New Insights into Oxidative and Reductive Stress Responses and Their Relation to the Anticancer Activity of Selenium-Containing Compounds as Hydrogen Selenide Donors
Abstract
Simple Summary
Abstract
1. Introduction
2. Redox Network in Mammalian Cells
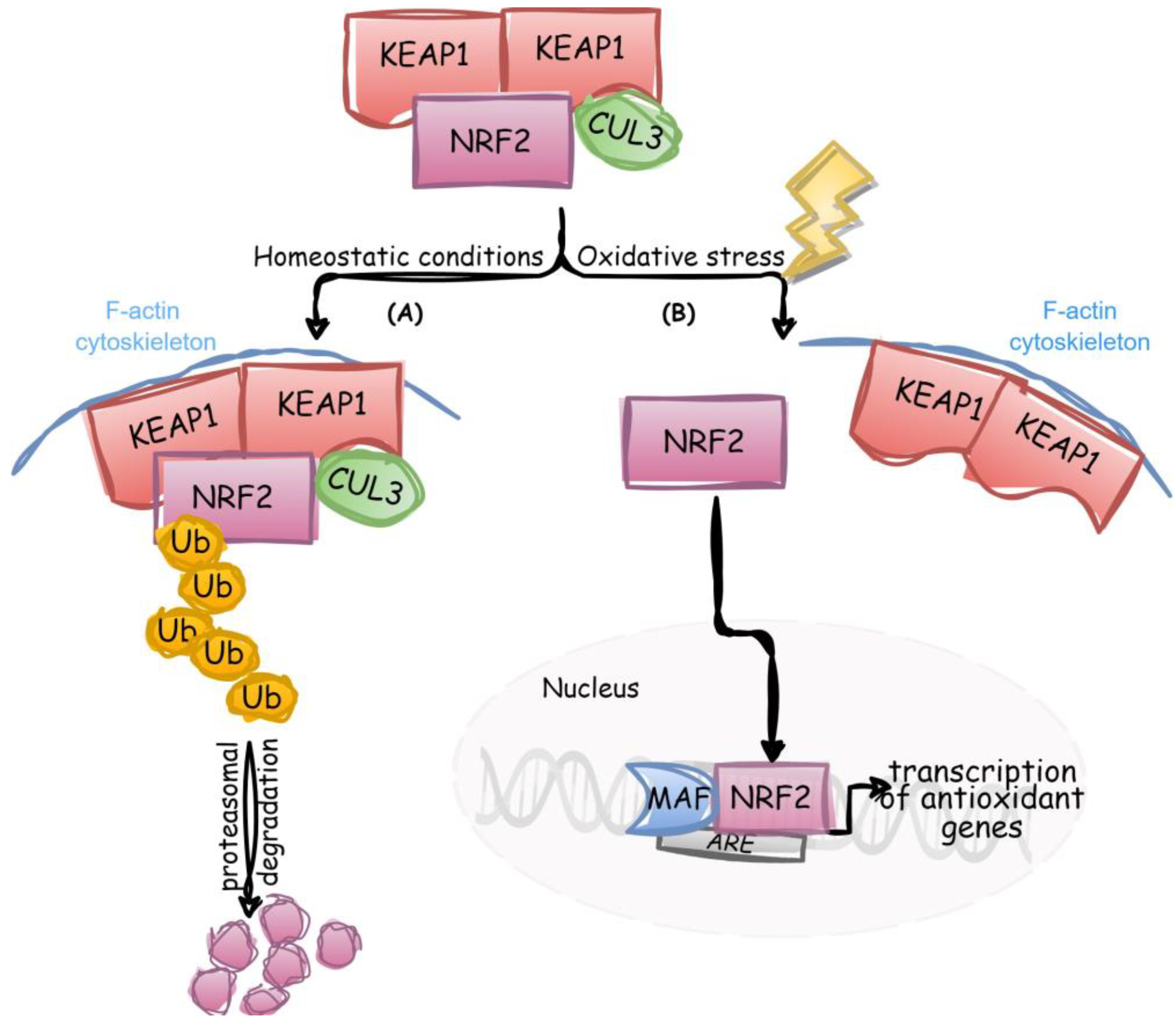
2.1. Detection and Response to Oxidative Stress
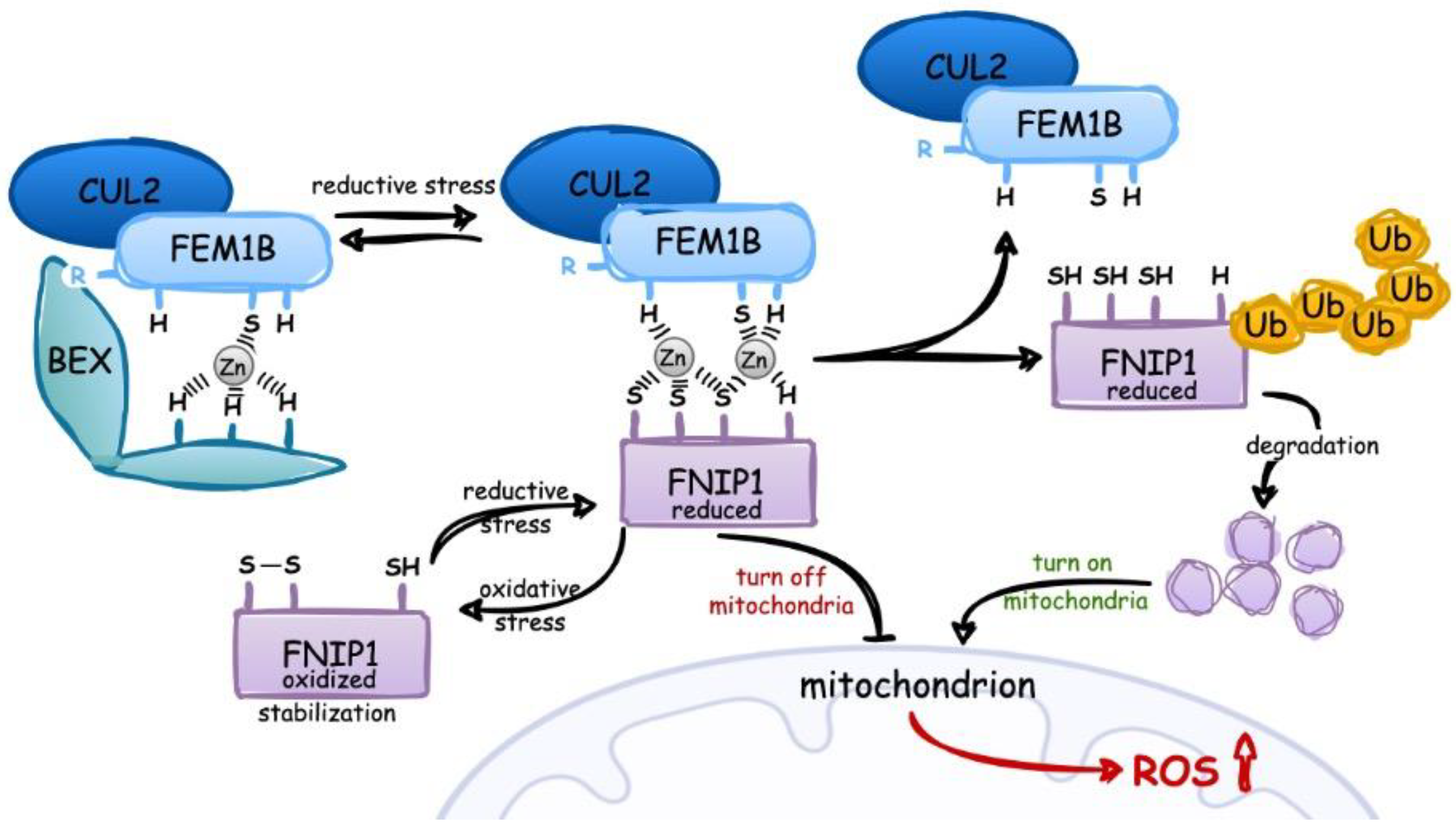
2.2. Detection and Response to Reductive Stress
3. The Biological Significance of Selenium Compounds
4. Selenium Compounds as H2Se Donors That May Affect Redox Homeostasis in Healthy Organisms and Cancers
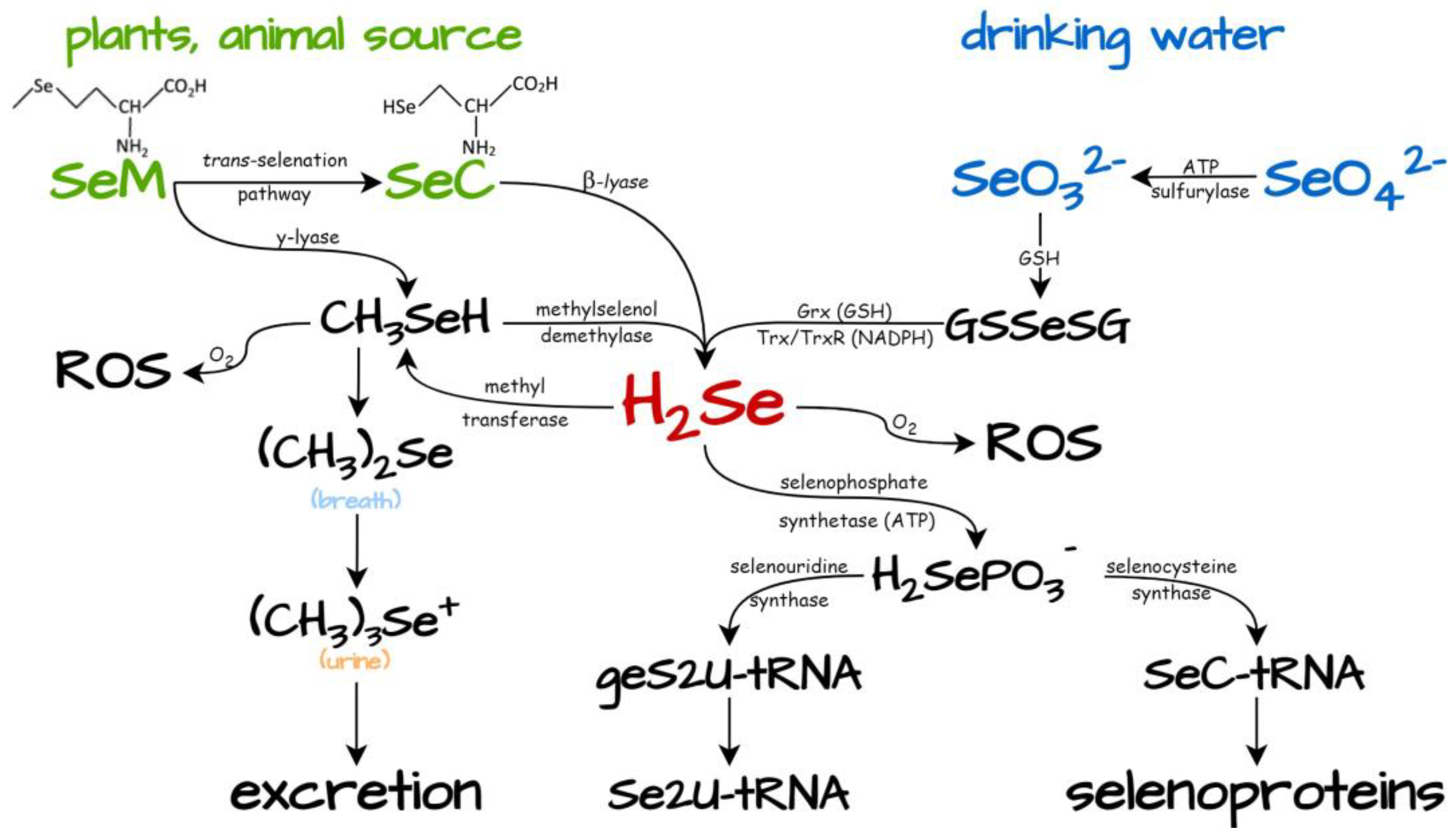
4.1. Dietary Selenium Compounds as Generators of H2Se
4.2. Chemically Synthesized Selenium Compounds as H2Se Suppliers
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quiles, J.L.; Sánchez-González, C.; Vera-Ramírez, L.; Giampieri, F.; Navarro-Hortal, M.D.; Xiao, J.; Llopis, J.; Battino, M.; Varela-López, A. Reductive Stress, Bioactive Compounds, Redox-Active Metals, and Dormant Tumor Cell Biology to Develop Redox-Based Tools for the Treatment of Cancer. Antioxid. Redox Signal. 2020, 33, 860–881. [Google Scholar] [CrossRef] [PubMed]
- Sabharwal, S.S.; Schumacker, P.T. Mitochondrial ROS in cancer: Initiators, amplifiers or an Achilles’ heel? Nat. Rev. Cancer 2014, 14, 709–721. [Google Scholar] [CrossRef]
- Xiao, W.; Loscalzo, J. Metabolic Responses to Reductive Stress. Antioxid. Redox Signal. 2020, 32, 1330–1347. [Google Scholar] [CrossRef]
- Yang, Y.; Sauve, A.A. NAD+ metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta 2016, 1864, 1787–1800. [Google Scholar] [CrossRef]
- Sarsour, E.H.; Kumar, M.G.; Chaudhuri, L.; Kalen, A.L.; Goswami, P.C.; Liu, G.-Y.; Sun, Y.-Z.; Zhou, N.; Du, X.-M.; Yang, J.; et al. Redox Control of the Cell Cycle in Health and Disease. Antioxid. Redox Signal. 2009, 11, 2985–3011. [Google Scholar] [CrossRef]
- Handy, D.; Loscalzo, J. Redox Regulation of Mitochondrial Function. Antioxid. Redox Signal. 2012, 16, 1323–1367. [Google Scholar] [CrossRef]
- Handy, D.E.; Loscalzo, J. Responses to reductive stress in the cardiovascular system. Free. Radic. Biol. Med. 2017, 109, 114–124. [Google Scholar] [CrossRef]
- Korge, P.; Calmettes, G.; Weiss, J.N. Increased reactive oxygen species production during reductive stress: The roles of mitochondrial glutathione and thioredoxin reductases. Biochim. Biophys. Acta 2015, 1847, 514–525. [Google Scholar] [CrossRef]
- Chun, K.-S.; Kim, D.-H.; Surh, Y.-J. Role of Reductive versus Oxidative Stress in Tumor Progression and Anticancer Drug Resistance. Cells 2021, 10, 758. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Abrahamse, H. Redox Potential of Antioxidants in Cancer Progression and Prevention. Antioxidants 2020, 9, 1156. [Google Scholar] [CrossRef]
- Ma, W.-X.; Li, C.-Y.; Tao, R.; Wang, X.-P.; Yan, L.-J. Reductive Stress-Induced Mitochondrial Dysfunction and Cardiomyopathy. Oxidative Med. Cell. Longev. 2020, 2020, 5136957. [Google Scholar] [CrossRef]
- Tretter, V.; Hochreiter, B.; Zach, M.L.; Krenn, K.; Klein, K.U. Understanding Cellular Redox Homeostasis: A Challenge for Precision Medicine. Int. J. Mol. Sci. 2021, 23, 106. [Google Scholar] [CrossRef]
- Zou, Z.; Chang, H.; Li, H.; Wang, S. Induction of Reactive Oxygen Species: An Emerging Approach for Cancer Therapy. Apoptosis 2017, 22, 1321–1335. [Google Scholar] [CrossRef]
- Zhang, L.; Tew, K.D. Chapter Ten-Reductive stress in cancer. Adv. Cancer Res. 2021, 152, 383–413. [Google Scholar]
- Rankin, E.B.; Giaccia, A.J. Hypoxic control of metastasis. Science 2016, 352, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.P.; Gandin, V. Selenium compounds as therapeutic agents in cancer. Biochim. Biophys. Acta BBA-Gen. Subj. 2015, 1850, 1642–1660. [Google Scholar] [CrossRef] [PubMed]
- Radomska, D.; Czarnomysy, D.; Radomski, D.; Bielawski, K. Selenium Compounds as Novel Potential Anticancer Agents. Int. J. Mol. Sci. 2021, 22, 1009. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Choi, M.C.; Park, J.M.; Chung, A.S. Antitumor Effects of Selenium. Int. J. Mol. Sci. 2021, 22, 11844. [Google Scholar] [CrossRef]
- Weekley, C.M.; Harris, H.H. Which form is that? The importance of selenium speciation and metabolism in the prevention and treatment of disease. Chem. Soc. Rev. 2013, 42, 8870–8894. [Google Scholar] [CrossRef]
- Gores, G.J.; Flarsheim, C.E.; Dawson, T.L.; Nieminen, A.L.; Herman, B.; Lemasters, J.J. Swelling, reductive stress, and cell death during chemical hypoxia in hepatocytes. Am. J. Physiol. Physiol. 1989, 257, C347–C354. [Google Scholar] [CrossRef]
- Paniker, N.; Srivastava, S.; Beutler, E. Glutathione metabolism of the red cells effect of glutathione reductase deficiency on the stimulation of hexose monophosphate shunt under oxidative stress. Biochim. Biophys. Acta BBA-Gen. Subj. 1970, 215, 456–460. [Google Scholar] [CrossRef]
- Holmström, K.M.; Finkel, T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 411–421. [Google Scholar] [CrossRef]
- Buettner, G.R. Superoxide Dismutase in Redox Biology: The Roles of Superoxide and Hydrogen Peroxide. Anti-Cancer Agents Med. Chem. 2011, 11, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, N.S.; Connell, P.; Christians, E.S.; Yan, L.-J.; Taylor, R.P.; Orosz, A.; Zhang, X.Q.; Stevenson, T.J.; Peshock, R.M.; Leopold, J.A.; et al. Human αB-Crystallin Mutation Causes Oxido-Reductive Stress and Protein Aggregation Cardiomyopathy in Mice. Cell 2007, 130, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Itoh, K.; Takahashi, S.; Sato, H.; Yanagawa, T.; Katoh, Y.; Bannai, S.; Yamamoto, M. Transcription Factor Nrf2 Coordinately Regulates a Group of Oxidative Stress-inducible Genes in Macrophages. J. Biol. Chem. 2000, 275, 16023–16029. [Google Scholar] [CrossRef]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016, 7, 11624. [Google Scholar] [CrossRef]
- Suzuki, T.; Hidaka, T.; Kumagai, Y.; Yamamoto, M. Environmental pollutants and the immune response. Nat. Immunol. 2020, 21, 1486–1495. [Google Scholar] [CrossRef]
- Rodríguez-Colman, M.J.; Schewe, M.; Meerlo, M.; Stigter, E.; Gerrits, J.; Pras-Raves, M.; Sacchetti, A.; Hornsveld, M.; Oost, K.C.; Snippert, H.J.; et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 2017, 543, 424–427. [Google Scholar] [CrossRef]
- Sena, L.A.; Chandel, N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 2012, 48, 158–167. [Google Scholar] [CrossRef]
- Padmanabhan, B.; Tong, K.I.; Ohta, T.; Nakamura, Y.; Scharlock, M.; Ohtsuji, M.; Kang, M.-I.; Kobayashi, A.; Yokoyama, S.; Yamamoto, M. Structural Basis for Defects of Keap1 Activity Provoked by Its Point Mutations in Lung Cancer. Mol. Cell 2006, 21, 689–700. [Google Scholar] [CrossRef]
- Shibata, T.; Ohta, T.; Tong, K.I.; Kokubu, A.; Odogawa, R.; Tsuta, K.; Asamura, H.; Yamamoto, M.; Hirohashi, S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA 2008, 105, 13568–13573. [Google Scholar] [CrossRef]
- Romero, R.; Sayin, V.I.; Davidson, S.M.; Bauer, M.R.; Singh, S.X.; Leboeuf, S.E.; Karakousi, T.R.; Ellis, D.C.; Bhutkar, A.; Sánchez-Rivera, F.J.; et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017, 23, 1362–1368. [Google Scholar] [CrossRef]
- Buckley, S.M.; Aranda-Orgilles, B.; Strikoudis, A.; Apostolou, E.; Loizou, E.; Moran-Crusio, K.; Farnsworth, C.L.; Koller, A.A.; Dasgupta, R.; Silva, J.C.; et al. Regulation of Pluripotency and Cellular Reprogramming by the Ubiquitin-Proteasome System. Cell Stem Cell 2012, 11, 783–798. [Google Scholar] [CrossRef] [PubMed]
- Akopian, D.; Rape, M. Principles of Ubiquitin-Dependent Signaling. Annu. Rev. Cell Dev. Biol. 2018, 34, 137–162. [Google Scholar] [CrossRef]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nature 2016, 18, 579–586. [Google Scholar] [CrossRef]
- Rape, M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2017, 19, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Lo, S.-C.; Cross, J.V.; Templeton, D.J.; Hannink, M. Keap1 Is a Redox-Regulated Substrate Adaptor Protein for a Cul3-Dependent Ubiquitin Ligase Complex. Mol. Cell. Biol. 2004, 24, 10941–10953. [Google Scholar] [CrossRef] [PubMed]
- Bellezza, I.; Giambanco, I.; Minelli, A.; Donato, R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2018, 1865, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Katoh, Y.; Kusunoki, H.; Itoh, K.; Tanaka, T.; Yamamoto, M. Keap1 Recruits Neh2 through Binding to ETGE and DLG Motifs: Characterization of the Two-Site Molecular Recognition Model. Mol. Cell. Biol. 2006, 26, 2887–2900. [Google Scholar] [CrossRef] [PubMed]
- Eggler, A.L.; Liu, G.; Pezzuto, J.M.; Van Breemen, R.B.; Mesecar, A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA 2005, 102, 10070–10075. [Google Scholar] [CrossRef]
- Kobayashi, M.; Li, L.; Iwamoto, N.; Nakajima-Takagi, Y.; Kaneko, H.; Nakayama, Y.; Eguchi, M.; Wada, Y.; Kumagai, Y.; Yamamoto, M. The Antioxidant Defense System Keap1-Nrf2 Comprises a Multiple Sensing Mechanism for Responding to a Wide Range of Chemical Compounds. Mol. Cell. Biol. 2009, 29, 493–502. [Google Scholar] [CrossRef]
- Suzuki, T.; Muramatsu, A.; Saito, R.; Iso, T.; Shibata, T.; Kuwata, K.; Kawaguchi, S.-I.; Iwawaki, T.; Adachi, S.; Suda, H.; et al. Molecular Mechanism of Cellular Oxidative Stress Sensing by Keap1. Cell Rep. 2019, 28, 746–758.e4. [Google Scholar] [CrossRef]
- Furukawa, M.; Xiong, Y. BTB Protein Keap1 Targets Antioxidant Transcription Factor Nrf2 for Ubiquitination by the Cullin 3-Roc1 Ligase. Mol. Cell. Biol. 2005, 25, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G. Von Hippel-Lindau disease. Annu. Rev. Pathol. 2007, 2, 145–173. [Google Scholar] [CrossRef]
- Lu, H.; Samanta, D.; Xiang, L.; Zhang, H.; Hu, H.; Chen, I.; Bullen, J.W.; Semenza, G.L. Chemotherapy triggers HIF-1–dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype. Proc. Natl. Acad. Sci. USA 2015, 112, E4600–E4609. [Google Scholar] [CrossRef]
- Wakabayashi, N.; Itoh, K.; Wakabayashi, J.; Motohashi, H.; Noda, S.; Takahashi, S.; Imakado, S.; Kotsuji, T.; Otsuka, F.; Roop, D.R.; et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003, 35, 238–245. [Google Scholar] [CrossRef]
- Gnarra, J.R.; Ward, J.M.; Porter, F.D.; Wagner, J.R.; Devor, D.E.; Grinberg, A.; Emmert-Buck, M.R.; Westphal, H.; Klausner, R.D.; Linehan, W.M. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc. Natl. Acad. Sci. USA 1997, 94, 9102–9107. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar] [CrossRef]
- Velichkova, M.; Hasson, T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol. Cell. Biol. 2005, 25, 4501–4513. [Google Scholar] [CrossRef]
- Bellezza, I.; Mierla, A.L.; Minelli, A. Nrf2 and NF-κB and their concerted modulation in cancer pathogenesis and progression. Cancer 2010, 2, 483–497. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Q.; Zhou, L.; Xie, N.; Nice, E.C.; Zhang, H.; Huang, C.; Lei, Y. Cancer drug resistance: Redox resetting renders a way. Oncotarget 2016, 7, 42740–42761. [Google Scholar] [CrossRef] [PubMed]
- McClung, J.P.; Roneker, C.A.; Mu, W.; Lisk, D.J.; Langlais, P.; Liu, F.; Lei, X.G. Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. Proc. Natl. Acad. Sci. USA 2004, 101, 8852–8857. [Google Scholar] [CrossRef] [PubMed]
- Dialynas, G.; Shrestha, O.K.; Ponce, J.M.; Zwerger, M.; Thiemann, D.A.; Young, G.H.; Moore, S.A.; Yu, L.; Lammerding, J.; Wallrath, L.L. Myopathic lamin mutations cause reductive stress and activate the Nrf2/Keap-1 pathway. PLoS Genet. 2015, 11, e1005231. [Google Scholar] [CrossRef] [PubMed]
- Bjelakovic, G.; Nikolova, D.; Gluud, L.L.; Simonetti, R.G.; Gluud, C. Mortality in randomized trials of antioxidant supplements for primary and secondary prevention: Systematic review and meta-analysis. JAMA 2007, 297, 842–857. [Google Scholar] [CrossRef] [PubMed]
- Manford, A.G.; Rodríguez-Pérez, F.; Shih, K.Y.; Shi, Z.; Berdan, C.A.; Choe, M.; Titov, D.V.; Nomura, D.K.; Rape, M. A cellular mechanism to detect and alleviate reductive stress. Cell 2020, 183, 46–61.e21. [Google Scholar] [CrossRef]
- Manford, A.G.; Mena, E.L.; Shih, K.Y.; Gee, C.L.; McMinimy, R.; Martínez-González, B.; Sherriff, R.; Lew, B.; Zoltek, M.; Rodríguez-Pérez, F.; et al. Structural basis and regulation of the reductive stress response. Cell 2021, 184, 5375–5390.e16. [Google Scholar] [CrossRef]
- Henning, N.J.; Manford, A.G.; Spradlin, J.N.; Brittain, S.M.; Zhang, E.; McKenna, J.M.; Tallarico, J.A.; Schirle, M.; Rape, M.; Nomura, D.K. Discovery of a Covalent FEM1B Recruiter for Targeted Protein Degradation Applications. J. Am. Chem. Soc. 2022, 144, 701–708. [Google Scholar] [CrossRef]
- Gao, X.; Wei, K.; Hu, B.; Xu, K.; Tang, B. Ascorbic acid induced HepG2 cells’ apoptosis via intracellular reductive stress. Theranostics 2019, 9, 4233–4240. [Google Scholar] [CrossRef]
- Kipp, A.P.; Frombach, J.; Deubel, S.; Brigelius-Flohé, R. Selenoprotein W as Biomarker for the Efficacy of Selenium Compounds to Act as Source for Selenoprotein Biosynthesis. Methods Enzymol. 2013, 527, 87–112. [Google Scholar] [PubMed]
- Ganyc, D.; Self, W.T. High affinity selenium uptake in a keratinocyte model. FEBS Lett. 2007, 582, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, T.; Kurata, H.; Yamada, K. Study of mammalian selenocysteyl-tRNA synthesis with [75Se]HSe−. FEBS Lett. 1991, 289, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Turanov, A.A.; Xu, X.-M.; Carlson, B.A.; Yoo, M.-H.; Gladyshev, V.N.; Hatfield, D.L. Biosynthesis of Selenocysteine, the 21st Amino Acid in the Genetic Code, and a Novel Pathway for Cysteine Biosynthesis. Adv. Nutr. Int. Rev. J. 2011, 2, 122–128. [Google Scholar] [CrossRef]
- Combs, G.F., Jr. Biomarkers of Selenium Status. Nutrients 2015, 7, 2209–2236. [Google Scholar] [CrossRef]
- Sierant, M.; Leszczynska, G.; Sadowska, K.; Komar, P.; Radzikowska-Cieciura, E.; Sochacka, E.; Nawrot, B. Escherichia coli tRNA 2-selenouridine synthase (SelU) converts S2U-RNA to Se2U-RNA via S-geranylated-intermediate. FEBS Lett. 2018, 592, 2248–2258. [Google Scholar] [CrossRef]
- Szczupak, P.; Sierant, M.; Wielgus, E.; Radzikowska-Cieciura, E.; Kulik, K.; Krakowiak, A.; Kuwerska, P.; Leszczynska, G.; Nawrot, B. Escherichia coli tRNA 2-Selenouridine Synthase (SelU): Elucidation of Substrate Specificity to Understand the Role of S-Geranyl-tRNA in the Conversion of 2-Thio- into 2-Selenouridines in Bacterial tRNA. Cells 2022, 11, 1522. [Google Scholar] [CrossRef]
- Sun, H.; Sheng, J.; Hassan, A.E.; Jiang, S.; Gan, J.; Huang, Z. Novel RNA base pair with higher specificity using single seleniumatom. Nucleic Acids Res. 2012, 40, 5171–5179. [Google Scholar] [CrossRef]
- Sanmartín, C.; Plano, D.; Sharma, A.K.; Palop, J.A. Selenium compounds, apoptosis and other types of cell death: An overview for cancer therapy. Int. J. Mol. Sci. 2012, 13, 9649–9672. [Google Scholar] [CrossRef]
- Ip, C.; Thompson, H.J.; Zhu, Z.; Ganther, H.E. In vitro and in vivo studies of methylseleninic acid: Evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention. Cancer Res. 2000, 60, 2882–2886. [Google Scholar]
- Lu, J.; Jiang, C. Selenium and Cancer Chemoprevention: Hypotheses Integrating the Actions of Selenoproteins and Selenium Metabolites in Epithelial and Non-Epithelial Target Cells. Antioxid. Redox Signal. 2005, 7, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Varlamova, E.G.; Turovsky, E.A. The main cytotoxic effects of methylseleninic acid on various cancer cells. Int. J. Mol. Sci. 2021, 22, 6614. [Google Scholar] [CrossRef]
- Pan, X.; Song, X.; Wang, C.; Cheng, T.; Luan, D.; Xu, K.; Tang, B. H2Se Induces Reductive Stress in HepG2 Cells and Activates Cell Autophagy by Regulating the Redox of HMGB1 Protein under Hypoxia. Theranostics 2019, 9, 1794–1808. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Ge, L.; Pan, X.; Xu, K.; Liu, X.; Tang, B. A highly selective near-infrared fluorescent probe for imaging H2Se in living cells and in vivo. Chem. Sci. 2015, 7, 1051–1056. [Google Scholar] [CrossRef]
- Kong, F.; Zhao, Y.; Liang, Z.; Liu, X.; Pan, X.; Luan, D.; Xu, K.; Tang, B. Highly Selective Fluorescent Probe for Imaging H2Se in Living Cells and in Vivo Based on the Disulfide Bond. Anal. Chem. 2017, 89, 688–693. [Google Scholar] [CrossRef]
- Nuttall, K.L.; Allen, F.S. Kinetics of the reaction between hydrogen selenide ion and oxygen. Inorg. Chim. Acta 1984, 91, 243–246. [Google Scholar] [CrossRef]
- Cupp-Sutton, K.A.; Ashby, M.T. Biological Chemistry of Hydrogen Selenide. Antioxidants 2016, 5, 42. [Google Scholar] [CrossRef]
- Cao, W.; Li, X.; Zheng, S.; Zheng, W.; Wong, Y.-S.; Chen, T. Selenocysteine derivative overcomes TRAIL resistance in melanoma cells: Evidence for ROS-dependent synergism and signaling crosstalk. Oncotarget 2014, 5, 7431–7445. [Google Scholar] [CrossRef]
- Zhang, K.; Su, J.; Chen, D.; Lin, B.; Wu, Y.; Wang, Y.; Lei, J.; Zheng, R.; Zhu, B.; Li, Y. L-Selenocysteine induced HepG-2 cells apoptosis through reactive oxygen species-mediated signaling pathway. Mol. Biol. Rep. 2022, 49, 8381–8390. [Google Scholar] [CrossRef]
- Gandin, V.; Khalkar, P.; Braude, J.; Fernandes, A.P. Organic selenium compounds as potential chemotherapeutic agents for improved cancer treatment. Free. Radic. Biol. Med. 2018, 127, 80–97. [Google Scholar] [CrossRef]
- Weekley, C.; Aitken, J.B.; Musgrave, I.; Harris, H.H. Methylselenocysteine Treatment Leads to Diselenide Formation in Human Cancer Cells: Evidence from X-ray Absorption Spectroscopy Studies. Biochemistry 2012, 51, 736–738. [Google Scholar] [CrossRef] [PubMed]
- Weekley, C.M.; Aitken, J.B.; Vogt, S.; Finney, L.A.; Paterson, D.J.; de Jonge, M.D.; Howard, D.L.; Musgrave, I.F.; Harris, H.H. Uptake, Distribution, and Speciation of Selenoamino Acids by Human Cancer Cells: X-ray Absorption and Fluorescence Methods. Biochemistry 2011, 50, 1641–1650. [Google Scholar] [CrossRef]
- Roman, M.; Jitaru, P.; Barbante, C. Selenium biochemistry and its role for human health. Metallomics 2013, 6, 25–54. [Google Scholar] [CrossRef]
- Kim, E.; Sohn, S.; Kwon, H.; Kim, S.; Kim, M. Sodium selenite induces superoxide-mediated mitochondrial damage and subsequent autophagic cell death in malignant glioma cells. Cancer Res. 2007, 67, 6314. [Google Scholar] [CrossRef] [PubMed]
- Xiang, N.; Zhao, R.; Zhong, W. Sodium selenite induces apoptosis by generation of superoxide via the mitochondrial-dependent pathway in human prostate cancer cells. Cancer Chemother. Pharmacol. 2009, 63, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Olm, E.; Fernandes, A.P.; Hebert, C.; Rundlöf, A.-K.; Larsen, E.H.; Danielsson, O.; Björnstedt, M. Extracellular thiol-assisted selenium uptake dependent on the xc− cystine transporter explains the cancer-specific cytotoxicity of selenite. Proc. Natl. Acad. Sci. USA 2009, 106, 11400–11405. [Google Scholar] [CrossRef]
- Weekley, C.M.; Aitken, J.B.; Vogt, S.; Finney, L.A.; Paterson, D.J.; de Jonge, M.D.; Howard, D.L.; Witting, P.K.; Musgrave, I.F.; Harris, H.H. Metabolism of Selenite in Human Lung Cancer Cells: X-Ray Absorption and Fluorescence Studies. J. Am. Chem. Soc. 2011, 133, 18272–18279. [Google Scholar] [CrossRef]
- Chen, P.; Wang, L.; Li, N.; Liu, Q.; Ni, J. Comparative proteomics analysis of sodium selenite induced apoptosis in human prostate cancer cells. Metallomics 2013, 5, 541. [Google Scholar] [CrossRef]
- Cheng, R.; Kong, F.; Tong, L.; Liu, X.; Xu, K.; Tang, B. Simultaneous Detection of Mitochondrial Hydrogen Selenide and Superoxide Anion in HepG2 Cells under Hypoxic Conditions. Anal. Chem. 2018, 90, 8116–8122. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. High Dose Inorganic Selenium for Preventing Chemotherapy Induced Peripheral Neuropathy (SELENIUM). Phase 3, Recruiting. Available online: https://clinicaltrials.gov/ct2/show/NCT04201561 (accessed on 17 December 2019).
- ClinicalTrials.gov. Sodium Selenite and Radiation Therapy in Treating Patients with Metastatic Cancer. Phase 1, Completed. Available online: https://clinicaltrials.gov/ct2/show/NCT02184533 (accessed on 9 July 2014).
- ClinicalTrials.gov. The Use of Selenium to Treat Secondary Lymphedema—Breast Cancer. Phase 2, Completed. Available online: https://clinicaltrials.gov/ct2/show/NCT00188604 (accessed on 13 August 2005).
- ClinicalTrials.gov. Sodium Selenite as a Cytotoxic Agent in Advanced Carcinoma (SECAR). Phase 2, Unknown. Available online: https://clinicaltrials.gov/ct2/show/NCT01959438 (accessed on 10 October 2013).
- ClinicalTrials.gov. Phase I Sodium Selenite in Combination with Docetaxel in Castration-resistant Prostate Cancer. Phase 1, Terminated. Available online: https://clinicaltrials.gov/ct2/show/NCT01155791 (accessed on 2 July 2010).
- Chuai, H.; Zhang, S.-Q.; Bai, H.; Li, J.; Wang, Y.; Sun, J.; Wen, E.; Zhang, J.; Xin, M. Small molecule selenium-containing compounds: Recent development and therapeutic applications. Eur. J. Med. Chem. 2021, 223, 113621. [Google Scholar] [CrossRef]
- Lu, J.; Jiang, C.; Kaeck, M.; Ganther, H.; Vadhanavikit, S.; Clement, I.P.; Thompson, H. Dissociation of the genotoxic and growth inhibitory effects of selenium. Biochem. Pharmacol. 1995, 50, 213–219. [Google Scholar] [CrossRef]
- Peyroche, G.; Saveanu, C.; Dauplais, M.; Lazard, M.; Beuneu, F.; Decourty, L.; Malabat, C.; Jacquier, A.; Blanquet, S.; Plateau, P. Sodium selenide toxicity is mediated by O2-dependent DNA breaks. PLoS ONE 2012, 7, e36343. [Google Scholar] [CrossRef] [PubMed]
- Brozmanová, J.; Mániková, D.; Vlcková, V.; Chovanec, M. Selenium: A double-edged sword for defense and offence in cancer. Arch. Toxicol. 2010, 84, 919–938. [Google Scholar] [CrossRef] [PubMed]
- Ringuet, M.T.; Hunne, B.; Lenz, M.; Bravo, D.M.; Furness, J.B. Analysis of bioavailability and induction of glutathione peroxidase by dietary nanoelemental, organic and inorganic selenium. Nutrients 2021, 13, 1073. [Google Scholar] [CrossRef]
- Kunwar, A.; Mishra, B.; Barik, A.; Kumbhare, L.B.; Pandey, R.; Jain, V.K.; Priyadarsini, K.I. 3,3′-Diselenodipropionic acid, an efficient peroxyl radical scavenger and a GPx mimic, protects erythrocytes (RBCs) from AAPH-induced hemolysis. Chem. Res. Toxicol. 2007, 20, 1482–1487. [Google Scholar] [CrossRef]
- Domínguez-Álvarez, E.; Plano, D.; Font, M.; Calvo, A.; Prior, C.; Jacob, C.; Palop, J.A.; Sanmartín, C. Synthesis and antiproliferative activity of novel selenoester derivatives. Eur. J. Med.Chem. 2014, 73, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M.; Spengler, G.; Sanmartín, C.; Marć, M.A.; Handzlik, J.; Domínguez-Álvarez, E. Selenoesters andselenoanhydrides as novel multidrug resistance reversing agents: Aconfirmation study in a colon cancer MDR cell line. Bioorg. Med.Chem. Lett. 2017, 27, 797–802. [Google Scholar] [CrossRef]
- Kharma, A.; Misak, A.; Grman, M.; Brezova, V.; Kurakova, L.; Baráth, P.; Jacob, C.; Chovanec, M.; Ondrias, K.; Domínguez-Álvarez, E. Release of reactive seleniumspecies from phthalic selenoanhydride in the presence of hydrogensulfide and glutathione with implications for cancer research. New J. Chem. 2019, 43, 11771–11783. [Google Scholar] [CrossRef]
- Li, L.; Whiteman, M.; Guan, Y.Y.; Neo, K.L.; Cheng, Y.; Lee, S.W.; Zhao, Y.; Baskar, R.; Tan, C.H.; Moore, P.K. Characterization of a novel, water-soluble hydrogen sulfide-releasing molecule (GYY4137): New insights into the biology of hydrogen sulfide. Circulation 2008, 117, 2351–2360. [Google Scholar] [CrossRef]
- Newton, T.D.; Pluth, M.D. Development of a hydrolysis-based small-molecule hydrogen selenide (H2Se) donor. Chem. Sci. 2019, 10, 10723–10727. [Google Scholar] [CrossRef]
- Newton, T.D.; Bolton, S.G.; Garcia, A.C.; Chouinard, J.E.; Golledge, S.L.; Zakharov, L.N.; Pluth, M.D. Hydrolysis-Based Small-Molecule Hydrogen Selenide (H2Se) Donors for Intracellular H2Se Delivery. J. Am. Chem. Soc. 2021, 143, 19542–19550. [Google Scholar] [CrossRef]
- Levinn, C.M.; Cerda, M.M.; Pluth, M.D. Activatable Small-Molecule Hydrogen Sulfide Donors. Antioxid. Redox Signal. 2020, 32, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Huang, H.; Jiang, C.; Cheng, L.; Sang, Y.; Cai, X.; Dong, Y.; Sun, L.; Wen, X.; Xi, Z.; et al. Cysteine-Activated Small-Molecule H2Se Donors Inspired by Synthetic H2S Donors. J. Am. Chem. Soc. 2022, 144, 3957–3967. [Google Scholar] [CrossRef] [PubMed]
- Krakowiak, A.; Czernek, L.; Pichlak, M.; Kaczmarek, R. Intracellular HINT1-Assisted Hydrolysis of Nucleoside 5′-O-Selenophosphate Leads to the Release of Hydrogen Selenide That Exhibits Toxic Effects in Human Cervical Cancer Cells. Int. J. Mol. Sci. 2022, 23, 607. [Google Scholar] [CrossRef]
- Martin, J.; St-Pierre, M.V.; Dufour, J.F. Hit proteins, mitochondria and cancer. Biochim. Biophys. Acta 2011, 1807, 626–632. [Google Scholar] [CrossRef]
- Krakowiak, A.; Pawłowska, R.; Kocoń-Rębowska, B.; Dolot, R.; Stec, W.J. Interactions of cellular histidine triad nucleotide binding protein 1 with nucleosides 5’-O-monophosphorothioate and their derivatives-Implication for desulfuration process in the cell. Biochim. Biophys. Acta 2014, 1840, 3357–3366. [Google Scholar] [CrossRef]
- Krakowiak, A.; Piotrzkowska, D.; Kocoń-Rębowska, B.; Kaczmarek, R.; Maciaszek, A. The role of the Hint1 protein in the metabolism of phosphorothioate oligonucleotides drugs and prodrugs, and the release of H2S under cellular conditions. Biochem. Pharmacol. 2019, 163, 250–259. [Google Scholar] [CrossRef]
- Lin, V.S.; Lippert, A.R.; Chang, C.J. Cell-trappable fluorescent probes for endogenous hydrogen sulfide signaling and imaging H2O2-dependent H2S production. Proc. Natl. Acad. Sci. USA 2013, 110, 7131–7135. [Google Scholar] [CrossRef]
- Ozga, M.; Dolot, R.; Janicka, M.; Kaczmarek, R.; Krakowiak, A. Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5′-O-Phosphorothioates to Nucleoside 5′-O-Phosphates. J. Biol. Chem. 2010, 285, 40809–40818. [Google Scholar] [CrossRef] [PubMed]
- Hankins, R.A.; Carter, M.E.; Zhu, C.; Chen, C.; Lukesh, J.C. 3rd. Enol-mediated delivery of H2Se from γ-keto selenides: Mechanistic insight and evaluation. Chem. Sci. 2022, 13, 13094–13099. [Google Scholar] [CrossRef]
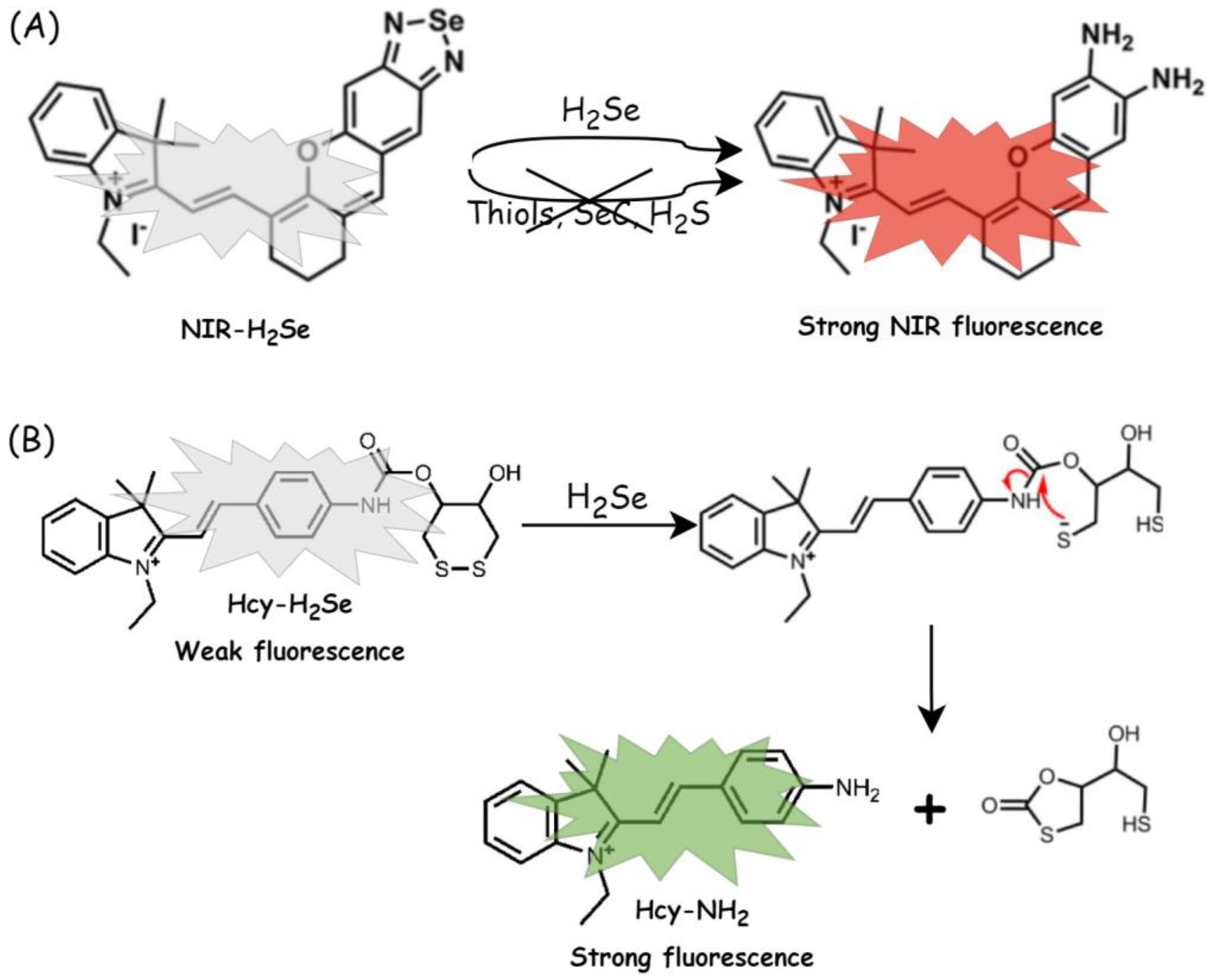
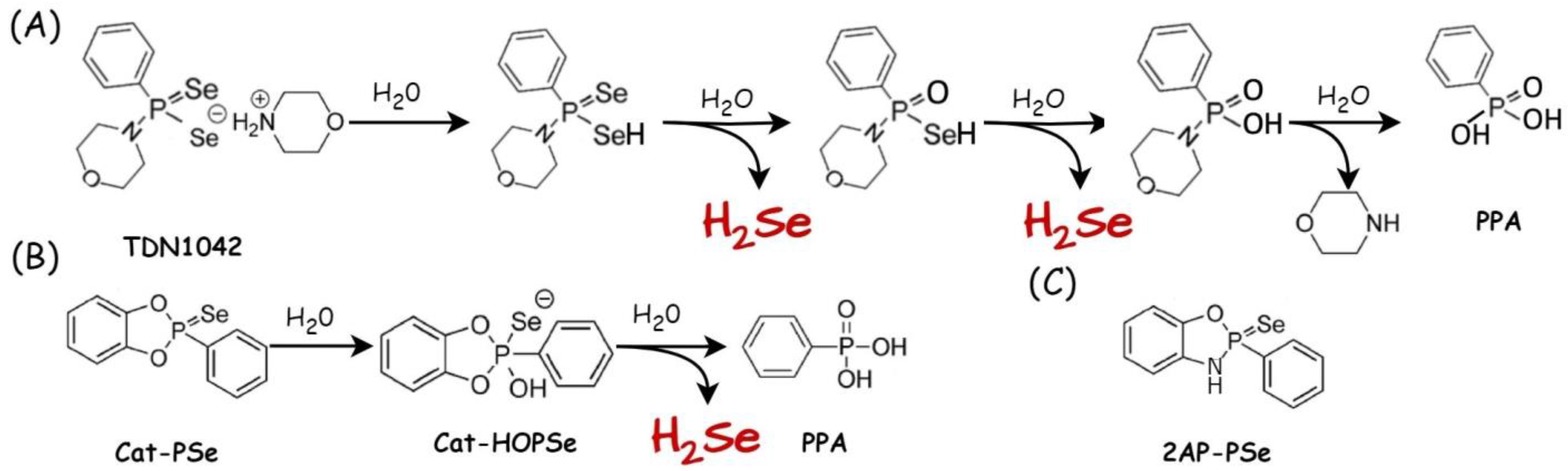
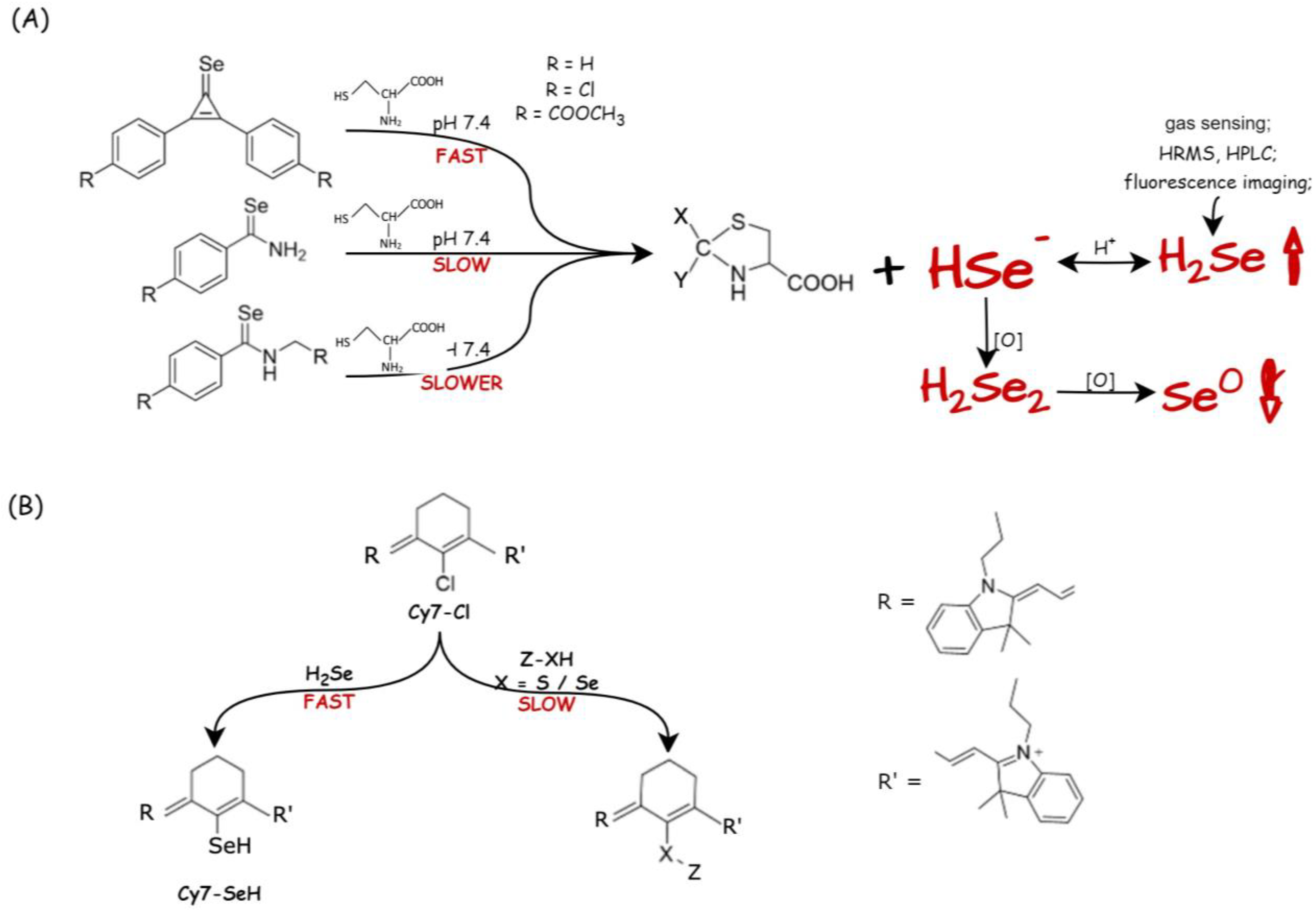
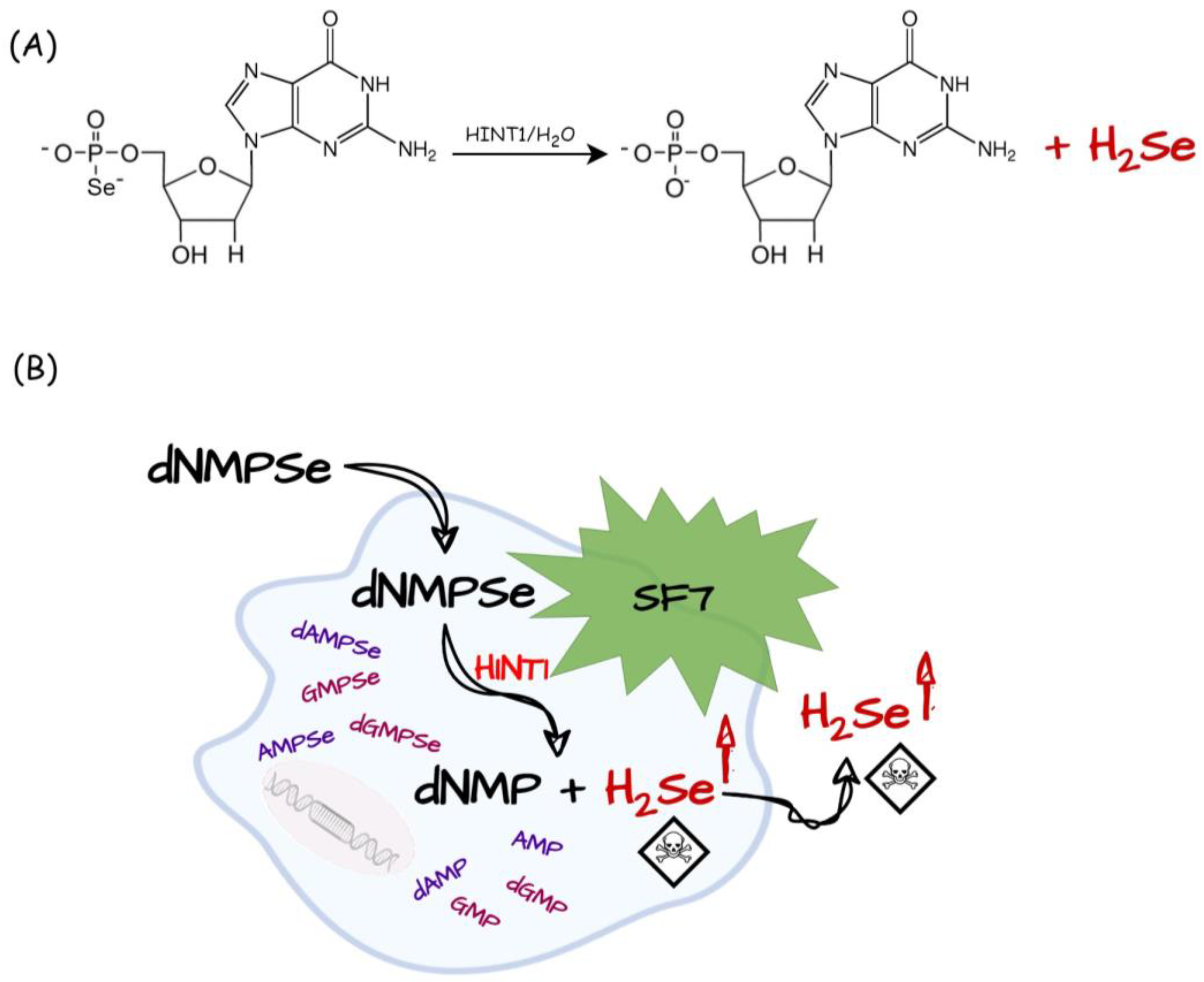

| Selenium Compound | Detection Method of H2Se Release | Mechanism of H2Se Release | Biological Model | Ref. |
|---|---|---|---|---|
| Na2SeO3 (under clinical trials) | Fluorescence imaging NIR-H2Se | Enzymatic: Grx (GSH), Trx, TrxR (NADPH) | HepG2 cells (cytotoxicity, reductive stress, H2Se release), mice | [74,75] |
| Fluorescence imaging Hcy-H2Se | HepG2 (H2Se release) | [76] | ||
| Fluorescence imaging Mito-N-D-MSN (nanoprobes mitochondria-targeted) | HepG2 (H2Se release) | [90] | ||
| TDN1042 (P=Se motif) | 31P and 77Se NMR and electrophilic trapping reagent | Acidic conditions | - | [106] |
| 2AP-PSe, Cat-PSe (P=Se motif) | 31P and 77Se NMR and electrophilic trapping reagent; colorimetric detection with NBD-Cl | pH 7.2 | HeLa cells (antioxidant activity) | [107] |
| selenocyclopropenones and arylselenoamides (C=Se motif) | H2Se-selective gas detector; electrophilic trapping reagent and HRMS analysis; Cy7-CI trapping; fluorescence imaging (NIR-H2Se) | Cys at pH 7.4 | HeLa cells (Cys-mediated H2Se release) | [109] |
| dGMPSe | Fluorescence imaging SF7 | Enzymatic: HINT1 | HeLa cells (cytotoxicity and H2Se release) | [110] |
| γ-keto selenides | Trapping reagent and HRMS | neutral to slightly basic conditions | HeLa and HCT116 cells (cytotoxicity) | [116] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krakowiak, A.; Pietrasik, S. New Insights into Oxidative and Reductive Stress Responses and Their Relation to the Anticancer Activity of Selenium-Containing Compounds as Hydrogen Selenide Donors. Biology 2023, 12, 875. https://doi.org/10.3390/biology12060875
Krakowiak A, Pietrasik S. New Insights into Oxidative and Reductive Stress Responses and Their Relation to the Anticancer Activity of Selenium-Containing Compounds as Hydrogen Selenide Donors. Biology. 2023; 12(6):875. https://doi.org/10.3390/biology12060875
Chicago/Turabian StyleKrakowiak, Agnieszka, and Sylwia Pietrasik. 2023. "New Insights into Oxidative and Reductive Stress Responses and Their Relation to the Anticancer Activity of Selenium-Containing Compounds as Hydrogen Selenide Donors" Biology 12, no. 6: 875. https://doi.org/10.3390/biology12060875
APA StyleKrakowiak, A., & Pietrasik, S. (2023). New Insights into Oxidative and Reductive Stress Responses and Their Relation to the Anticancer Activity of Selenium-Containing Compounds as Hydrogen Selenide Donors. Biology, 12(6), 875. https://doi.org/10.3390/biology12060875






