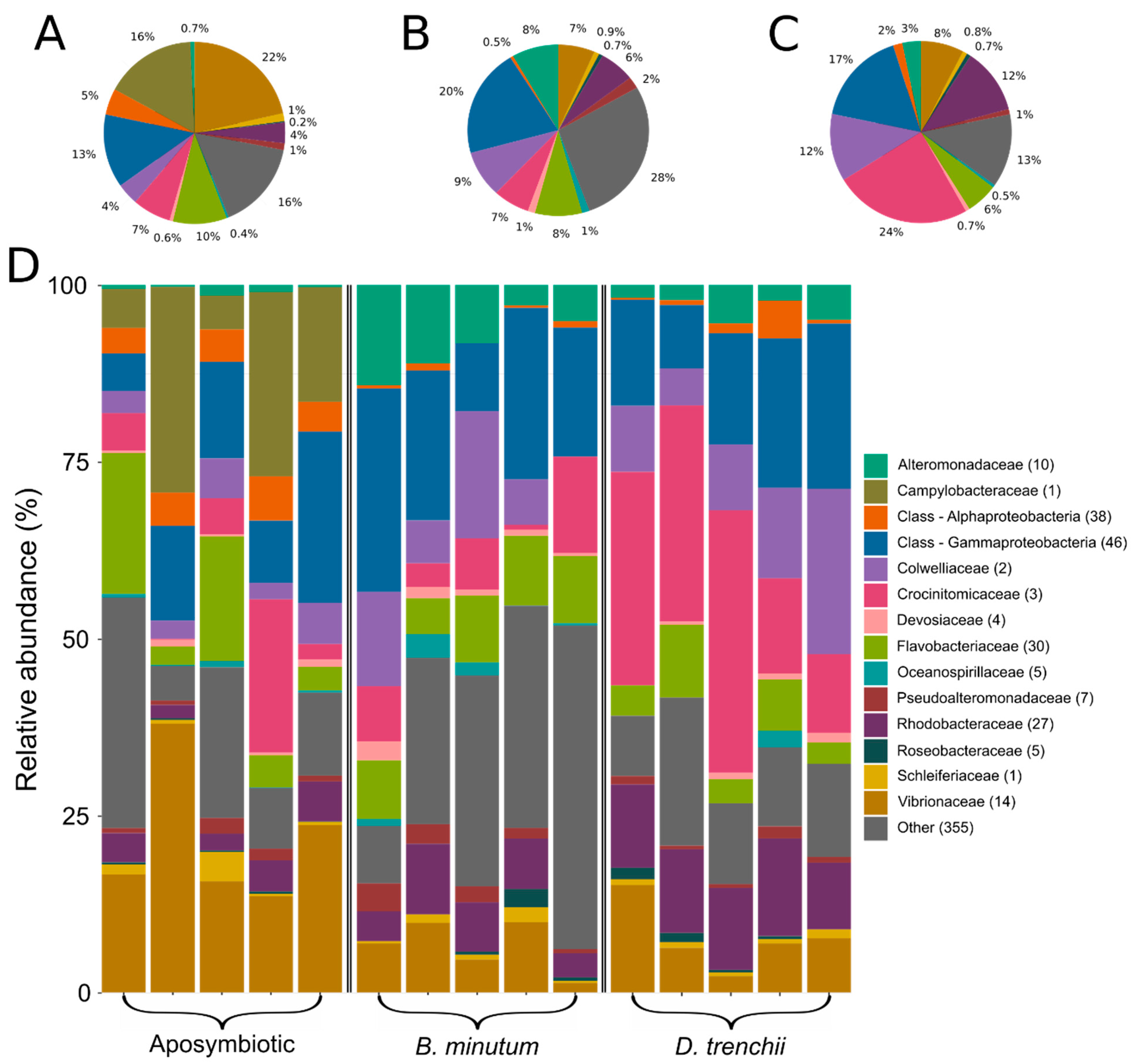
3.1. Bacterial Community of Aiptasia in Different Symbiotic States
3.2. Contrasting Microbiota between Symbiotic States
- (i)
- Aposymbiotic anemones versus anemones harbouring homologous symbionts
- (ii)
- Aposymbiotic anemones versus anemones harbouring heterologous symbionts
- (iii)
- Anemones harbouring homologous symbionts versus heterologous symbionts
3.3. BVOC Emissions Are Affected by Symbiont Identity
3.4. Contrasting Volatilomes between Symbiotic States
3.5. Correlation of Microbiome and Volatilome
4. Discussion
4.1. Symbiosis and Symbiont Type Are Correlated to Changes in the Holobiont Microbiome
4.2. Symbiosis and Symbiont Types Induce Changes in the Holobiont Volatilome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bouchet, P. The magnitude of marine biodiversity. In The Exploration of Marine Biodiversity: Scientific and Technological Challenges; Duarte, C.M., Ed.; Fundacion BBVA, Pub.: Bilbao, Spain, 2006; pp. 31–64. [Google Scholar]
- Davy, S.K.; Allemand, D.; Weis, V.M. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 2012, 76, 229–261. [Google Scholar] [CrossRef] [PubMed]
- Rosset, S.L.; Oakley, C.A.; Ferrier-Pagès, C.; Suggett, D.J.; Weis, V.M.; Davy, S.K. The molecular language of the cnidarian–dinoflagellate symbiosis. Trends Microbiol. 2021, 29, 320–333. [Google Scholar] [CrossRef]
- Roth, M.S. The engine of the reef: Photobiology of the coral-algal symbiosis. Front. Microbiol. 2014, 5, 422. [Google Scholar] [CrossRef] [PubMed]
- Suggett, D.J.; Smith, D.J. Coral bleaching patterns are the outcome of complex biological and environmental networking. Glob. Chang. Biol. 2020, 26, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.P.; Kerry, J.T.; Connolly, S.R.; Alvarez-Romero, J.G.; Eakin, C.M.; Heron, S.F.; Gonzalez, M.A.; Moneghetti, J. Emergent properties in the responses of tropical corals to recurrent climate extremes. Curr. Biol. 2021, 31, 5393–5399.e3. [Google Scholar] [CrossRef]
- Glynn, P.W. Coral reef bleaching: Ecological perspectives. Coral Reefs 2012, 12, 1–17. [Google Scholar] [CrossRef]
- Hoegh-Guldberg, O.; Poloczanska, E.S.; Skirving, W.; Dove, S. Coral reef ecosystems under climate change and ocean acidification. Front. Mar. Sci. 2017, 4, 158. [Google Scholar] [CrossRef]
- LaJeunesse, T.C.; Parkinson, J.E.; Gabrielson, P.W.; Jeong, H.J.; Reimer, J.D.; Voolstra, C.R.; Santos, S.R. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 2018, 28, 2570–2580.e6. [Google Scholar] [CrossRef]
- Nitschke, M.R.; Craveiro, S.C.; Brandao, C.; Fidalgo, C.; Serodio, J.; Calado, A.J.; Frommlet, J.C. Description of Freudenthalidium gen. nov. and Halluxium gen. nov. to formally recognize clades Fr3 and H as genera in the family Symbiodiniaceae (Dinophyceae). J. Phycol. 2020, 56, 923–940. [Google Scholar] [CrossRef]
- LaJeunesse, T.C.; Wiedenmann, J.; Casado-Amezua, P.; D’Ambra, I.; Turnham, K.E.; Nitschke, M.R. Revival of Philozoon Geddes for host-specialized dinoflagellates, ‘zooxanthellae’, in animals from coastal temperate zones of northern and southern hemispheres. Eur. J. Phycol. 2022, 57, 166–180. [Google Scholar] [CrossRef]
- Suggett, D.J.; Goyen, S.; Evenhuis, C.; Szabo, M.; Pettay, D.T.; Warner, M.E.; Ralph, P.J. Functional diversity of photobiological traits within the genus Symbiodinium appears to be governed by the interaction of cell size with cladal designation. New Phytol. 2015, 208, 370–381. [Google Scholar] [CrossRef]
- Nitschke, M.R.; Rosset, S.L.; Oakley, C.A.; Gardner, S.G.; Camp, E.F.; Suggett, D.J.; Davy, S.K. The diversity and ecology of Symbiodiniaceae: A traits-based review. Adv. Mar. Biol. 2022, 92, 55–127. [Google Scholar] [CrossRef] [PubMed]
- Thornhill, D.J.; Xiang, Y.; Pettay, D.T.; Zhong, M.; Santos, S.R. Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Mol. Ecol. 2013, 22, 4499–4515. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.C.; Romanski, A.M. Multiple symbiotic partnerships are common in scleractinian corals, but not in octocorals: Comment on Goulet (2006). Mar. Ecol. Prog. Ser. 2007, 335, 237–242. [Google Scholar] [CrossRef]
- Boulotte, N.M.; Dalton, S.J.; Carroll, A.G.; Harrison, P.L.; Putnam, H.M.; Peplow, L.M.; van Oppen, M.J.H. Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J. 2016, 10, 2693–2701. [Google Scholar] [CrossRef] [PubMed]
- Cunning, R.; Silverstein, R.N.; Baker, A.C. Investigating the causes and consequences of symbiont shuffling in a multi-partner reef coral symbiosis under environmental change. Proc. R. Soc. B Biol. Sci. 2015, 282, 20141725. [Google Scholar] [CrossRef]
- Matthews, J.L.; Crowder, C.M.; Oakey, C.A.; Lutz, A.; Roessner, U.; Meyer, E.; Grossman, A.R.; Weis, V.M.; Davy, S.K. Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc. Natl. Acad. Sci. USA 2017, 114, 13194–13199. [Google Scholar] [CrossRef]
- Matthews, J.L.; Oakley, C.A.; Lutz, A.; Hillyer, K.E.; Roessner, U.; Grossman, A.R.; Weis, V.M.; Davy, S.K. Partner switching and metabolic flux in a model cnidarian–dinoflagellate symbiosis. Proc. R. Soc. B Biol. Sci. 2018, 285, 20182336. [Google Scholar] [CrossRef]
- Medrano, E.; Merselis, D.G.; Bellantuono, A.J.; Rodriguez-Lenetty, M. Proteomic basis of symbiosis: A heterologous partner fails to duplicate homologous colonization in a novel cnidarian-symbiodiniaceae mutualism. Front. Microbiol. 2019, 10, 1153. [Google Scholar] [CrossRef]
- Sproles, A.E.; Oakley, C.A.; Matthews, J.L.; Peng, L.; Owen, J.G.; Grossman, A.R.; Weis, V.M.; Davy, S.K. Proteomics quantifies protein expression changes in a model cnidarian colonised by a thermally tolerant but suboptimal symbiont. ISME J. 2019, 13, 2334–2345. [Google Scholar] [CrossRef]
- Tsang Min Ching, S.J.; Chan, W.Y.; Perez-Gonzalez, A.; Hillyer, K.E.; Buerger, P.; van Oppen, M.J.H. Colonization and metabolite profiles of homologous, heterologous and experimentally evolved algal symbionts in the sea anemone Exaiptasia diaphana. ISME Commun. 2022, 2, 30. [Google Scholar] [CrossRef]
- Bosch, T.C.G.; Mcfall-Ngai, M.J. Metaorganisms as the new frontier. Zoology 2011, 114, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Van Oppen, M.J.H.; Blackall, L. Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 2019, 17, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Osman, E.O.; Suggett, D.J.; Voolstra, C.R.; Pettay, D.T.; Clark, D.R.; Pogoreutz, C.; Sampayo, E.M.; Warner, M.E.; Smith, D.J. Coral microbiome composition along the northern Red Sea suggests high plasticity of bacterial and specificity of endosymbiotic dinoflagellate communities. Microbiome 2020, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Reshef, L.; Koren, O.; Loya, Y.; Zilber-Rosenberg, I.; Rosenbreg, E. The coral probiotic hypothesis. Environ. Microbiol. 2006, 8, 2068–2073. [Google Scholar] [CrossRef]
- Raina, J.B.; Tapiolas, D.; Willis, B.L.; Bourne, D.G. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 2009, 75, 3492–3501. [Google Scholar] [CrossRef]
- Krediet, C.J.; Ritchie, K.B.; Paul, V.J.; Teplitski, M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122328. [Google Scholar] [CrossRef]
- Pogoreutz, C.; Oakley, C.A.; Radecker, N.; Cardenas, A.; Perna, G.; Xiang, N.; Peng, L.; Davy, S.K.; Ngugi, D.K.; Voolstra, C.R. Coral holobiont cues prime Endozoicomonas for a symbiotic lifestyle. ISME J. 2022, 16, 1883–1895. [Google Scholar] [CrossRef]
- Voolstra, C.R.; Ziegler, M. Adapting with microbial help: Microbiome flexibility facilitates rapid responses to environmental change. BioEssays 2020, 42, e2000004. [Google Scholar] [CrossRef]
- Röthig, T.; Costa, R.M.; Simona, F.; Baumgarten, S.; Torres, A.F.; Radhakrishnan, A.; Aranda, M.; Voolstra, C.R. Distinct bacterial communities associated with the coral model Aiptasia in aposymbiotic and symbiotic states with Symbiodinium. Front. Mar. Sci. 2016, 3, 234. [Google Scholar] [CrossRef]
- Voolstra, C.R.; Suggett, D.J.; Peixoto, R.; Parkinson, J.; Quigley, K.; Silveira, C.B.; Sweet, M.J.; Muller, E.M.; Barshis, D.J.; Bourne, D.; et al. Extending the natural adaptive capacity of coral holobionts. Nat. Rev. Earth Environ. 2021, 2, 747–762. [Google Scholar] [CrossRef]
- Rothig, T.; Ochsenkuhn, M.A.; Roik, A.; van der Merwe, R.; Voolstra, C.R. Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol. Ecol. 2016, 25, 1308–1323. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, M.; Seneca, F.O.; Yum, L.K.; Palumbi, S.R.; Voolstra, C.R. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat. Commun. 2017, 8, 14213. [Google Scholar] [CrossRef]
- Costa, R.M.; Cárdenas, A.; Loussert-Fonta, C.; Toullec, G.; Meibom, A.; Voolstra, C.R. Surface topography, bacterial carrying capacity, and the prospect of microbiome manipulation in the sea anemone coral model Aiptasia. Front. Microbiol. 2021, 12, 637834. [Google Scholar] [CrossRef]
- Mansurova, M.; Ebert, B.E.; Blank, L.M.; Ibanez, A.J. A breath of information: The volatilome. Curr. Genet. 2018, 64, 959–964. [Google Scholar] [CrossRef]
- Tahir, H.A.S.; Gu, Q.; Wu, H.; Raza, W.; Hanif, A.; Wu, L.; Colman, M.V.; Gao, X. Plant growth promotion by volatile organic compounds produced by Bacillus subtilis SYST2. Front. Microbiol. 2017, 8, 171. [Google Scholar] [CrossRef]
- Werner, S.; Polle, A.; Brinkmann, N. Belowground communication: Impacts of volatile organic compounds (VOCs) from soil fungi on other soil-inhabiting organisms. Appl. Microbiol. Biotechnol. 2016, 100, 8651–8665. [Google Scholar] [CrossRef] [PubMed]
- Lawson, C.A.; Possell, M.; Seymour, J.R.; Raina, J.B.; Suggett, D.J. Coral endosymbionts (Symbiodiniaceae) emit species-specific volatilomes that shift when exposed to thermal stress. Sci. Rep. 2019, 9, 17395. [Google Scholar] [CrossRef]
- AJürgens, A.; Wee, S.L.; Shuttleworth, A.; Johnson, S.D. Chemical mimicry of insect oviposition sites: A global analysis of convergence in angiosperms. Ecol. Lett. 2013, 16, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, T.D. Pheromones and animal behaviour. In Communication by Smell and Taste; Cambridge University Press: Cambridge, UK, 2003; pp. 359–370. [Google Scholar]
- Lawson, C.A.; Raina, J.B.; Deschaseaux, E.; Hrebien, V.; Possell, M.; Seymour, J.R.; Suggett, D.J. Heat stress decreases the diversity, abundance and functional potential of coral gas emissions. Glob. Chang. Biol. 2021, 27, 879–891. [Google Scholar] [CrossRef]
- Wuerz, M.; Lawson, C.A.; Ueland, M.; Oakley, C.A.; Grossman, A.R.; Weis, V.M.; Suggett, D.J.; Davy, S.K. Symbiosis induces unique volatile profiles in the model cnidarian Aiptasia. J. Exp. Biol. 2022, 225, jeb244600. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, M.; Wysocki, C.J.; Leyden, J.J.; Spielman, A.I.; Sun, X.; Preti, G. Analyses of volatile organic compounds from human skin. Br. J. Dermatol. 2008, 159, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Guenther, A.; Geron, C.; Pierce, T.; Lamb, B.; Harley, P.; Fall, R. Natural emissions of non-methane volatile organic compounds, carbon monoxide, and oxides of nitrogen from North America. Atmos. Environ. 2000, 34, 2205–2230. [Google Scholar] [CrossRef]
- Tyc, O.; Song, C.; Dickschat, J.S.; Vos, M.; Garbeva, P. The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol. 2017, 25, 280–292. [Google Scholar] [CrossRef]
- Raguso, R.A. Wake up and smell the roses: The ecology and evolution of floral scent. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 549–569. [Google Scholar] [CrossRef]
- Dani, K.G.S.; Loreto, F. Plant volatiles as regulators of hormone homeostasis. New Phytol. 2022, 234, 804–812. [Google Scholar] [CrossRef]
- Monson, R.K. Volatile organic compound emissions from terrestrial ecosystems: A primary biological control over atmospheric chemistry. Isr. J. Chem. 2002, 42, 29–42. [Google Scholar] [CrossRef]
- Steinke, M.; Randell, L.; Dumbrell, A.J.; Saha, M. Volatile Biomarkers for Aquatic Ecological Research. Adv. Ecol. Res. 2018, 59, 75–92. [Google Scholar] [CrossRef]
- Baumgarten, S.; Simakov, O.; Esherick, L.Y.; Liew, Y.J.; Lehnert, E.M.; Michell, C.T.; Li, Y.; Hambleton, E.A.; Guse, A.; Oates, M.E.; et al. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. USA 2015, 112, 11893–11898. [Google Scholar] [CrossRef]
- Rädecker, N.; Raina, J.B.; Pernice, M.; Perna, G.; Guagliardo, P.; Kilburn, M.R.; Aranda, M.; Voolstra, C.R. Using Aiptasia as a model to study metabolic interactions in Cnidarian-Symbiodinium symbioses. Front. Physiol. 2018, 9, 214. [Google Scholar] [CrossRef]
- Wolfowicz, I.; Baumgarten, S.; Voss, P.A.; Hambleton, E.A.; Voolstra, C.R.; Hatta, M.; Guse, A. Aiptasia sp. larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Sci. Rep. 2016, 6, 32366. [Google Scholar] [CrossRef]
- Matthews, J.L.; Sproles, A.E.; Oakley, C.A.; Grossman, A.R.; Weis, V.M.; Davy, S.K. Menthol-induced bleaching rapidly and effectively provides experimental aposymbiotic sea anemones (Aiptasia sp.) for symbiosis investigations. J. Exp. Biol. 2016, 219, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Gabay, Y.; Weis, V.M.; Davy, S.K. Symbiont identity influences patterns of symbiosis establishment, host growth, and asexual reproduction in a model cnidarian-dinoflagellate symbiosis. Biol. Bull. 2018, 234, 1–10. [Google Scholar] [CrossRef]
- Lehnert, E.M.; Burriesci, M.S.; Pringle, J.R. Developing the anemone Aiptasia as a tractable model for cnidarian-dinoflagellate symbiosis: Generating transcriptomic resources and profiling gene expression. BMC Genom. 2012, 13, 1–10. [Google Scholar] [CrossRef]
- Hartman, L.M.; van Oppen, M.J.H.; Blackall, L. The effect of thermal stress on the bacterial microbiome of Exaiptasia diaphana. Microorganisms 2020, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Davy, S.K.; Lucas, I.A.N.; Turner, J.R. Uptake and persistence of homologous and heterologous zooxanthellae in the temperate sea anemone Cereus pedunculatus (Pennant). Biol. Bull. 1997, 192, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Illumina. 16S metagenomic sequencing library preparation. 2013. Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 23 June 2023).
- Olander, A.; Lawson, C.A.; Possell, M.; Raina, J.B.; Ueland, M.; Suggett, D.J. Comparative volatilomics of coral endosymbionts from one- and comprehensive two-dimensional gas chromatography approaches. Mar. Biol. 2021, 168, 76. [Google Scholar] [CrossRef]
- Wenig, P.; Odermatt, J. OpenChrom: A cross-platform open source software for the mass spectrometric analysis of chromatographic data. BMC Bioinform. 2010, 11, 405. [Google Scholar] [CrossRef]
- Guitton, Y.; Tremblay-Franco, M.; Le Corguille, G.; Martin, J.F.; Petera, M.; Roger-Mele, P.; Delabriere, A.; Goulitquer, S.; Monsoor, M.; Duperier, C.; et al. Create, run, share, publish, and reference your LC–MS, FIA–MS, GC–MS, and NMR data analysis workflows with the Workflow4Metabolomics 3.0 Galaxy online infrastructure for metabolomics. Int. J. Biochem. Cell Biol. 2017, 93, 89–101. [Google Scholar] [CrossRef]
- Wehrens, R.; Weingart, G.; Mattivi, F. MetaMS: An open-source pipeline for GC-MS-based untargeted metabolomics. J. Chromatogr. B 2014, 966, 109–116. [Google Scholar] [CrossRef]
- Giacomoni, F.; Le Corguillé, G.; Monsoor, M.; Landi, M.; Pericard, P.; Pétéra, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.F.; Jacob, D.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 2015, 31, 1493–1495. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible, and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Vergauwen, G.; Dhondt, B.; Van Deun, J.; De Smedt, E.; Berx, G.; Timmerman, E.; Gevaert, K.; Miinalainen, I.; Cocquyt, V.; Braems, G.; et al. Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci. Rep. 2017, 7, 2704. [Google Scholar] [CrossRef] [PubMed]
- Cella, J.A.; Carpenter, J.C. Procedures for the preparation of silanols. J. Organomet. Chem. 1994, 480, 23–26. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: http://www.R-project.org (accessed on 1 June 2019).
- Wilkinson, S.P.; Davy, S.K.; Bunce, M.; Stat, M. Taxonomic identification of environmental DNA with informatic sequence classification trees. PeerJ 2018, preprints. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Edgar, R. SINTAX: A simple non-Bayesian taxonomy classifier for 16S and ITS sequences. Biorxiv 2016, 074161. [Google Scholar]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 2016, 10, 946–963. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Volume 35, Available online: http://link.springer.com/10.1007/978-0-387-98141-3 (accessed on 1 June 2019).
- Oksanen, A.J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchen, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2020. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 1 June 2019).
- Ahn, J.; Hayes, R.B. Environmental influences on the human microbiome and implications for noncommunicable disease. Annu. Rev. Public Health 2021, 42, 277–292. [Google Scholar] [CrossRef]
- Hacquard, S.; Wang, E.; Slater, H.; Martin, F. Impact of global change on the plant microbiome. New Phytol. 2022, 234, 1907–1909. [Google Scholar] [CrossRef]
- Pootakham, W.; Mhuantong, W.; Yoocha, T.; Putchim, L.; Jomchai, N.; Sonthirod, C.; Naktang, C.; Kongkachana, W.; Tangphatsornruang, S. Heat-induced shift in coral microbiome reveals several members of the Rhodobacteraceae family as indicator species for thermal stress in Porites lutea. Microbiologyopen 2019, 8, e935. [Google Scholar] [CrossRef]
- Hartman, L.M.; van Oppen, M.J.H.; Blackall, L.L. Microbiota characterization of Exaiptasia diaphana from the Great Barrier Reef. Anim. Microbiome. 2020, 2, 10. [Google Scholar] [CrossRef]
- Dungan, A.M.; Hartman, L.M.; Tortorelli, G.; Belderok, R.; Lamb, A.M.; Pisan, L.; McFadden, G.I.; Blackall, L.L.; van Oppen, M.J.H. Exaiptasia diaphana from the great barrier reef: A valuable resource for coral symbiosis research. Symbiosis 2020, 80, 195–206. [Google Scholar] [CrossRef]
- Gevers, D.; Knight, R.; Petrosino, J.F.; Huang, K.; McGuire, A.L.; Birren, B.W.; Nelson, K.E.; White, O.; Methe, B.A.; Huttenhower, C. The human microbiome project: A community resource for the healthy human microbiome. PLoS Biol. 2012, 10, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Rook, G.A.W.; Raison, C.L.; Lowry, C.A. Microbial ‘old friends’, immunoregulation and socioeconomic status. Clin. Exp. Immunol. 2014, 177, 1–12. [Google Scholar] [CrossRef]
- Deng, F.; Li, Y.; Zhao, J. The gut microbiome of healthy long-living people. Aging 2019, 11, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Ben-Haim, Y.; Rosenberg, E. A novel Vibrio sp. pathogen of the coral Pocillopora damicornis. Mar. Biol. 2002, 141, 47–55. [Google Scholar] [CrossRef]
- Ben-Haim, Y.; Thompson, F.L.; Thompson, C.C.; Cnockaert, M.C.; Hoste, B.; Swings, J.; Rosenberg, E. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 2003, 53, 309–315. [Google Scholar] [CrossRef]
- Ben-Haim, Y.; Banim, E.; Kushmaro, A.; Loya, Y.; Rosenberg, E. Inhibition of photosynthesis and bleaching of zooxanthellae by the coral pathogen Vibrio shiloi. Environ. Microbiol. 1999, 1, 223–229. [Google Scholar] [CrossRef]
- Wilson, B.; Aeby, G.S.; Work, T.M.; Bourne, D.G. Bacterial communities associated with healthy and Acropora white syndrome-affected corals from American Samoa. FEMS Microbiol. Ecol. 2012, 80, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Dasti, J.I.; Tareen, A.M.; Lugert, R.; Zautner, A.E.; Groβ, U. Campylobacter jejuni: A brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int. J. Med. Microbiol. 2010, 300, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Ortega, J.; Thomé, P.E. Contrasting antibacterial capabilities of the surface mucus layer from three symbiotic cnidarians. Front. Mar. Sci. 2018, 5, 392. [Google Scholar] [CrossRef]
- Crossland, C.J.; Barnes, D.J.; Borowitzka, M.A. Diurnal lipid and mucus production in the staghorn coral Acropora acuminata. Mar. Biol. 1980, 60, 81–90. [Google Scholar] [CrossRef]
- Oakley, C.A.; Ameismeier, M.F.; Peng, L.; Weis, V.M.; Grossman, A.R.; Davy, S.K. Symbiosis induces widespread changes in the proteome of the model cnidarian Aiptasia. Cell. Microbiol. 2016, 18, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Munoz, R.; Rosselló-Móra, R.; Amann, R. Revised phylogeny of Bacteroidetes and proposal of sixteen new taxa and two new combinations including Rhodothermaeota phyl. nov. Syst. Appl. Microbiol. 2016, 39, 281–296. [Google Scholar] [CrossRef]
- Ratiu, I.A.; Ligor, T.; Bocos-Bintintan, V.; Mayhew, C.A.; Buszewski, B. Volatile organic compounds in exhaled breath as fingerprints of lung cancer, asthma and COPD. J. Clin. Med. 2021, 10, 32. [Google Scholar] [CrossRef]
- Laothawornkitkul, J.; Taylor, J.E.; Paul, N.D.; Hewitt, C.N. Biogenic volatile organic compounds in the Earth system. New Phytol. 2009, 183, 27–51. [Google Scholar] [CrossRef]
- Lawson, C.A.; Camp, E.; Davy, S.K.; Ferrier-Pages, C.; Matthews, J.; Suggett, D.J. Informing coral reef conservation through metabolomic approaches. In Coral Reef Conservation and Restoration in the Omics Age. Coral Reefs of the World; Van Oppen, M.J.H., Aranda Lastra, M., Eds.; Springer: Cham, Switzerland, 2022; Volume 15. [Google Scholar] [CrossRef]
- Bouwmeester, H.; Schuurink, R.C.; Bleeker, P.M.; Schiestl, F. The role of volatiles in plant communication. Plant J. 2019, 100, 892–907. [Google Scholar] [CrossRef]
- Yi, H.S.; Heil, M.; Adame-Álvarez, R.M.; Ballhorn, D.J.; Ryu, C.M. Airborne induction and priming of plant defenses against a bacterial pathogen. Plant Physiol. 2009, 151, 2152–2161. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [PubMed]
- Kuek, F.W.I.; Motti, C.A.; Zhang, J.; Cooke, I.R.; Todd, J.D.; Miller, D.J.; Bourne, D.G.; Raina, J.B. DMSP production by coral-associated bacteria. Front. Mar. Sci. 2022, 9, 1–12. [Google Scholar] [CrossRef]
- Hopkins, F.E.; Bell, T.G.; Yang, M.; Suggett, D.J.; Steinke, M. Air exposure of coral is a significant source of dimethylsulfide (DMS) to the atmosphere. Sci. Rep. 2016, 6, 36031. [Google Scholar] [CrossRef] [PubMed]
- Otte, M.L.; Wilson, G.; Morris, J.T.; Moran, B.M. Dimethylsulphoniopropionate (DMSP) and related compounds in higher plants. J. Exp. Bot. 2004, 55, 1919–1925. [Google Scholar] [CrossRef] [PubMed]
- Trevena, A.J.; Jones, G.B.; Wright, S.W.; Van Den Enden, R.L. Profiles of DMSP, algal pigments, nutrients and salinity in pack ice from eastern Antarctica. J. Sea Res. 2000, 43, 265–273. [Google Scholar] [CrossRef]
- Sunda, W.; Kieber, D.J.; Kiene, R.P.; Huntsman, S. An antioxidant function for DMSP and DMS in marine algae. Nature 2002, 418, 317–320. [Google Scholar] [CrossRef]
- Rodrigues, T.B.; Baker, C.R.; Walker, A.P.; McDowell, N.; Rogers, A.; Higuchi, N.; Chambers, J.Q.; Jardine, K.J. Stimulation of isoprene emissions and electron transport rates as key mechanisms of thermal tolerance in the tropical species Vismia guianensis. Glob. Chang. Biol. 2020, 26, 5928–5941. [Google Scholar] [CrossRef]
- Velikova, V.; Edreva, A.; Loreto, F. Endogenous isoprene protects Phragmites australis leaves against singlet oxygen. Physiol. Plant. 2004, 122, 219–225. [Google Scholar] [CrossRef]
- Laothawornkitkul, J.; Paul, N.D.; Vickers, C.E.; Possell, M.; Taylor, J.E.; Mullineaux, P.M.; Hewitt, C.N. Isoprene emissions influence herbivore feeding decisions. Plant Cell Environ. 2008, 31, 1410–1415. [Google Scholar] [CrossRef]
- Exton, D.A.; Suggett, D.J.; Steinke, M. Chlorophyll-normalized isoprene production in laboratory cultures of marine microalgae and implications for global models. Limnol. Oceanogr. 2013, 58, 1301–1311. [Google Scholar] [CrossRef]
- Pophof, B.; Stange, G.; Abrell, L. Volatile organic compounds as signals in a plant-herbivore system: Electrophysiological responses in olfactory sensilla of the moth Cactoblastis cactorum. Chem. Senses 2005, 30, 51–68. [Google Scholar] [CrossRef]
- Brambilla, A.; Sommer, A.; Ghirardo, A.; Wenig, M.; Knappe, C.; Weber, B.; Amesmaier, M.; Lenk, M.; Schnitzler, J.P.; Volt, A.C. Immunity-associated volatile emissions of β-ionone and nonanal propagate defence responses in neighbouring barley plants. J. Exp. Bot. 2022, 73, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Fontan, A.; Audino, P.G.; Martinez, A.; Alzogaray, R.A.; Zerb, E.N.; Camps, F.; Cork, A. Attractant volatiles released by female and male Triatoma infestans (Hemiptera: Reduviidae), a vector of chagas disease: Chemical analysis and behavioral bioassay. J. Med. Entomol. 2002, 39, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Siljander, E.; Gries, R.; Khaskin, G.; Gries, G. Identification of the airborne aggregation pheromone of the common bed bug, Cimex lectularius. J. Chem. Ecol. 2008, 34, 708–718. [Google Scholar] [CrossRef]
- Shirasu, M.; Ito, S.; Itoigawa, A.; Hayakara, T.; Kinoshita, K.; Munechika, I.; Imai, H.; Touhara, K. Key male glandular odorants attracting female ring-tailed lemurs. Curr. Biol. 2020, 30, 2131–2138. [Google Scholar] [CrossRef]
- Zagrobelny, M.; Simonsen, H.T.; Olsen, C.E.; Bak, S.; Møller, B.L. Volatiles from the burnet moth Zygaena filipendulae (Lepidoptera) and associated flowers, and their involvement in mating communication. Physiol. Entomol. 2015, 40, 284–295. [Google Scholar] [CrossRef]
- Laznik, Z.; Trdan, S. Attraction behaviors of entomopathogenic nematodes (Steinernematidae and Heterorhabditidae) to synthetic volatiles emitted by insect damaged potato tubers. J. Chem. Ecol. 2016, 42, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.M.; Bartels, E.; Baums, I.B. Bleaching causes loss of disease resistance within the threatened coral species Acropora cervicornis. ELife 2018, 7, e35066. [Google Scholar] [CrossRef]
- Leal, M.C.; Hoadley, K.; Pettay, D.T.; Grajales, A.; Calado, R.; Warner, M.E. Symbiont type influences trophic plasticity of a model cnidarian-dinoflagellate symbiosis. J. Exp. Biol. 2015, 218, 858–863. [Google Scholar] [CrossRef]
- Sproles, A.E.; Oakley, C.A.; Krueger, T.; Grossman, A.R.; Weis, V.M.; Meibom, A.; Davy, S.K. Sub-cellular imaging shows reduced photosynthetic carbon and increased nitrogen assimilation by the non-native endosymbiont Durusdinium trenchii in the model cnidarian Aiptasia. Environ. Microbiol. 2020, 22, 3741–3753. [Google Scholar] [CrossRef]
- Mashini, A.G.; Oakley, C.A.; Grossman, A.R.; Weis, V.M.; Davy, S.K. Immunolocalization of metabolite transporter proteins in a model cnidarian-dinoflagellate symbiosis. Appl. Environ. Microbiol. 2022, 88, e0041222. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wuerz, M.; Lawson, C.A.; Oakley, C.A.; Possell, M.; Wilkinson, S.P.; Grossman, A.R.; Weis, V.M.; Suggett, D.J.; Davy, S.K. Symbiont Identity Impacts the Microbiome and Volatilome of a Model Cnidarian-Dinoflagellate Symbiosis. Biology 2023, 12, 1014. https://doi.org/10.3390/biology12071014
Wuerz M, Lawson CA, Oakley CA, Possell M, Wilkinson SP, Grossman AR, Weis VM, Suggett DJ, Davy SK. Symbiont Identity Impacts the Microbiome and Volatilome of a Model Cnidarian-Dinoflagellate Symbiosis. Biology. 2023; 12(7):1014. https://doi.org/10.3390/biology12071014
Chicago/Turabian StyleWuerz, Maggie, Caitlin A. Lawson, Clinton A. Oakley, Malcolm Possell, Shaun P. Wilkinson, Arthur R. Grossman, Virginia M. Weis, David J. Suggett, and Simon K. Davy. 2023. "Symbiont Identity Impacts the Microbiome and Volatilome of a Model Cnidarian-Dinoflagellate Symbiosis" Biology 12, no. 7: 1014. https://doi.org/10.3390/biology12071014
APA StyleWuerz, M., Lawson, C. A., Oakley, C. A., Possell, M., Wilkinson, S. P., Grossman, A. R., Weis, V. M., Suggett, D. J., & Davy, S. K. (2023). Symbiont Identity Impacts the Microbiome and Volatilome of a Model Cnidarian-Dinoflagellate Symbiosis. Biology, 12(7), 1014. https://doi.org/10.3390/biology12071014






