Comparative Genomic Analysis of Warthog and Sus Scrofa Identifies Adaptive Genes Associated with African Swine Fever
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation for Genome Assembly
2.2. Sample Preparation for Demographic Histories
2.3. DNA and RNA Extraction and Sequencing
2.4. Reads Alignment to the Reference Genome Sus Scrofa 11.1
2.5. Genome Assembly
2.6. Evaluation of Genome Assembly
2.7. Read Alignment and Variant Calling of DNA Sequence Reads
2.8. Annotation of Assembled Genomes
2.9. Annotation of Merged Genomes
2.10. Non-Coding Genes and Coding Gene Prediction
2.11. Function Annotation
2.12. The Identification of Presence–Absence Variation (PAV)
2.13. Phylogeny Construction and Estimate of Divergence Time
2.14. Gene Family Expansion and Contraction
2.15. Phylogenetic Analysis
2.16. Selection Signature Analysis
2.17. Gene Functional Analysis
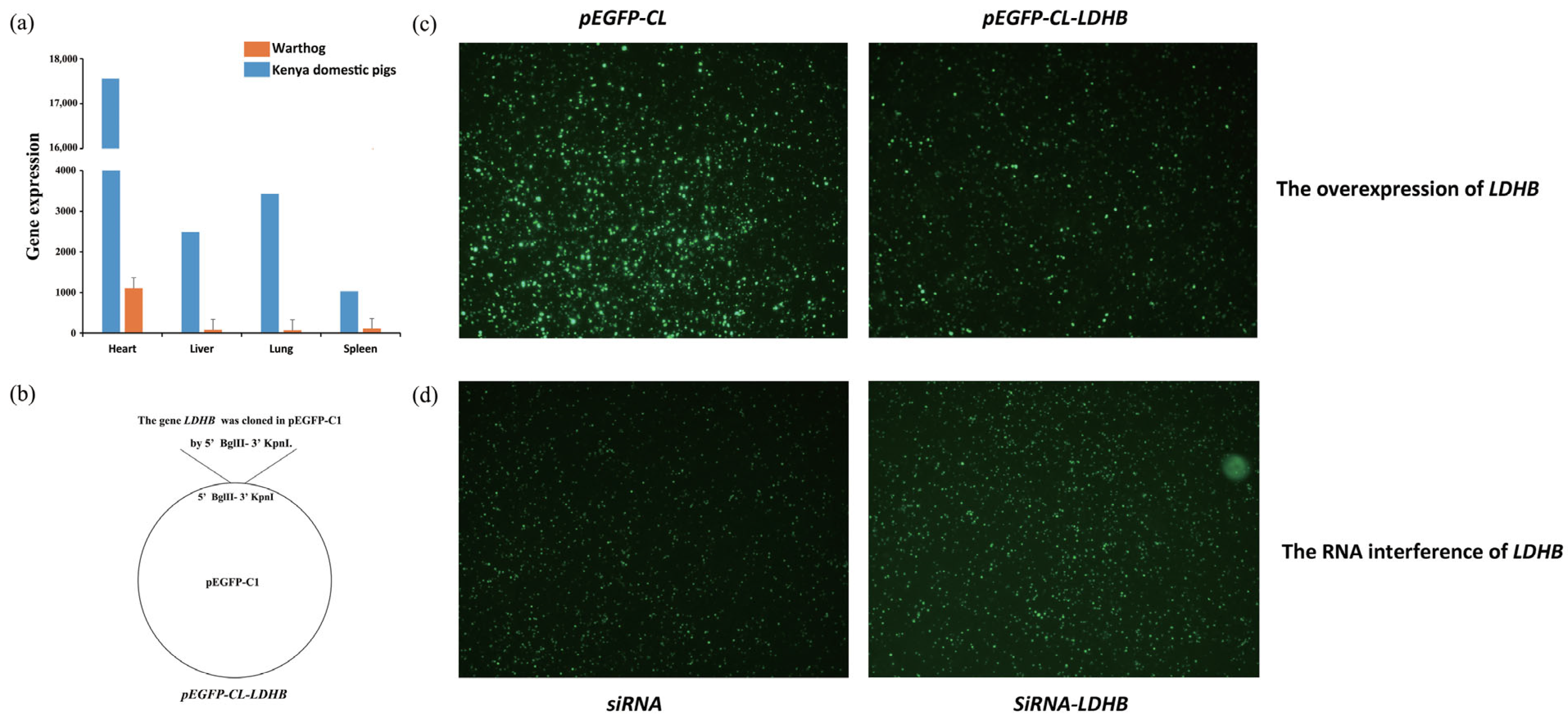
2.18. The Expression of Fluorescent Proteins in 3D4/21 Cells after Plasmid Transfection
3. Results
3.1. De Novo Assembly of Warthog and the Kenyan Domestic Pig Genomes
3.2. Annotation of the Assembled Genomes
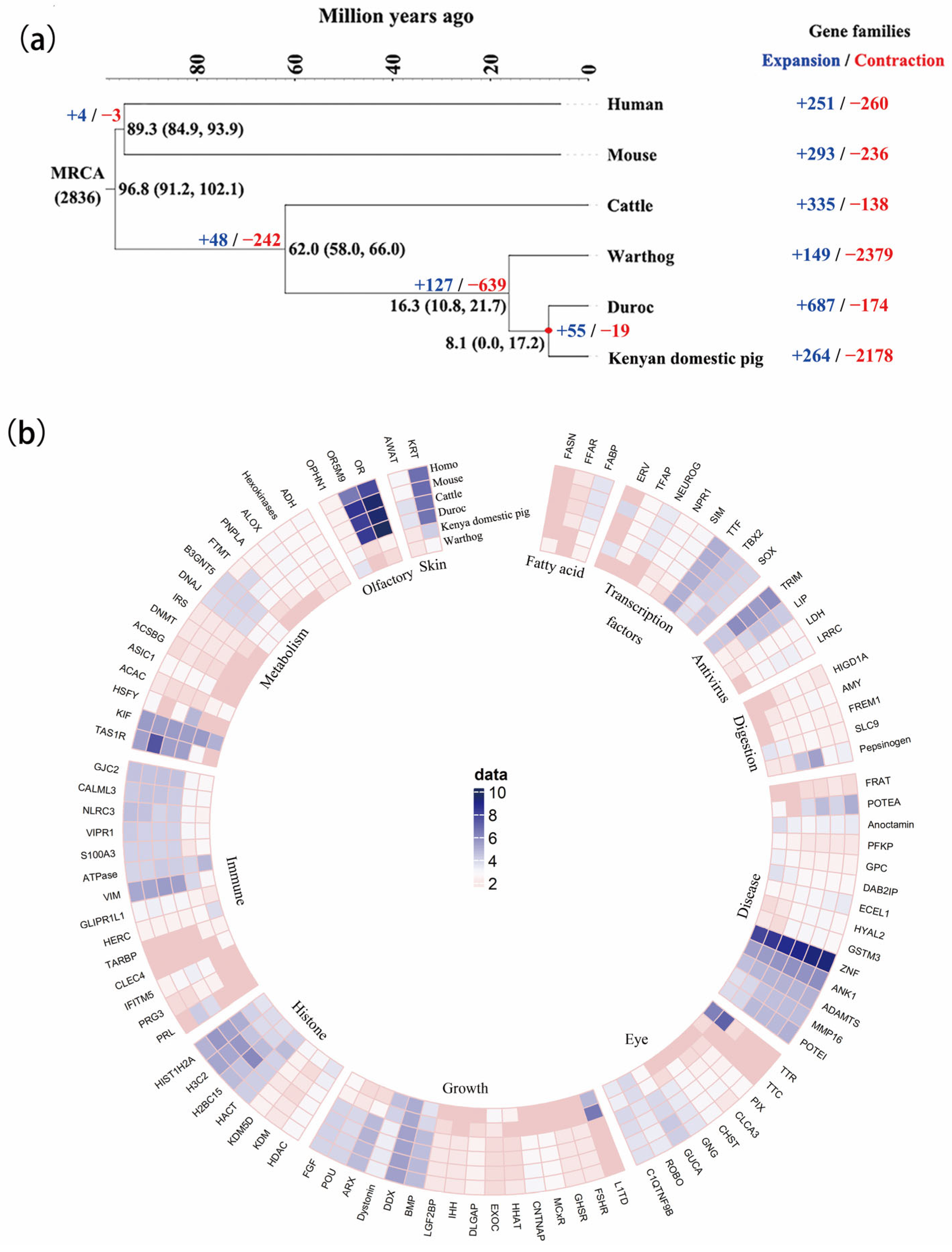
3.3. The Expansion and Contraction of Gene Family Evolution
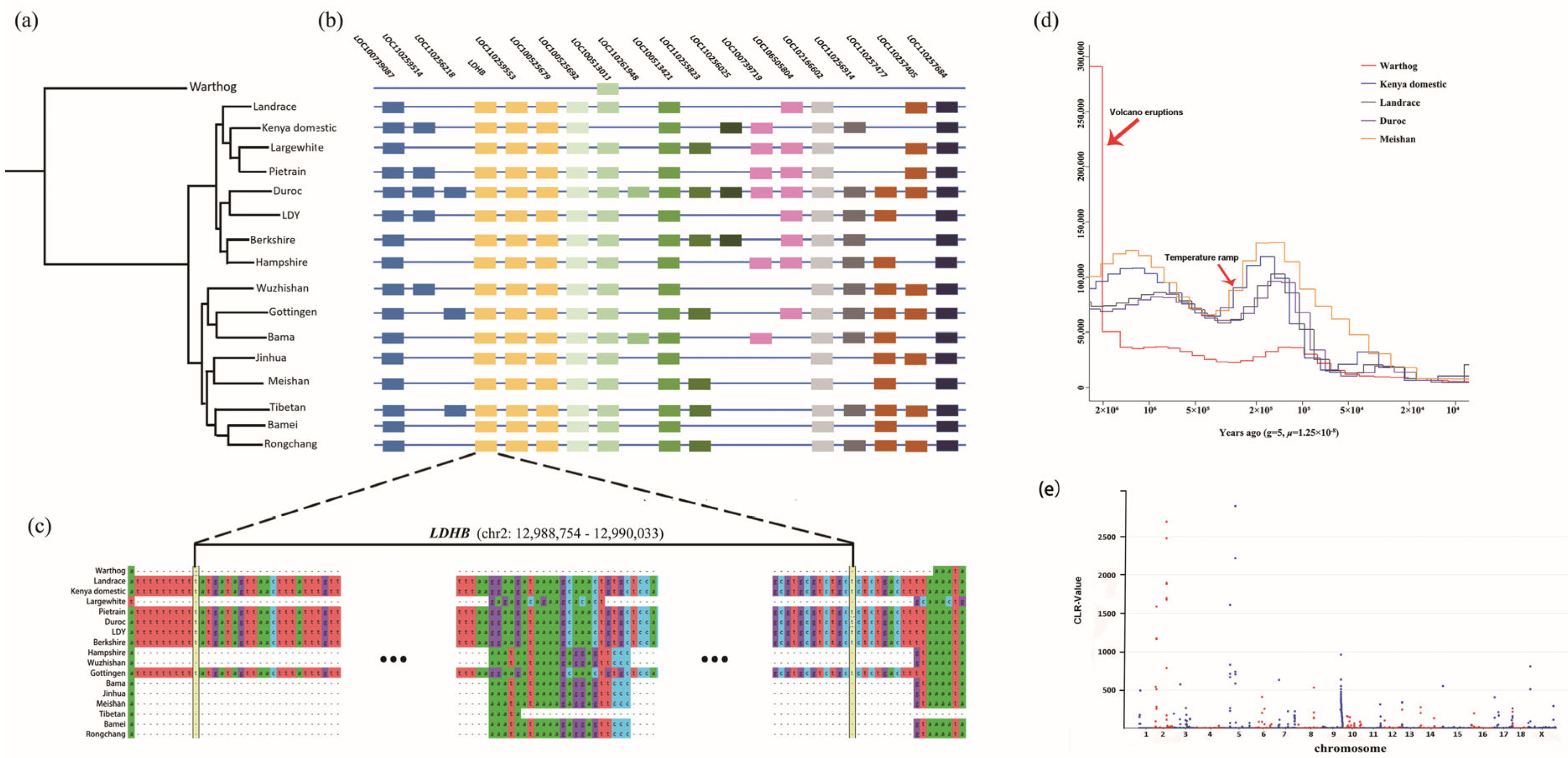
3.4. Phylogenic Analysis and Demographic History
3.5. Signature Analysis of Selection
3.6. Massive Presence-Absence Sequences among the Suidae Genomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frantz, L.; Meijaard, E.; Gongora, J.; Haile, J.; Groenen, M.A.; Larson, G. The Evolution of Suidae. Annu. Rev. Anim. Biosci. 2016, 4, 61–85. [Google Scholar] [CrossRef] [PubMed]
- Heuschele, W.P.; Coggins, L. Epizootiology of African swine fever virus in warthogs. Bull. Epizoot. Dis. Afr. 1969, 17, 179–183. [Google Scholar] [PubMed]
- Dixon, L.; Escribano, J.; Martins, C.; Rock, D.; Salas, M.; Wilkinson, P. Asfarviridae in Virus Taxonomy; Viiith Report of the ICTV; Elsevier: London, UK, 2005. [Google Scholar]
- Eustace Montgomery, R. On A Form of Swine Fever Occurring in British East Africa (Kenya Colony). J. Comp. Pathol. Ther. 1921, 34, 159–191. [Google Scholar] [CrossRef]
- Zhou, X.; Li, N.; Luo, Y.; Liu, Y.; Miao, F.; Chen, T.; Zhang, S.; Cao, P.; Li, X.; Tian, K.; et al. Emergence of African Swine Fever in China, 2018. Transbound. Emerg. Dis. 2018, 65, 1482–1484. [Google Scholar] [CrossRef] [PubMed]
- Frezal, C.; Gay, S.H.; Nenert, C. The Impact of the African Swine Fever Outbreak in China on Global Agricultural Markets; OECD: Paris, France, 2021. [Google Scholar] [CrossRef]
- Ewers, R.M.; Nathan, S.; Lee, P.A.K. African swine fever ravaging Borneo’s wild pigs. Nature 2021, 593, 37. [Google Scholar] [CrossRef]
- Chen, W.; Zhao, D.; He, X.; Liu, R.; Wang, Z.; Zhang, X.; Li, F.; Shan, D.; Chen, H.; Zhang, J.; et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci. 2020, 63, 623–634. [Google Scholar] [CrossRef]
- Urbano, A.C.; Ferreira, F. African swine fever control and prevention: An update on vaccine development. Emerg. Microbes Infect. 2022, 11, 2021–2033. [Google Scholar] [CrossRef]
- Xie, H.B.; Yan, C.; Adeola, A.C.; Wang, K.; Huang, C.P.; Xu, M.M.; Qiu, Q.; Yin, X.; Fan, C.Y.; Ma, Y.F.; et al. African Suid Genomes Provide Insights into the Local Adaptation to Diverse African Environments. Mol. Biol. Evol. 2022, 39, msac256. [Google Scholar] [CrossRef]
- Brian, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. In Proceedings of the Conference 9th Annual Genomics of Energy & Environment Meeting, Walnut Creek, CA, USA, 17–20 March 2014. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 8 January 2019).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Paulino, D.; Warren, R.L.; Vandervalk, B.P.; Raymond, A.; Jackman, S.D.; Birol, I. Sealer: A scalable gap-closing application for finishing draft genomes. BMC Bioinform. 2015, 16, 230. [Google Scholar] [CrossRef]
- Xu, G.-C.; Xu, T.-J.; Zhu, R.; Zhang, Y.; Li, S.-Q.; Wang, H.-W.; Li, J.-T. LR_Gapcloser: A tiling path-based gap closer that uses long reads to complete genome assembly. GigaScience 2019, 8, giy157. [Google Scholar] [CrossRef]
- Xu, M.; Guo, L.; Gu, S.; Wang, O.; Zhang, R.; Peters, B.A.; Fan, G.; Liu, X.; Xu, X.; Deng, L.; et al. TGS-GapCloser: A fast and accurate gap closer for large genomes with low coverage of error-prone long reads. GigaScience 2020, 9, giaa094. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Saha, S.; Bridges, S.; Magbanua, Z.V.; Peterson, D.G. Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 2008, 36, 2284–2294. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Bergman, C.M.; Quesneville, H. Discovering and detecting transposable elements in genome sequences. Brief. Bioinform. 2007, 8, 382–392. [Google Scholar] [CrossRef]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef] [PubMed]
- Lomsadze, A.; Ter-Hovhannisyan, V.; Chernoff, Y.O.; Borodovsky, M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005, 33, 6494–6506. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simao, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef]
- Bruna, T.; Hoff, K.J.; Lomsadze, A.; Stanke, M.; Borodovsky, M. BRAKER2: Automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinform. 2021, 3, lqaa108. [Google Scholar] [CrossRef]
- Gotoh, O. Direct mapping and alignment of protein sequences onto genomic sequence. Bioinformatics 2008, 24, 2438–2444. [Google Scholar] [CrossRef]
- Bruna, T.; Lomsadze, A.; Borodovsky, M. GeneMark-EP+: Eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genom. Bioinform. 2020, 2, lqaa026. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Yang, Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997, 13, 555–556. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef]
- Berlin, K.; Koren, S.; Chin, C.S.; Drake, J.P.; Landolin, J.M.; Phillippy, A.M. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 2015, 33, 623–630. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Schiffels, S.; Durbin, R. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 2014, 46, 919–925. [Google Scholar] [CrossRef]
- DeGiorgio, M.; Huber, C.D.; Hubisz, M.J.; Hellmann, I.; Nielsen, R. SweepFinder2: Increased sensitivity, robustness and flexibility. Bioinformatics 2016, 32, 1895–1897. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Teklue, T.; Wang, T.; Luo, Y.; Hu, R.; Sun, Y.; Qiu, H.J. Generation and Evaluation of an African Swine Fever Virus Mutant with Deletion of the CD2v and UK Genes. Vaccines 2020, 8, 763. [Google Scholar] [CrossRef] [PubMed]
- Borca, M.V.; O’Donnell, V.; Holinka, L.G.; Sanford, B.; Azzinaro, P.A.; Risatti, G.R.; Gladue, D.P. Development of a fluorescent ASFV strain that retains the ability to cause disease in swine. Sci. Rep. 2017, 7, 46747. [Google Scholar] [CrossRef]
- Xu, H.; Ghishan, F.K.; Kiela, P.R. SLC9 Gene Family: Function, Expression, and Regulation. Compr. Physiol. 2018, 8, 555–583. [Google Scholar] [CrossRef]
- Tao, R.J.; Luo, X.L.; Xu, W.; Mao, B.; Dai, R.X.; Li, C.W.; Yu, L.; Gu, F.; Liang, S.; Lu, H.W.; et al. Viral infection in community acquired pneumonia patients with fever: A prospective observational study. J. Thorac. Dis. 2018, 10, 4387–4395. [Google Scholar] [CrossRef]
- Marmol-Sanchez, E.; Ramayo-Caldas, Y.; Quintanilla, R.; Cardoso, T.F.; Gonzalez-Prendes, R.; Tibau, J.; Amills, M. Co-expression network analysis predicts a key role of microRNAs in the adaptation of the porcine skeletal muscle to nutrient supply. J. Anim. Sci. Biotechnol. 2020, 11, 10. [Google Scholar] [CrossRef]
- Kim, K.S.; Seibert, J.T.; Edea, Z.; Graves, K.L.; Kim, E.S.; Keating, A.F.; Baumgard, L.H.; Ross, J.W.; Rothschild, M.F. Characterization of the acute heat stress response in gilts: III. Genome-wide association studies of thermotolerance traits in pigs. J. Anim. Sci. 2018, 96, 2074–2085. [Google Scholar] [CrossRef]
- Fliegauf, M.; Bryant, V.L.; Frede, N.; Slade, C.; Woon, S.-T.; Lehnert, K.; Winzer, S.; Bulashevska, A.; Scerri, T.; Leung, E.; et al. Haploinsufficiency of the NF-κB1 Subunit p50 in Common Variable Immunodeficiency. Am. J. Hum. Genet. 2015, 97, 389–403. [Google Scholar] [CrossRef]
- Fan, W.; Jiao, P.; Zhang, H.; Chen, T.; Zhou, X.; Qi, Y.; Sun, L.; Shang, Y.; Zhu, H.; Hu, R.; et al. Inhibition of African Swine Fever Virus Replication by Porcine Type I and Type II Interferons. Front. Microbiol. 2020, 11, 1203. [Google Scholar] [CrossRef]
- Lee, S.; Ishitsuka, A.; Noguchi, M.; Hirohama, M.; Fujiyasu, Y.; Petric, P.; Schwemmle, M.; Staeheli, P.; Nagata, K.; Kawaguchi, A. Influenza restriction factor MxA functions as inflammasome sensor in the respiratory epithelium. Sci. Immunol. 2019, 4, eaau4643. [Google Scholar] [CrossRef]
- Wang, J.; Rajbhandari, P.; Damianov, A.; Han, A.; Sallam, T.; Waki, H.; Villanueva, C.J.; Lee, S.D.; Nielsen, R.; Mandrup, S.; et al. RNA-binding protein PSPC1 promotes the differentiation-dependent nuclear export of adipocyte RNAs. J. Clin. Investig. 2017, 127, 987–1004. [Google Scholar] [CrossRef]
- Maslin, M.A.; Shultz, S.; Trauth, M.H. A synthesis of the theories and concepts of early human evolution. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140064. [Google Scholar] [CrossRef]
- DeMenocal, P.B. Anthropology. Climate and human evolution. Science 2011, 331, 540–542. [Google Scholar] [CrossRef]
- Chaudhry, S.I.; Hooper, S.; Nye, E.; Williamson, P.; Harrington, K.; Sahai, E. Autocrine IL-1β-TRAF6 signalling promotes squamous cell carcinoma invasion through paracrine TNFα signalling to carcinoma-associated fibroblasts. Oncogene 2013, 32, 747–758. [Google Scholar] [CrossRef]
- Hamilton, A.C.; Taylor, D. History of climate and forests in tropical Africa during the last 8 million years. Clim. Chang. 1991, 19, 65–78. [Google Scholar] [CrossRef]
- Amills, M.; Ramírez, O.; Galman-Omitogun, O.; Clop, A. Domestic Pigs in Africa. Afr. Archaeol. Rev. 2013, 30, 73–82. [Google Scholar] [CrossRef]
- Noce, A.; Amills, M.; Manunza, A.; Muwanika, V.; Muhangi, D.; Aliro, T.; Mayega, J.; Ademun, R.; Sànchez, A.; Egbhalsaied, S.; et al. East African pigs have a complex Indian, Far Eastern and Western ancestry. Anim. Genet. 2015, 46, 433–436. [Google Scholar] [CrossRef]
- Vansina, J. New Linguistic Evidence and ‘the Bantu Expansion’. J. Afr. Hist. 1995, 36, 173–195. [Google Scholar] [CrossRef]
- Reymond, A.; Meroni, G.; Fantozzi, A.; Merla, G.; Cairo, S.; Luzi, L.; Riganelli, D.; Zanaria, E.; Messali, S.; Cainarca, S.; et al. The tripartite motif family identifies cell compartments. EMBO J. 2001, 20, 2140–2151. [Google Scholar] [CrossRef]
- Rajsbaum, R.; Stoye, J.P.; O’Garra, A. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur. J. Immunol. 2008, 38, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Uchil, P.D.; Quinlan, B.D.; Chan, W.T.; Luna, J.M.; Mothes, W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008, 4, e16. [Google Scholar] [CrossRef] [PubMed]
- Uchil, P.D.; Hinz, A.; Siegel, S.; Coenen-Stass, A.; Pertel, T.; Luban, J.; Mothes, W. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J. Virol. 2013, 87, 257–272. [Google Scholar] [CrossRef]
- Revilla, Y.; Callejo, M.; Rodríguez, J.M.; Culebras, E.; Nogal, M.L.; Salas, M.L.; Viñuela, E.; Fresno, M. Inhibition of nuclear factor kappaB activation by a virus-encoded IkappaB-like protein. J. Biol. Chem. 1998, 273, 5405–5411. [Google Scholar] [CrossRef] [PubMed]
- Okoth, E.; Gallardo, C.; Macharia, J.M.; Omore, A.; Pelayo, V.; Bulimo, D.W.; Arias, M.; Kitala, P.; Baboon, K.; Lekolol, I.; et al. Comparison of African swine fever virus prevalence and risk in two contrasting pig-farming systems in South-west and Central Kenya. Prev. Vet. Med. 2013, 110, 198–205. [Google Scholar] [CrossRef]
- Mujibi, F.D.; Okoth, E.; Cheruiyot, E.K.; Onzere, C.; Bishop, R.P.; Fevre, E.M.; Thomas, L.; Masembe, C.; Plastow, G.; Rothschild, M. Genetic diversity, breed composition and admixture of Kenyan domestic pigs. PLoS ONE 2018, 13, e0190080. [Google Scholar] [CrossRef]
- Fan, S.; Wu, K.; Zhao, M.; Yuan, J.; Ma, S.; Zhu, E.; Chen, Y.; Ding, H.; Yi, L.; Chen, J. LDHB inhibition induces mitophagy and facilitates the progression of CSFV infection. Autophagy 2021, 17, 2305–2324. [Google Scholar] [CrossRef]
- Decking, S.M.; Bruss, C.; Babl, N.; Bittner, S.; Klobuch, S.; Thomas, S.; Feuerer, M.; Hoffmann, P.; Dettmer, K.; Oefner, P.J.; et al. LDHB Overexpression Can Partially Overcome T Cell Inhibition by Lactic Acid. Int. J. Mol. Sci. 2022, 23, 5970. [Google Scholar] [CrossRef]
- Zdralevic, M.; Brand, A.; Di Ianni, L.; Dettmer, K.; Reinders, J.; Singer, K.; Peter, K.; Schnell, A.; Bruss, C.; Decking, S.M.; et al. Double genetic disruption of lactate dehydrogenases A and B is required to ablate the “Warburg effect” restricting tumor growth to oxidative metabolism. J. Biol. Chem. 2018, 293, 15947–15961. [Google Scholar] [CrossRef]
- Santos, A.A.; Penha, H.A.; Bellec, A.; Munhoz, C.d.F.; Pedrosa-Harand, A.; Bergès, H.; Vieira, M.L.C. Begin at the beginning: A BAC-end view of the passion fruit (Passiflora) genome. BMC Genom. 2014, 15, 816. [Google Scholar] [CrossRef]
- Chen, B.; Zhou, Z.; Ke, Q.; Wu, Y.; Bai, H.; Pu, F.; Xu, P. The sequencing and de novo assembly of the Larimichthys crocea genome using PacBio and Hi-C technologies. Sci. Data 2019, 6, 188. [Google Scholar] [CrossRef]
- Zhao, P.; Li, J.; Kang, H.; Wang, H.; Fan, Z.; Yin, Z.; Wang, J.; Zhang, Q.; Wang, Z.; Liu, J.-F. Structural Variant Detection by Large-scale Sequencing Reveals New Evolutionary Evidence on Breed Divergence between Chinese and European Pigs. Sci. Rep. 2016, 6, 18501. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Zhao, P.; Yu, Y.; Feng, W.; Du, H.; Yu, J.; Kang, H.; Zheng, X.; Wang, Z.; Liu, G.E.; Ernst, C.W.; et al. Evidence of evolutionary history and selective sweeps in the genome of Meishan pig reveals its genetic and phenotypic characterization. Gigascience 2018, 7, giy058. [Google Scholar] [CrossRef]
- Zhao, Z.; Fu, Y.X.; Hewett-Emmett, D.; Boerwinkle, E. Investigating single nucleotide polymorphism (SNP) density in the human genome and its implications for molecular evolution. Gene 2003, 312, 207–213. [Google Scholar] [CrossRef]
| The Number of Raw Base (bp) | The Number of Raw Reads | Coverage Depth | Read Length | |
|---|---|---|---|---|
| Warthog | 180,321,392,700 | 1,202,142,618 | 69× | 150 bp |
| Kenya domestic pig | 176,972,667,300 | 1,179,817,782 | 68× | 150 bp |
| Warthog | Kenya Domestic | |
|---|---|---|
| Assembled genome size (Gb) | 2.417 | 2.445 |
| number of ‘N’ (Mb) | 40.3 | 21.8 |
| N content of whole genome (%) | 1.64 | 0.883 |
| Contig N50 (kb) | 132.54 | 144.01 |
| Number of scaffold | 19,366 | 13,380 |
| Total scaffolds (>=1 Mb) | 275 | 138 |
| Total scaffolds (>=100 kb) | 493 | 262 |
| Total scaffolds (>=10 kb) | 2479 | 2253 |
| Total scaffolds (>=1 kb) | 20,477 | 14,837 |
| Scaffold N50 (Mb) | 13.75 | 30.52 |
| Scaffold N75 (Mb) | 6.1 | 15.47 |
| Scaffold N90 (Mb) | 1.6 | 5.24 |
| Average scaffold length (kb) | 120.017 | 166.2 |
| Longest scaffold (Mb) | 65.64 | 100 |
| Breeds | Project NO. | Breeds | Project NO. |
|---|---|---|---|
| Warthog | PRJNA691462 | Duroc (Sus scrofa11.1) | PRJNA13421 |
| Kenya domestic | PRJNA691462 | ½ Landrace- ¼ Duroc- ¼ Yorkshire (LDY) | PRJNA392765 |
| Bama | PRJNA478804 | Ellegaard Gottingen minipig | PRJNA176189 |
| Wuzhishan | PRJNA144099 | Tibetan | PRJNA186497 |
| Jinhua | PRJNA309108 | Hampshire | PRJNA309108 |
| Bamei | PRJNA309108 | Landrace | PRJNA309108 |
| Meishan | PRJNA309108 | LargeWhite | PRJNA309108 |
| Pietrain | PRJNA309108 | Berkshire | PRJNA309108 |
| Rongchang | PRJNA309108 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, W.; Zhou, L.; Zhao, P.; Du, H.; Diao, C.; Zhang, Y.; Liu, Z.; Jin, W.; Yu, J.; Han, J.; et al. Comparative Genomic Analysis of Warthog and Sus Scrofa Identifies Adaptive Genes Associated with African Swine Fever. Biology 2023, 12, 1001. https://doi.org/10.3390/biology12071001
Feng W, Zhou L, Zhao P, Du H, Diao C, Zhang Y, Liu Z, Jin W, Yu J, Han J, et al. Comparative Genomic Analysis of Warthog and Sus Scrofa Identifies Adaptive Genes Associated with African Swine Fever. Biology. 2023; 12(7):1001. https://doi.org/10.3390/biology12071001
Chicago/Turabian StyleFeng, Wen, Lei Zhou, Pengju Zhao, Heng Du, Chenguang Diao, Yu Zhang, Zhen Liu, Wenjiao Jin, Jian Yu, Jianlin Han, and et al. 2023. "Comparative Genomic Analysis of Warthog and Sus Scrofa Identifies Adaptive Genes Associated with African Swine Fever" Biology 12, no. 7: 1001. https://doi.org/10.3390/biology12071001
APA StyleFeng, W., Zhou, L., Zhao, P., Du, H., Diao, C., Zhang, Y., Liu, Z., Jin, W., Yu, J., Han, J., Okoth, E., Mrode, R., & Liu, J.-F. (2023). Comparative Genomic Analysis of Warthog and Sus Scrofa Identifies Adaptive Genes Associated with African Swine Fever. Biology, 12(7), 1001. https://doi.org/10.3390/biology12071001






