Remodelling of the Mitochondrial Bioenergetic Pathways in Human Cultured Fibroblasts with Carbohydrates
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture Conditions
2.2. Growth Rate
2.3. Copy Number of mtDNA
2.4. Reverse Transcriptase Real-Time Quantitative PCR
2.5. Western Blot Analysis
2.6. Enzyme Activity Measurements
2.7. Extracellular Flux Analysis
2.8. Statistical Analyses
3. Results
3.1. Growth Rate
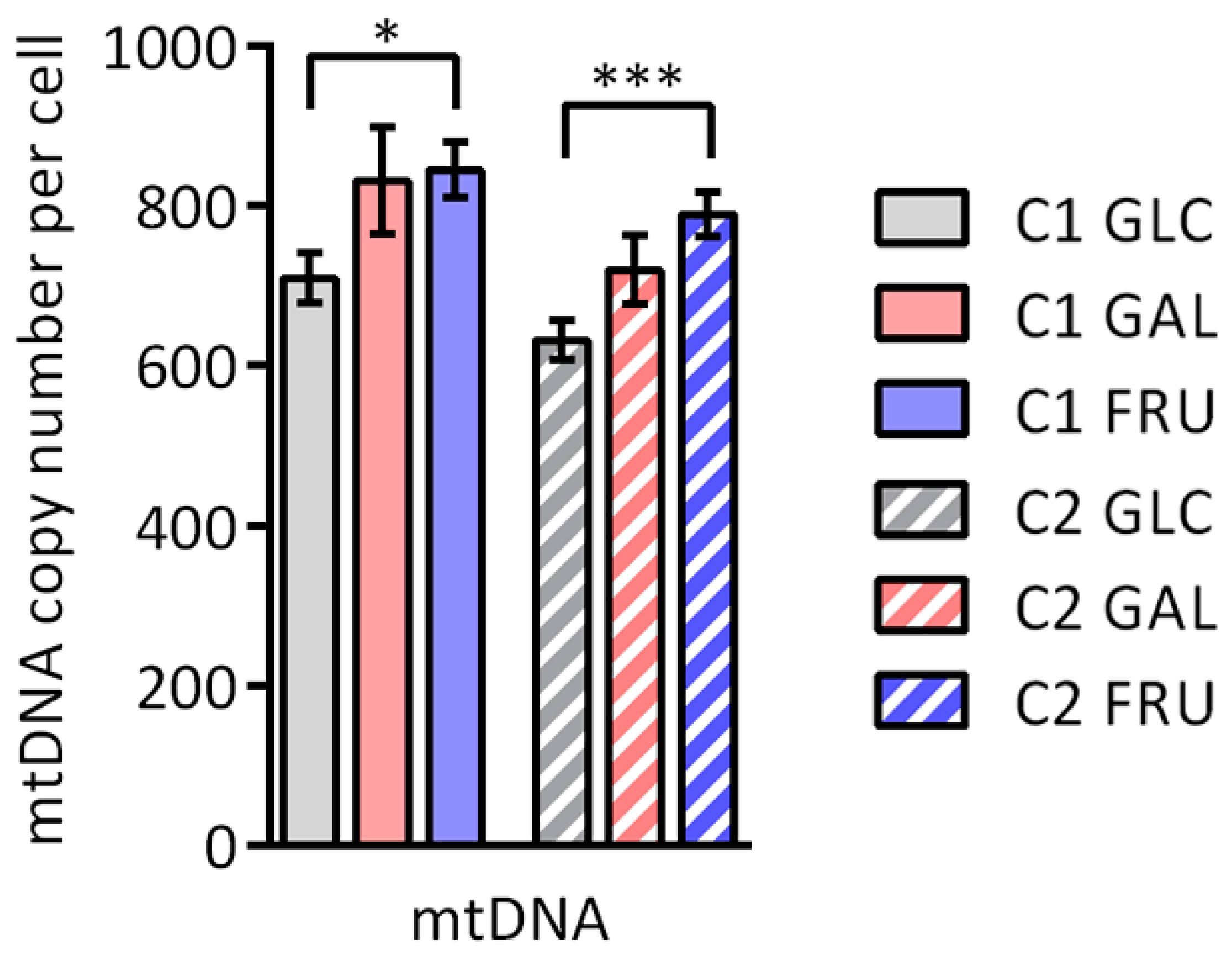
3.2. Copy Number of mtDNA
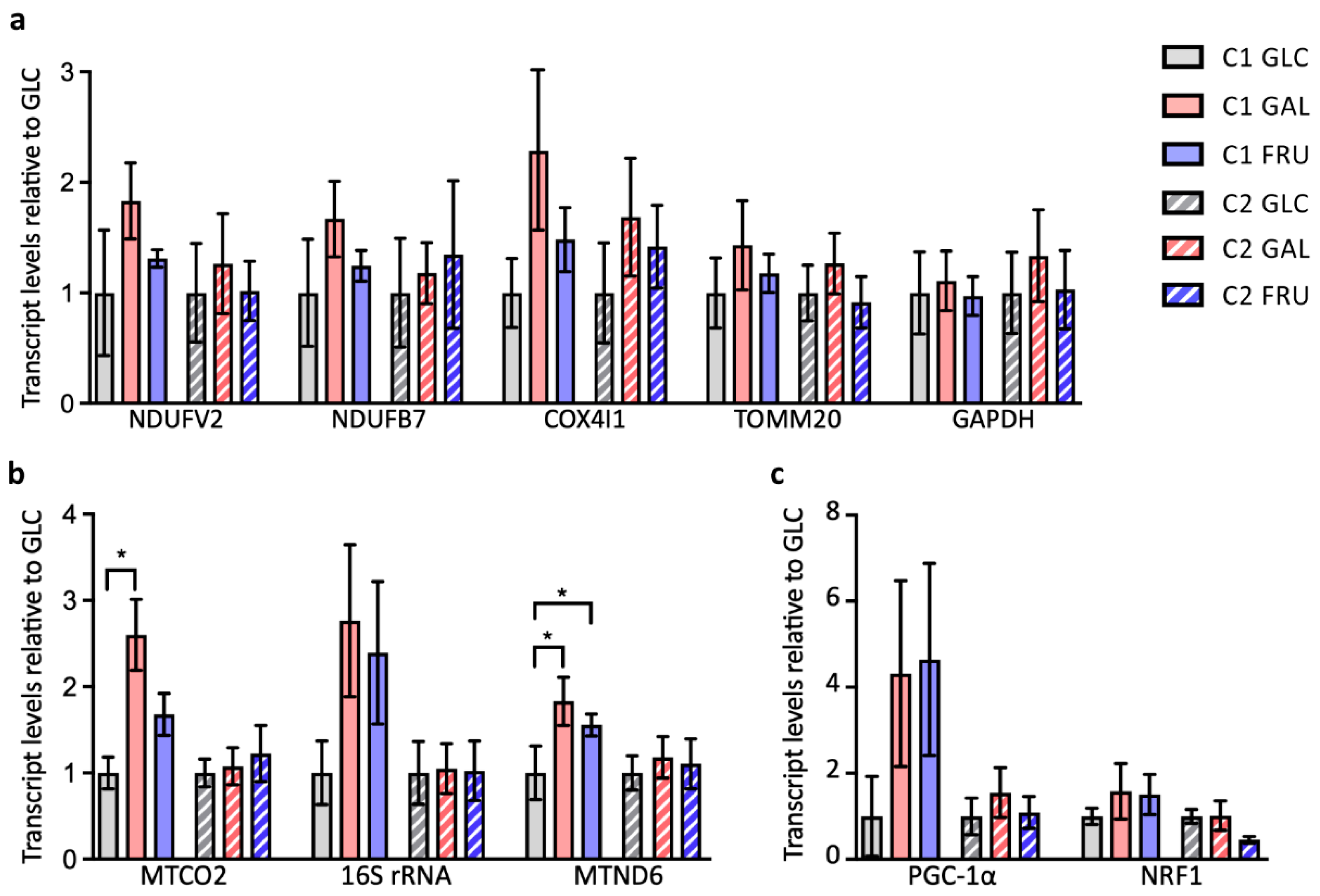
3.3. Levels of Nuclear and Mitochondrial Transcripts
3.4. Protein Levels of Enzymes Involved in Glycolysis
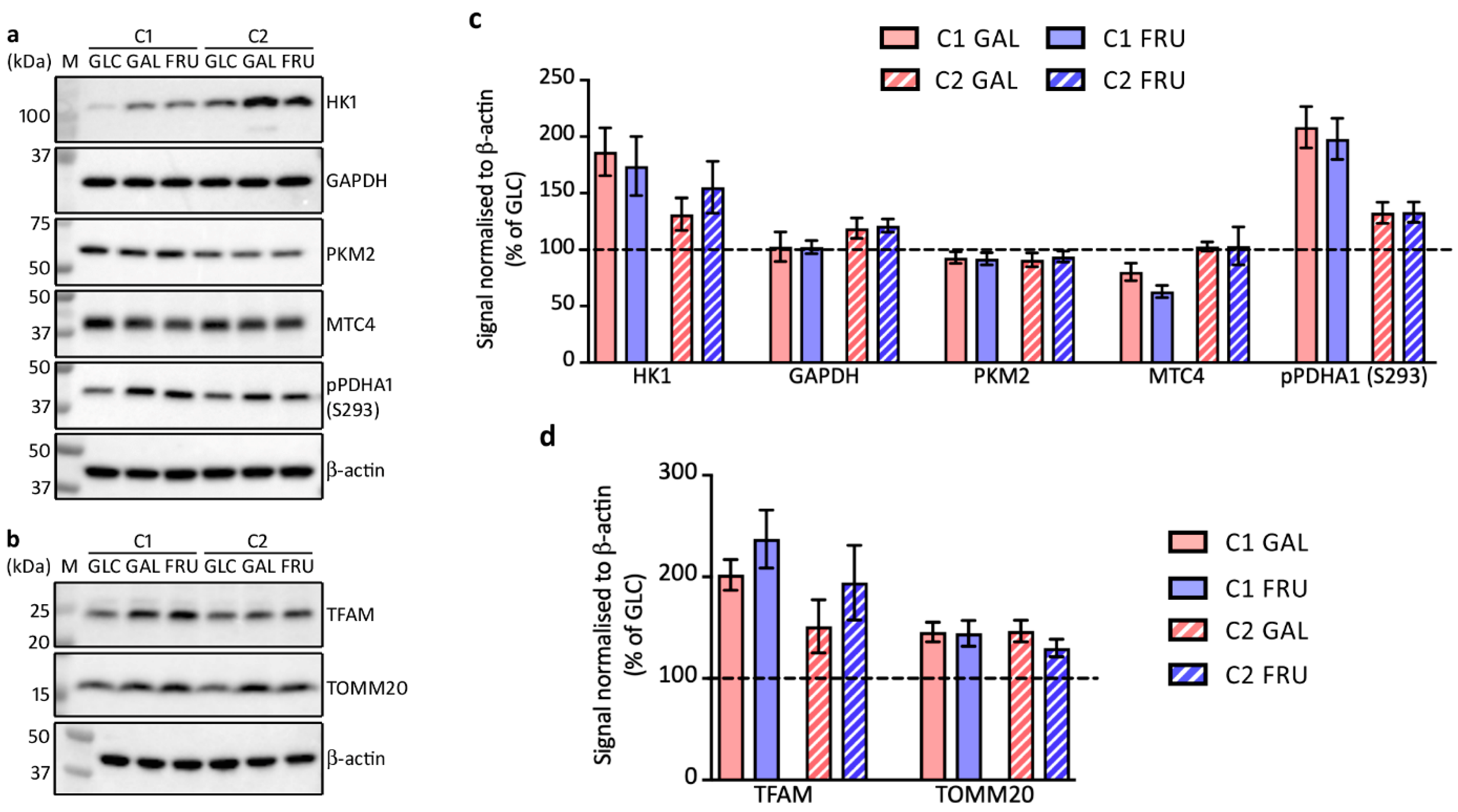
3.5. Protein Levels of Factors Involved in Mitochondrial Biogenesis
3.6. Protein Levels of OXPHOS Complex Subunits
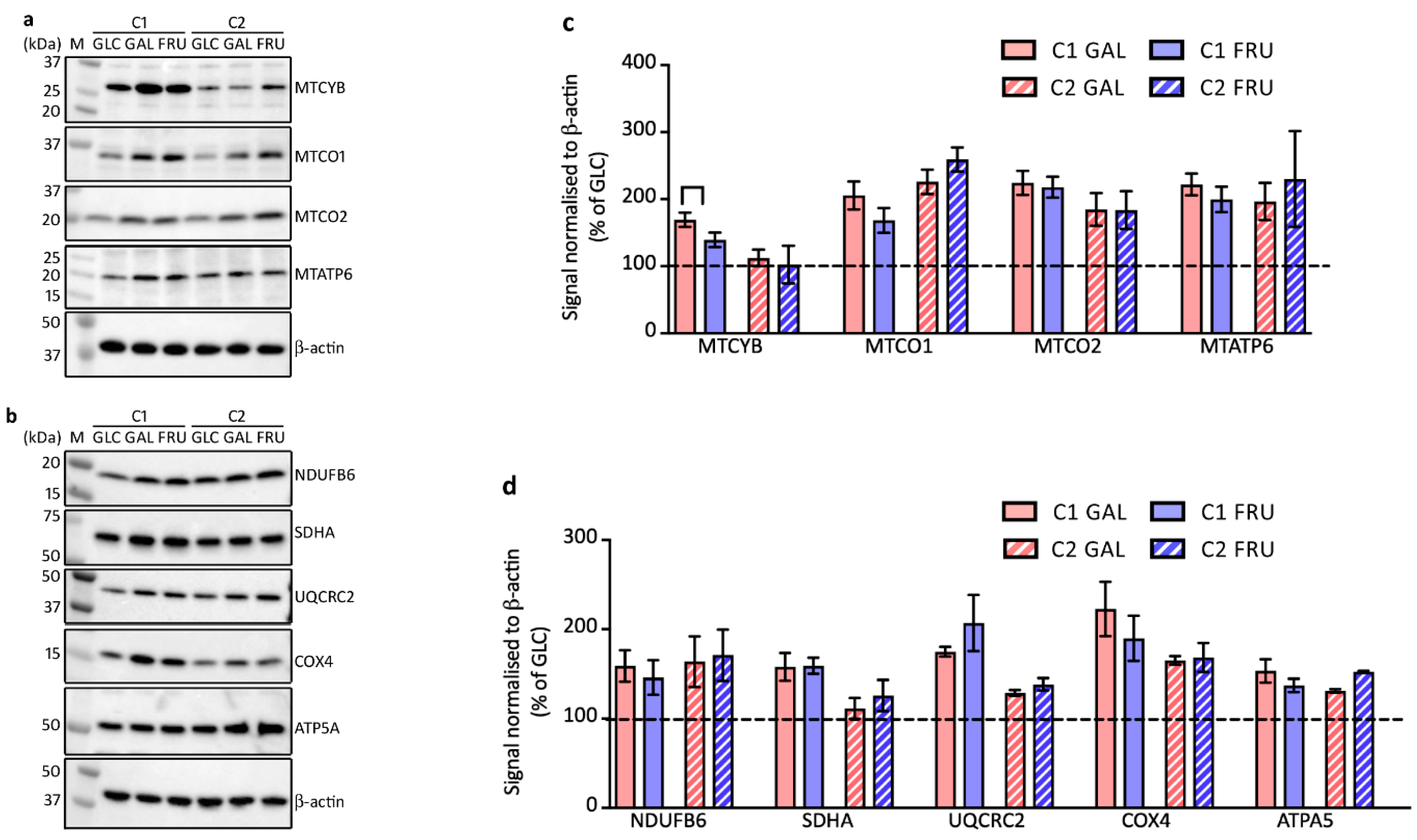
3.7. Levels of OXPHOS Complexes
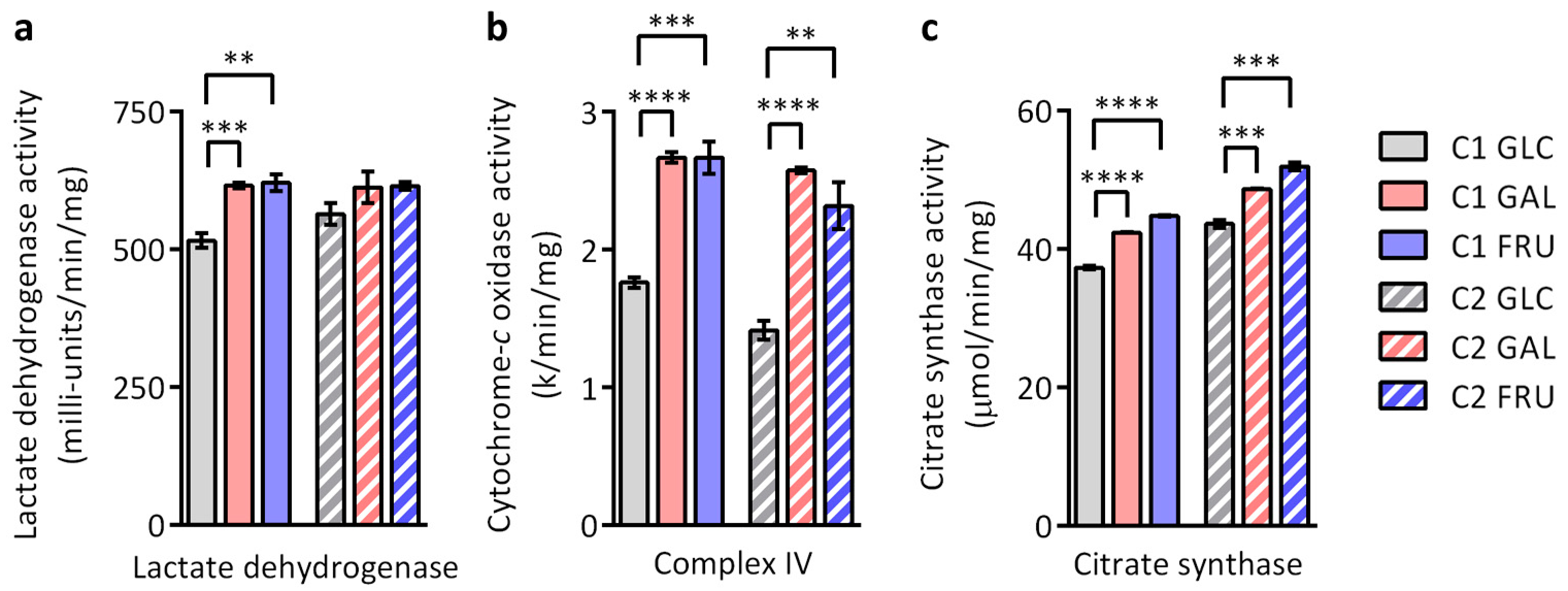
3.8. Activities of Lactate Dehydrogenase, OXPHOS Complex IV, and Citrate Synthase
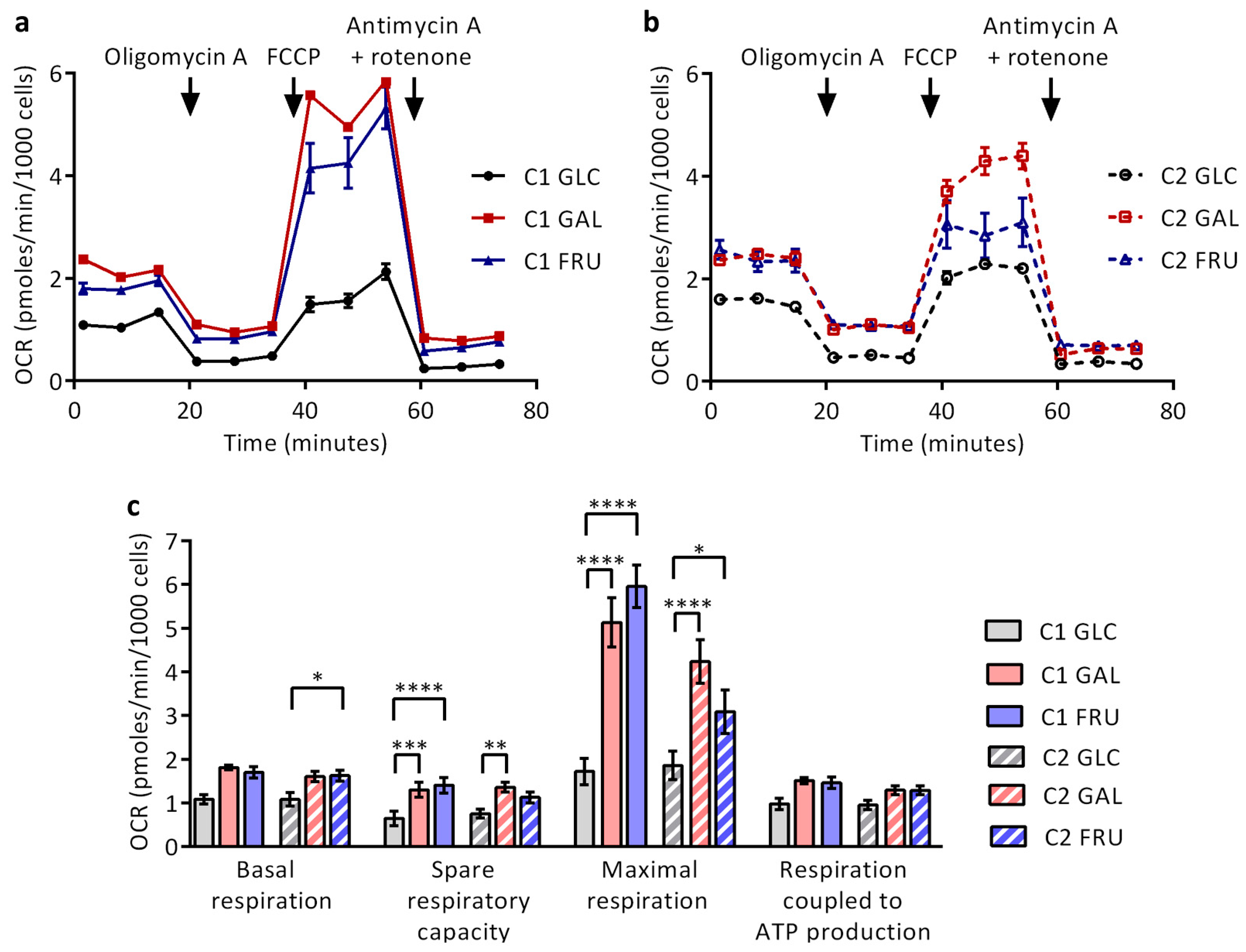
3.9. Rates of Mitochondrial Respiration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vercellino, I.; Sazanov, L.A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat. Rev. Mol. Cell. Biol. 2022, 23, 141–161. [Google Scholar] [CrossRef] [PubMed]
- Ibsen, K.H. The Crabtree effect: A review. Cancer Res. 1961, 21, 829–841. [Google Scholar]
- Suomalainen, A.; Battersby, B.J. Mitochondrial diseases: The contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell. Biol. 2018, 19, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, M.; Scheffler, I.E. Energy metabolism in respiration-deficient and wild type Chinese hamster fibroblasts in culture. J. Cell. Physiol. 1976, 89, 39–52. [Google Scholar] [CrossRef]
- Hayashi, J.I.; Ohta, S.; Kikuchi, A.; Takemitsu, M.; Goto, Y.I.; Nonaka, I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 1991, 88, 10614–10618. [Google Scholar] [CrossRef]
- Robinson, B.H.; Petrova-Benedict, R.; Buncic, J.R.; Wallace, D.C. Nonviability of Cells with Oxidative Defects in Galactose Medium: A Screening Test for Affected Patient Fibroblasts. Biochem. Med. Metab. Biol. 1992, 48, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Perli, E.; Giordano, C.; Tuppen, H.A.L.; Montopoli, M.; Montanari, A.; Orlandi, M.; Pisano, A.; Catanzaro, D.; Caparrotta, L.; Musumeci, B.; et al. Isoleucyl-tRNA synthetase levels modulate the penetrance of a homoplasmic m.4277T>C mitochondrial tRNA Ile lutation causing hypertrophic cardiomyopathy. Hum. Mol. Genet. 2012, 21, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Hofhaus, G.; Johns, D.R.; Hurko, O.; Attardi, G.; Chomyn, A. Respiration and growth defects in transmitochondrial cell lines carrying the 11778 mutation associated with Leber’s hereditary optic neuropathy. J. Biol. Chem. 1996, 271, 13155–13161. [Google Scholar] [CrossRef]
- Mortiboys, H.; Thomas, K.J.; Koopman, W.J.H.; Klaffke, S.; Abou-Sleiman, P.; Olpin, S.; Wood, N.W.; Willems, P.H.G.M.; Smeitink, J.A.M.; Cookson, M.R.; et al. Mitochondrial function and morphology are impaired in Parkin-mutant fibroblasts. Ann. Neurol. 2008, 64, 555–565. [Google Scholar] [CrossRef]
- Pitceathly, R.D.S.; Rahman, S.; Wedatilake, Y.; Polke, J.M.; Cirak, S.; Foley, A.R.; Sailer, A.; Hurles, M.E.; Stalker, J.; Hargreaves, I.; et al. NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Rep. 2013, 3, 1795–1805. [Google Scholar] [CrossRef]
- Reyes, A.; Melchionda, L.; Nasca, A.; Carrara, F.; Lamantea, E.; Zanolini, A.; Lamperti, C.; Fang, M.; Zhang, J.; Ronchi, D.; et al. RNASEH1 mutations impair mtDNA replication and cause adult-onset mitochondrial encephalomyopathy. Am. J. Hum. Genet. 2015, 97, 186–193. [Google Scholar] [CrossRef]
- Dykens, J.A.; Jamieson, J.; Marroquin, L.; Nadanaciva, S.; Billis, P.A.; Will, Y. Biguanide-induced mitochondrial dysfunction yields increased lactate production and cytotoxicity of aerobically-poised HepG2 cells and human hepatocytes in vitro. Toxicol. Appl. Pharmacol. 2008, 233, 203–210. [Google Scholar] [CrossRef]
- Gohil, V.M.; Sheth, S.A.; Nilsson, R.; Wojtovich, A.P.; Lee, J.H.; Perocchi, F.; Chen, W.; Clish, C.B.; Ayata, C.; Brookes, P.S.; et al. Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat. Biotechnol. 2010, 28, 249–255. [Google Scholar] [CrossRef]
- Carbó, J.M.; Cornet-Masana, J.M.; Cuesta-Casanovas, L.; Delgado-Martínez, J.; Banús-Mulet, A.; Clément-Demange, L.; Serra, C.; Catena, J.; Llebaria, A.; Esteve, J.; et al. A novel family of lysosomotropic tetracyclic compounds for treating leukemia. Cancers 2023, 15, 1912. [Google Scholar] [CrossRef]
- Marroquin, L.D.; Hynes, J.; Dykens, J.A.; Jamieson, J.D.; Will, Y. Circumventing the Crabtree effect: Replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol. Sci. 2007, 97, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Holden, H.M.; Rayment, I.; Thoden, J.B. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J. Biol. Chem. 2003, 278, 43885–43888. [Google Scholar] [CrossRef]
- Lyssiotis, C.A.; Cantley, L.C. F stands for fructose and fat. Nature 2013, 502, 181–182. [Google Scholar] [CrossRef] [PubMed]
- Reitzer, L.J.; Wice, B.M.; Kennel, D. Evidence That Glutamine, Not Sugar, Is the Major Energy Source for Cultured HeLa Cells. J. Biol. Chem. 1979, 254, 2669–2676. [Google Scholar] [CrossRef] [PubMed]
- Reitzer, L.J.; Wice, B.M.; Kennell, D. The pentose cycle. Control and essential function in HeLa cell nucleic acid synthesis. J. Biol. Chem. 1980, 255, 5616–5626. [Google Scholar] [CrossRef]
- Zielke, H.R.; Zielke, C.L.; Ozand, P.T. Glutamine: A major energy source for cultured mammalian cells. Fed. Proc. 1984, 43, 121–125. [Google Scholar]
- Ghesquière, B.; Wong, B.W.; Kuchnio, A.; Carmeliet, P. Metabolism of stromal and immune cells in health and disease. Nature 2014, 511, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, R.; Gilkerson, R.; Aggeler, R.; Yamagata, K.; Remington, S.J.; Capaldi, R.A. Energy Substrate Modulates Mitochondrial Structure and Oxidative Capacity in Cancer Cells. Cancer Res. 2004, 64, 985–993. [Google Scholar] [CrossRef]
- Palmfeldt, J.; Vang, S.; Stenbroen, V.; Pedersen, C.B.; Christensen, J.H.; Bross, P.; Gregersen, N. Mitochondrial proteomics on human fibroblasts for identification of metabolic imbalance and cellular stress. Proteome Sci. 2009, 7, 20. [Google Scholar] [CrossRef]
- Smolková, K.; Bellance, N.; Scandurra, F.; Génot, E.; Gnaiger, E.; Plecitá-Hlavatá, L.; Ježek, P.; Rossignol, R. Mitochondrial bioenergetic adaptations of breast cancer cells to aglycemia and hypoxia. J. Bioenerg. Biomembr. 2010, 42, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Aguer, C.; Gambarotta, D.; Mailloux, R.J.; Moffat, C.; Dent, R.; McPherson, R.; Harper, M.-E. Galactose enhances oxidative metabolism and reveals mitochondrial dysfunction in human primary muscle cells. PLoS ONE 2011, 6, e28536. [Google Scholar] [CrossRef]
- Kase, E.T.; Nikolić, N.; Bakke, S.S.; Bogen, K.K.; Aas, V.; Thoresen, G.H.; Rustan, A.C. Remodeling of oxidative energy metabolism by galactose improves glucose handling and metabolic switching in human skeletal muscle cells. PLoS ONE 2013, 8, e59972. [Google Scholar] [CrossRef]
- Elkalaf, M.; Anděl, M.; Trnka, J. Low glucose but not galactose enhances oxidative mitochondrial metabolism in C2C12 myoblasts and myotubes. PLoS ONE 2013, 8, e70772. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Huang, D.; McArthur, D.L.; Boros, L.G.; Nissen, N.; Heaney, A.P. Fructose induces transketolase flux to promote pancreatic cancer growth. Cancer Res. 2010, 70, 6368–6376. [Google Scholar] [CrossRef]
- Mulukutla, B.C.; Khan, S.; Lange, A.; Hu, W.S. Glucose metabolism in mammalian cell culture: New insights for tweaking vintage pathways. Trends Biotechnol. 2010, 28, 476–484. [Google Scholar] [CrossRef]
- Taanman, J.-W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta Bioenerg. 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Park, I.H.; Lerou, P.H.; Zhao, R.; Huo, H.; Daley, G.Q. Generation of human-induced pluripotent stem cells. Nat. Protoc. 2008, 3, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Cantanhede, G.I.; Liu, H.; Liu, H.; Balbuena Rodriguez, V.; Shiwen, X.; Ong, V.H.; Denton, C.P.; Ponticos, M.; Xiong, G.; Lima-Filho, J.L.; et al. Exploring metabolism in scleroderma reveals opportunities for pharmacological intervention for therapy in fibrosis. Front. Immunol. 2022, 13, 1004949. [Google Scholar] [CrossRef]
- Bai, R.-K.; Wong, L.-J.C. Simultaneous detection and quantification of mitochondrial DNA deletion(s), depletion, and over-replication in patients with mitochondrial disease. J. Mol. Diagn 2005, 7, 613–622. [Google Scholar] [CrossRef]
- Bradshaw, A.V.; Campbell, P.; Schapira, A.H.V.; Morris, H.R.; Taanman, J.W. The PINK1-Parkin mitophagy signalling pathway is not functional in peripheral blood mononuclear cells. PLoS ONE 2021, 16, e0259903. [Google Scholar] [CrossRef]
- Protasoni, M.; Kroon, A.M.; Taanman, J.-W. Mitochondria as oncotarget: A comparison between the tetracycline analogs doxycycline and COL-3. Oncotarget 2018, 9, 33818–33831. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H. Quantification of oxidative phosphorylation enzymes after blue native electrophoresis and two-dimensional resolution: Normal Complex I protein amounts in Parkinson’s disease conflict with reduced catalytic activities. Electrophoresis 1995, 16, 763–770. [Google Scholar] [CrossRef]
- Capaldi, R.A.; Marusich, M.F.; Taanman, J.-W. Mammalian cytochrome-c oxidase: Characterization of enzyme and immunological detection of subunits in tissue extracts and whole cells. Meth. Enzymol. 1995, 260, 117–132. [Google Scholar]
- Montoya, J.; Gaines, G.L.; Attardi, G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell 1983, 34, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Cho, J.; Cesare, A.J.; Griffith, J.D.; Attardi, G. Termination factor-mediated DNA Loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell 2005, 123, 1227–1240. [Google Scholar] [CrossRef]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef]
- Kelly, D.P.; Scarpulla, R.C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes. Dev. 2004, 18, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Ventura-Clapier, R.; Garnier, A.; Veksler, V. Transcriptional control of mitochondrial biogenesis: The central role of PGC-1α. Cardiovasc. Res. 2008, 79, 208–217. [Google Scholar] [CrossRef]
- Smith, T.A. Mammalian hexokinases and their abnormal expression in cancer. Br. J. Biomed. Sci. 2000, 57, 170–178. [Google Scholar]
- Barber, R.D.; Harmer, D.W.; Coleman, R.A.; Clark, B.J. GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genomics 2005, 21, 389–395. [Google Scholar] [CrossRef]
- Hitosugi, T.; Kang, S.; Vander Heiden, M.G.; Chung, T.-W.; Elf, S.; Lythgoe, K.; Dong, S.; Lonial, S.; Wang, X.; Chen, G.Z.; et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2009, 2, ra73. [Google Scholar] [CrossRef]
- Wang, H.-J.; Hsieh, Y.-J.; Cheng, W.-C.; Lin, C.-P.; Lin, Y.; Yang, S.-F.; Chen, C.-C.; Izumiya, Y.; Yu, J.-S.; Kung, H.-J.; et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α–mediated glucose metabolism. Proc. Natl. Acad. Sci. USA 2014, 111, 279–284. [Google Scholar] [CrossRef]
- Yang, Y.-C.; Chien, M.-H.; Liu, H.-Y.; Chang, Y.-C.; Chen, C.-K.; Lee, W.-J.; Kuo, T.-C.; Hsiao, M.; Hua, K.-T.; Cheng, T.-Y. Nuclear translocation of PKM2/AMPK complex sustains cancer stem cell populations under glucose restriction stress. Cancer Lett. 2018, 421, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Seifert, F.; Ciszak, E.; Korotchkina, L.; Golbik, R.; Spinka, M.; Dominiak, P.; Sidhu, S.; Brauer, J.; Patel, M.S.; Tittmann, K. Phosphorylation of serine 264 impedes active site accessibility in the E1 component of the human pyruvate dehydrogenase multienzyme complex. Biochemistry 2007, 46, 6277–6287. [Google Scholar] [CrossRef]
- Fan, J.; Kang, H.-B.; Shan, C.; Elf, S.; Lin, R.; Xie, J.; Gu, T.-L.; Aguiar, M.; Lonning, S.; Chung, T.-W.; et al. Tyr-301 phosphorylation inhibits pyruvate dehydrogenase by blocking substrate binding and promotes the Warburg effect. J. Biol. Chem. 2014, 289, 26533–26541. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Huang, L.; Whelchel, A.; Yuan, T.; Ma, X.; Cheng, R.; Takahashi, Y.; Karamichos, D.; Ma, J.-X. Peroxisome proliferator-activated receptor-α (PPARα) regulates wound healing and mitochondrial metabolism in the cornea. Proc. Natl. Acad. Sci. USA 2023, 120, e2217576120. [Google Scholar] [CrossRef]
- Gu, J.; Wu, M.; Guo, R.; Yan, K.; Lei, J.; Gao, N.; Yang, M. The Architecture of the mammalian respirasome. Nature 2016, 537, 639–643. [Google Scholar] [CrossRef]
- Hood, D.A.; Zak, R.; Pette, D. Chronic stimulation of rat skeletal muscle induces coordinate increases in mitochondrial and nuclear mRNAs of cytochrome-c-oxidase subunits. Eur. J. Biochem. 1989, 179, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Renner, K.; Amberger, A.; Konwalinka, G.; Kofler, R.; Gnaiger, E. Changes of Mitochondrial Respiration, Mitochondrial content and cell size after induction of apoptosis in leukemia cells. Biochim. Biophys. Acta Mol. Cell Res. 2003, 1642, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Larsen, S.; Nielsen, J.; Hansen, C.N.; Nielsen, L.B.; Wibrand, F.; Stride, N.; Schroder, H.D.; Boushel, R.; Helge, J.W.; Dela, F.; et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J. Physiol. 2012, 590, 3349–3360. [Google Scholar] [CrossRef] [PubMed]
- Dott, W.; Mistry, P.; Wright, J.; Cain, K.; Herbert, K.E. Modulation of mitochondrial bioenergetics in a skeletal muscle cell line model of mitochondrial toxicity. Redox Biol. 2014, 2, 224–233. [Google Scholar] [CrossRef] [PubMed]

| Cell Culture | Glucose | Galactose | Fructose |
|---|---|---|---|
| C1 (adult fibroblasts) | 37.2 ± 1.3 h | 122.0 ± 6.0 h | 40.8 ± 1.9 h |
| C2 (neonatal fibroblasts) | 25.3 ± 1.1 h | 66.2 ± 3.2 h | 31.6 ± 1.4 h |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Protasoni, M.; Taanman, J.-W. Remodelling of the Mitochondrial Bioenergetic Pathways in Human Cultured Fibroblasts with Carbohydrates. Biology 2023, 12, 1002. https://doi.org/10.3390/biology12071002
Protasoni M, Taanman J-W. Remodelling of the Mitochondrial Bioenergetic Pathways in Human Cultured Fibroblasts with Carbohydrates. Biology. 2023; 12(7):1002. https://doi.org/10.3390/biology12071002
Chicago/Turabian StyleProtasoni, Margherita, and Jan-Willem Taanman. 2023. "Remodelling of the Mitochondrial Bioenergetic Pathways in Human Cultured Fibroblasts with Carbohydrates" Biology 12, no. 7: 1002. https://doi.org/10.3390/biology12071002
APA StyleProtasoni, M., & Taanman, J.-W. (2023). Remodelling of the Mitochondrial Bioenergetic Pathways in Human Cultured Fibroblasts with Carbohydrates. Biology, 12(7), 1002. https://doi.org/10.3390/biology12071002







