N6-Methyladenosine Directly Regulates CD40L Expression in CD4+ T Lymphocytes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. meRIP Sequencing and qPCR
2.3. Flow Cytometry
2.4. Western Blot
2.5. Statistics
3. Results
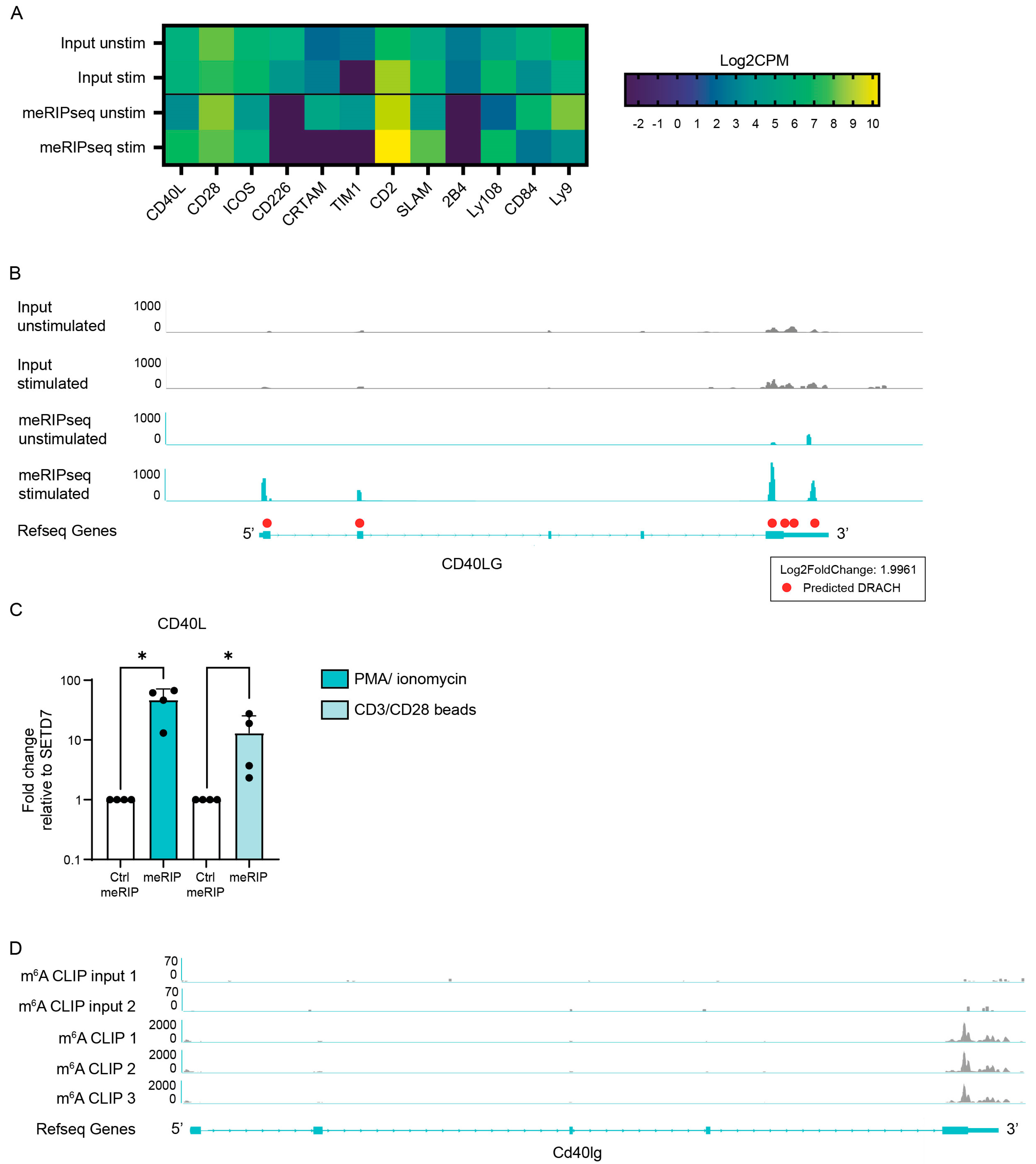
3.1. m6A Enrichment Is Increased on CD40L mRNA in Activated CD4+ T Lymphocytes
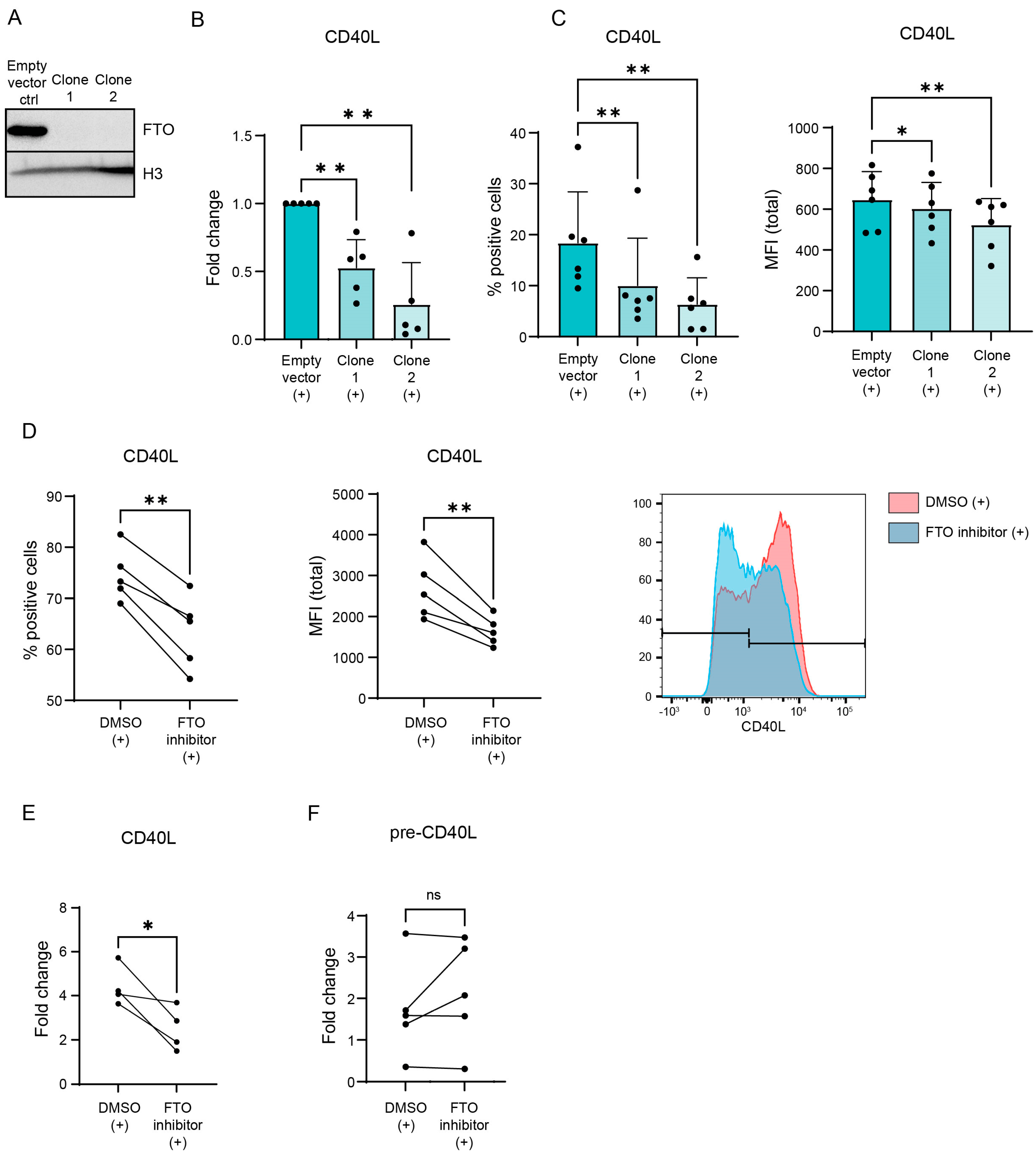
3.2. CD40L Expression Is Regulated via m6A ‘Eraser’ FTO in Activated CD4+ T Lymphocytes
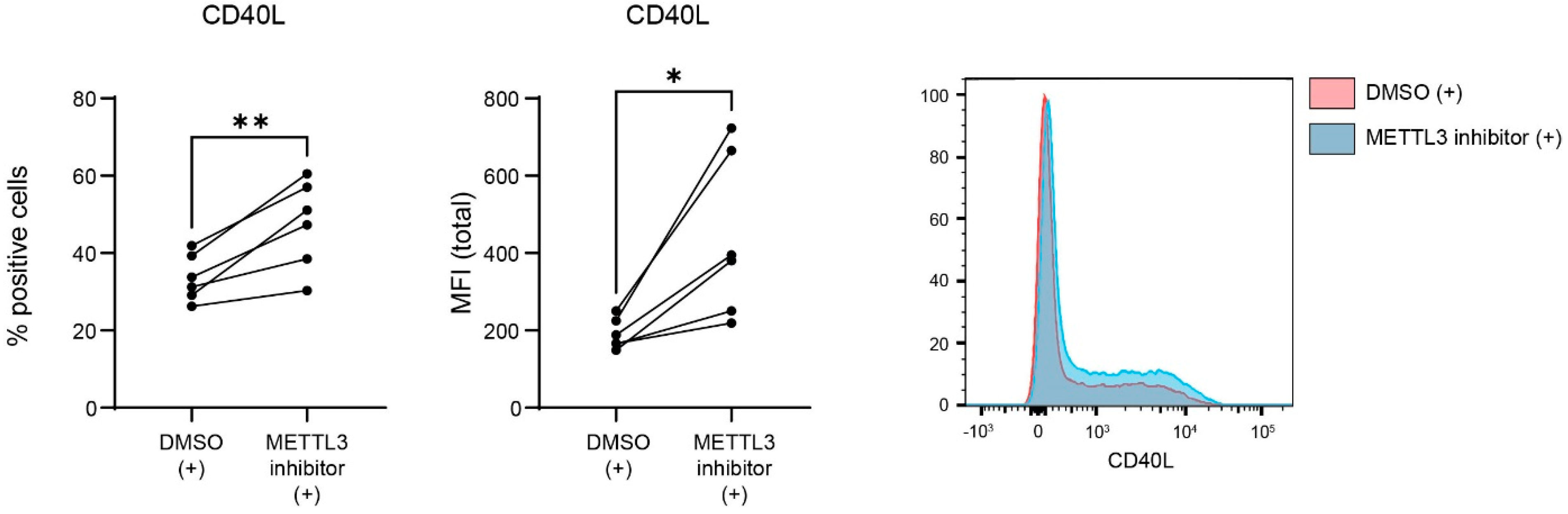
3.3. METTL3 Regulates CD40L Expression in Activated CD4+ T Lymphocytes
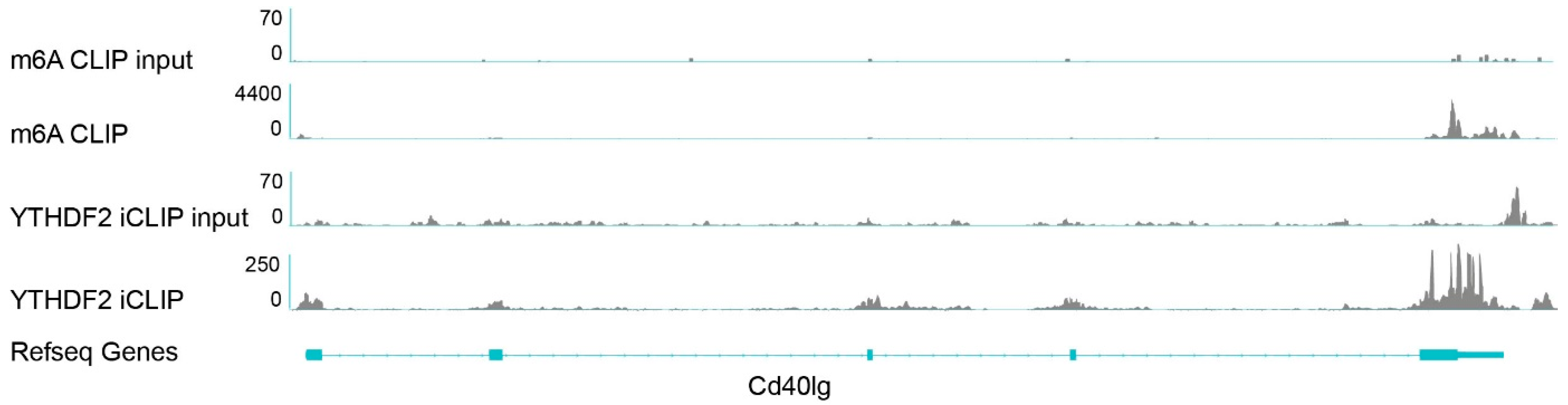
3.4. m6A Reader YTHDF2 Possiblybinds to m6A Sequences on Cd40lg mRNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Luo, G.-Z.; He, C. High-Resolution Mapping of N6-Methyladenosine in Transcriptome and Genome Using a Photo-Crosslinking-Assisted Strategy, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2015; Volume 560. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.; Rath-Shambaugh, M.; Ludwiczak, R.; Narayan, P.; Rottman, F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994, 269, 17697–17704. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2013, 10, 93–95. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell 2020, 181, 1582–1595.e18. [Google Scholar] [CrossRef]
- Patil, D.P.; Pickering, B.F.; Jaffrey, S.R. Reading m6A in the Transcriptome: m6A-Binding Proteins. Trends Cell Biol. 2017, 28, 113–127. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m 6 A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985.e5. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Yang, Y.; Guo, W.; Xu, L.; You, M.; Zhang, Y.-C.; Sun, Z.; Cui, X.; Yu, G.; Qi, Z.; et al. METTL3-dependent m6A modification programs T follicular helper cell differentiation. Nat. Commun. 2021, 12, 1333. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-B.; Tong, J.; Zhu, S.; Batista, P.J.; Duffy, E.E.; Zhao, J.; Bailis, W.; Cao, G.; Kroehling, L.; Chen, Y.; et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017, 548, 338–342. [Google Scholar] [CrossRef]
- Ding, C.; Xu, H.; Yu, Z.; Roulis, M.; Qu, R.; Zhou, J.; Oh, J.; Crawford, J.; Gao, Y.; Jackson, R.; et al. RNA m6A demethylase ALKBH5 regulates the development of γδ T cells. Proc. Natl. Acad. Sci. USA 2022, 119, e2203318119. [Google Scholar] [CrossRef]
- van Diemen, F.R.; Kruse, E.M.; Hooykaas, M.J.G.; Bruggeling, C.E.; Schürch, A.C.; van Ham, P.M.; Imhof, S.M.; Nijhuis, M.; Wiertz, E.J.H.J.; Lebbink, R.J. CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections. PLoS Pathog. 2016, 12, e1005701. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chatzigeorgiou, A.; Lyberi, M.; Chatzilymperis, G.; Nezos, A.; Kamper, E. CD40/CD40L signaling and its implication in health and disease. Biofactors 2009, 35, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Cheng, X.; Truong, B.; Sun, L.; Yang, X.; Wang, H. Molecular basis and therapeutic implications of CD40/CD40L immune checkpoint. Pharmacol. Ther. 2020, 219, 107709. [Google Scholar] [CrossRef] [PubMed]
- Karnell, J.L.; Rieder, S.A.; Ettinger, R.; Kolbeck, R. Targeting the CD40-CD40L pathway in autoimmune diseases: Humoral immunity and beyond. Adv. Drug Deliv. Rev. 2018, 141, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Ito-Kureha, T.; Leoni, C.; Borland, K.; Cantini, G.; Bataclan, M.; Metzger, R.N.; Ammann, G.; Krug, A.B.; Marsico, A.; Kaiser, S.; et al. The function of Wtap in N6-adenosine methylation of mRNAs controls T cell receptor signaling and survival of T cells. Nat. Immunol. 2022, 23, 1208–1221. [Google Scholar] [CrossRef]
- Peng, S.; Xiao, W.; Ju, D.; Sun, B.; Hou, N.; Liu, Q.; Wang, Y.; Zhao, H.; Gao, C.; Zhang, S.; et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 2019, 11, eaau7116. [Google Scholar] [CrossRef] [PubMed]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef]
- Li, J.; Gregory, R.I. Mining for METTL3 inhibitors to suppress cancer. Nat. Struct. Mol. Biol. 2021, 28, 460–462. [Google Scholar] [CrossRef]
- Zhang, Z.-W.; Teng, X.; Zhao, F.; Ma, C.; Zhang, J.; Xiao, L.-F.; Wang, Y.; Chang, M.; Tian, Y.; Li, C.; et al. METTL3 regulates m6A methylation of PTCH1 and GLI2 in Sonic hedgehog signaling to promote tumor progression in SHH-medulloblastoma. Cell Rep. 2022, 41, 111530. [Google Scholar] [CrossRef]
- Li, S.; Zou, D.; Chen, W.; Britz, G.W.; Liu, Z.; Weng, Y. METTL3 inhibition reduces N6-methyladenosine levels and prevents allogeneic CD4 + T-cell responses. Immunol. Cell Biol. 2022, 100, 718–730. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Kitagawa, M.; Suzuki, H.; Adachi, Y.; Nakamura, H.; Yoshino, S.; Sumida, T. Interferon-γ enhances interleukin 12 production in rheumatoid synovial cells via CD40-CD154 dependent and independent pathways. J. Rheumatol. 2001, 28, 1764–1771. [Google Scholar] [PubMed]
- Goules, A.; Tzioufas, A.G.; Manousakis, M.N.; Kirou, K.A.; Crow, M.K.; Routsias, J.G. Elevated levels of soluble CD40 ligand (sCD40L) in serum of patients with systemic autoimmune diseases. J. Autoimmun. 2006, 26, 165–171. [Google Scholar] [CrossRef]
- Zhang, S.; Qin, J.; Zhao, Y.; Wang, J.; Tian, Z. Identification of molecular patterns and diagnostic biomarkers in juvenile idiopathic arthritis based on the gene expression of m6A regulators. Front. Pediatr. 2022, 10, 930119b. [Google Scholar] [CrossRef]
- Luo, Q.; Fu, B.; Zhang, L.; Guo, Y.; Huang, Z.; Li, J. Decreased Peripheral Blood ALKBH5 Correlates with Markers of Autoimmune Response in Systemic Lupus Erythematosus. Dis. Markers 2020, 2020, 8193895. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Rao, J.; Zhang, L.; Fu, B.; Guo, Y.; Huang, Z.; Li, J. The study of METTL14, ALKBH5, and YTHDF2 in peripheral blood mononuclear cells from systemic lupus erythematosus. Mol. Genet. Genom. Med. 2020, 8, e1298. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Zhang, Y.-H.; Lei, S.-F. Genome-Wide Identification of N6-Methyladenosine (m6A) SNPs Associated With Rheumatoid Arthritis. Front. Genet. 2018, 9, 299. [Google Scholar] [CrossRef]
- Wang, J.; Yan, S.; Lu, H.; Wang, S.; Xu, D. METTL3 Attenuates LPS-Induced Inflammatory Response in Macrophages via NF-κB Signaling Pathway. Mediat. Inflamm. 2019, 2019, 3120391. [Google Scholar] [CrossRef]
- Mo, X.-B.; Lei, S.-F.; Qian, Q.-Y.; Guo, Y.-F.; Zhang, Y.-H.; Zhang, H. Integrative analysis revealed potential causal genetic and epigenetic factors for multiple sclerosis. J. Neurol. 2019, 266, 2699–2709. [Google Scholar] [CrossRef]
- Lu, T.X.; Zheng, Z.; Zhang, L.; Sun, H.-L.; Bissonnette, M.; Huang, H.; He, C. A New Model of Spontaneous Colitis in Mice Induced by Deletion of an RNA m6A Methyltransferase Component METTL14 in T Cells. Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 747–761. [Google Scholar] [CrossRef]
- McIntyre, A.B.R.; Gokhale, N.S.; Cerchietti, L.; Jaffrey, S.R.; Horner, S.M.; Mason, C.E. Limits in the detection of m6A changes using MeRIP/m6A-seq. Sci. Rep. 2020, 10, 6590. [Google Scholar] [CrossRef]
- Mauer, J.; Jaffrey, S.R. FTO, m6Am, and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett. 2018, 592, 2012–2022. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zeng, P.; Li, Y.-H.; Zhang, Z.; Cui, Q. SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016, 44, e91. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Vroonhoven, E.C.N.; Picavet, L.W.; Scholman, R.C.; van den Dungen, N.A.M.; Mokry, M.; Evers, A.; Lebbink, R.J.; Calis, J.J.A.; Vastert, S.J.; van Loosdregt, J. N6-Methyladenosine Directly Regulates CD40L Expression in CD4+ T Lymphocytes. Biology 2023, 12, 1004. https://doi.org/10.3390/biology12071004
van Vroonhoven ECN, Picavet LW, Scholman RC, van den Dungen NAM, Mokry M, Evers A, Lebbink RJ, Calis JJA, Vastert SJ, van Loosdregt J. N6-Methyladenosine Directly Regulates CD40L Expression in CD4+ T Lymphocytes. Biology. 2023; 12(7):1004. https://doi.org/10.3390/biology12071004
Chicago/Turabian Stylevan Vroonhoven, Ellen C. N., Lucas W. Picavet, Rianne C. Scholman, Noortje A. M. van den Dungen, Michal Mokry, Anouk Evers, Robert J. Lebbink, Jorg J. A. Calis, Sebastiaan J. Vastert, and Jorg van Loosdregt. 2023. "N6-Methyladenosine Directly Regulates CD40L Expression in CD4+ T Lymphocytes" Biology 12, no. 7: 1004. https://doi.org/10.3390/biology12071004
APA Stylevan Vroonhoven, E. C. N., Picavet, L. W., Scholman, R. C., van den Dungen, N. A. M., Mokry, M., Evers, A., Lebbink, R. J., Calis, J. J. A., Vastert, S. J., & van Loosdregt, J. (2023). N6-Methyladenosine Directly Regulates CD40L Expression in CD4+ T Lymphocytes. Biology, 12(7), 1004. https://doi.org/10.3390/biology12071004





