Genetic Diversity, Morphometric Characterization, and Conservation Reassessment of the Critically Endangered Freshwater Snail, Heleobia atacamensis, in the Atacama Saltpan, Northern Chile
Abstract
Simple Summary
Abstract
1. Introduction
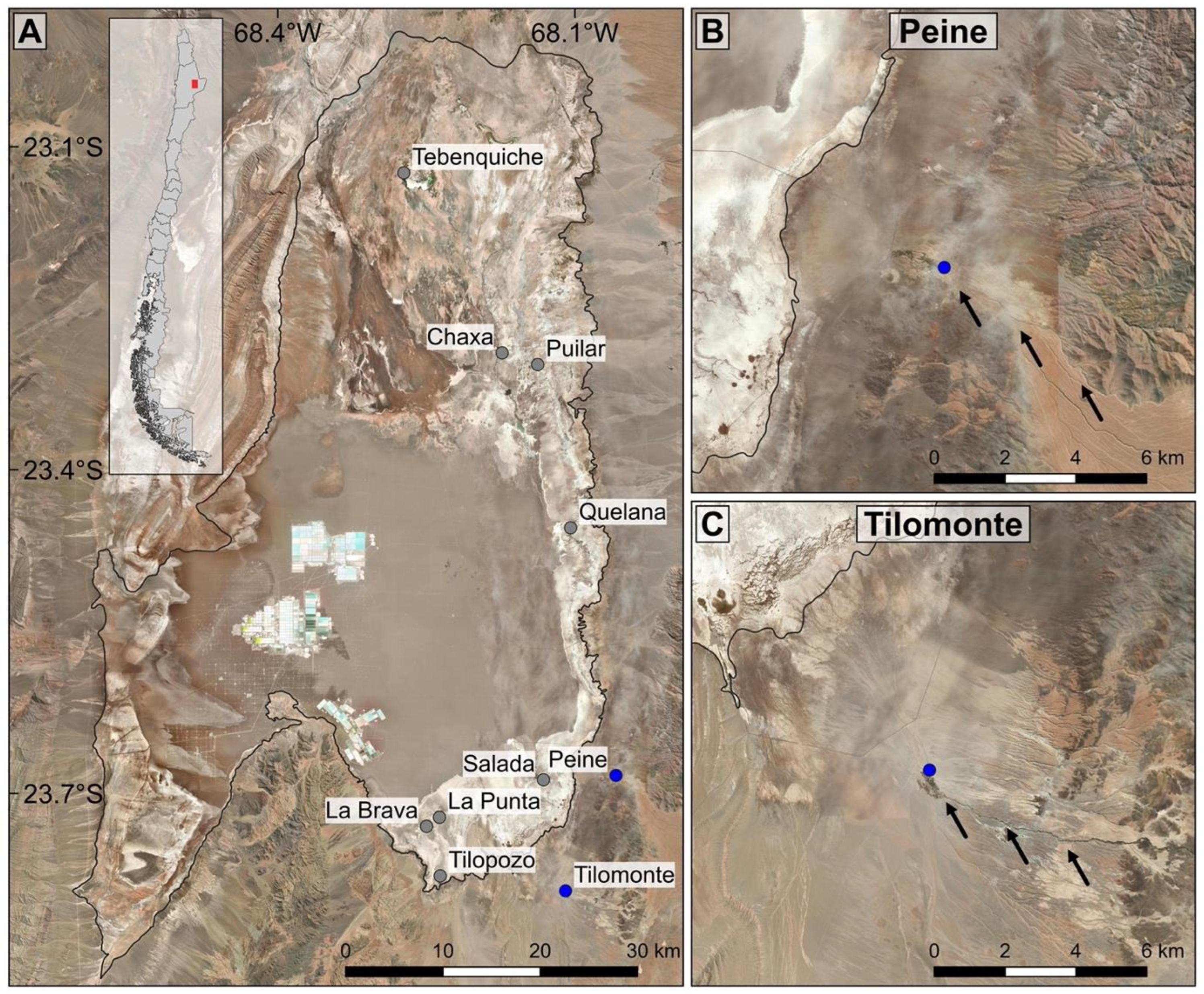
2. Materials and Methods
3. Results
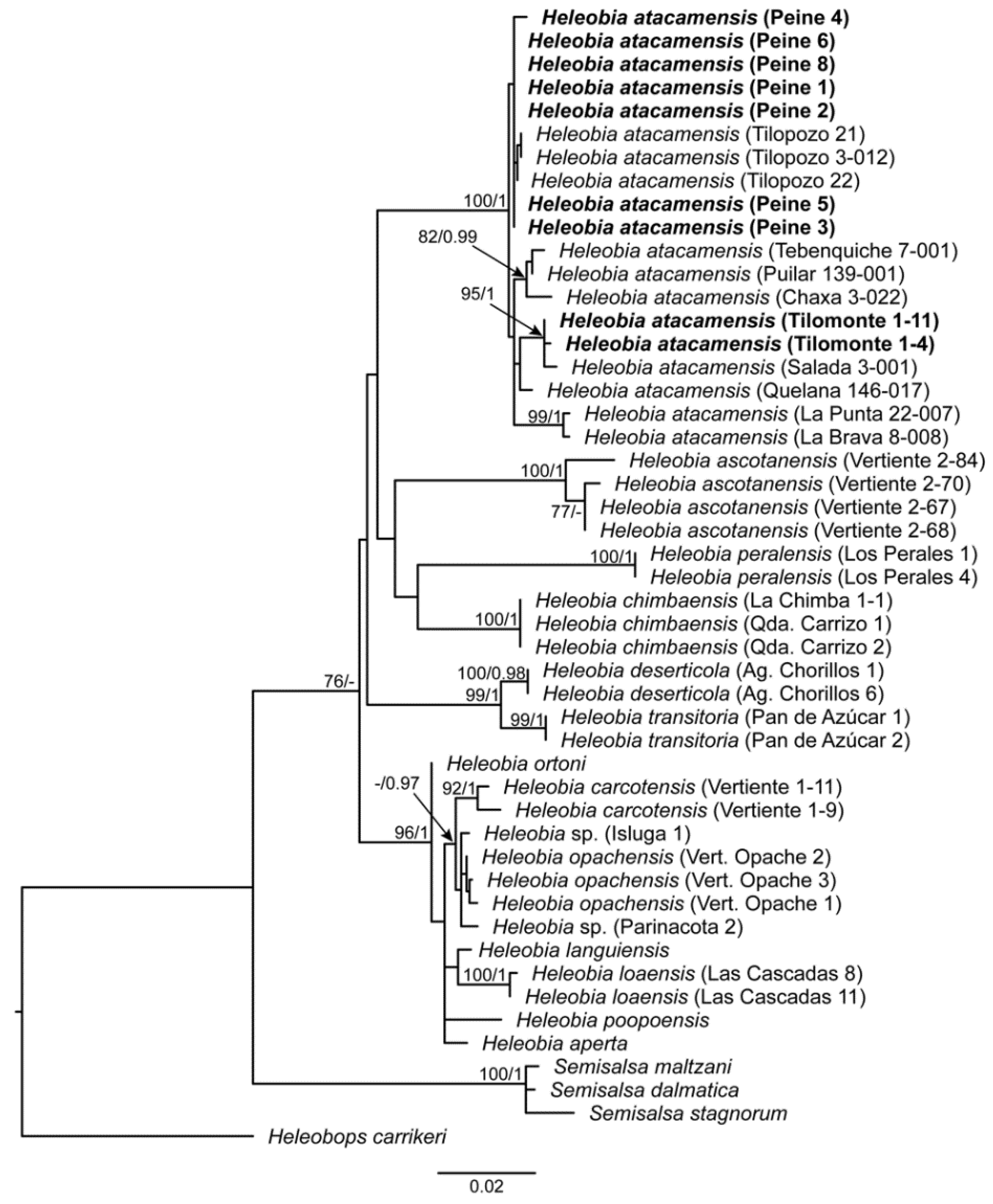
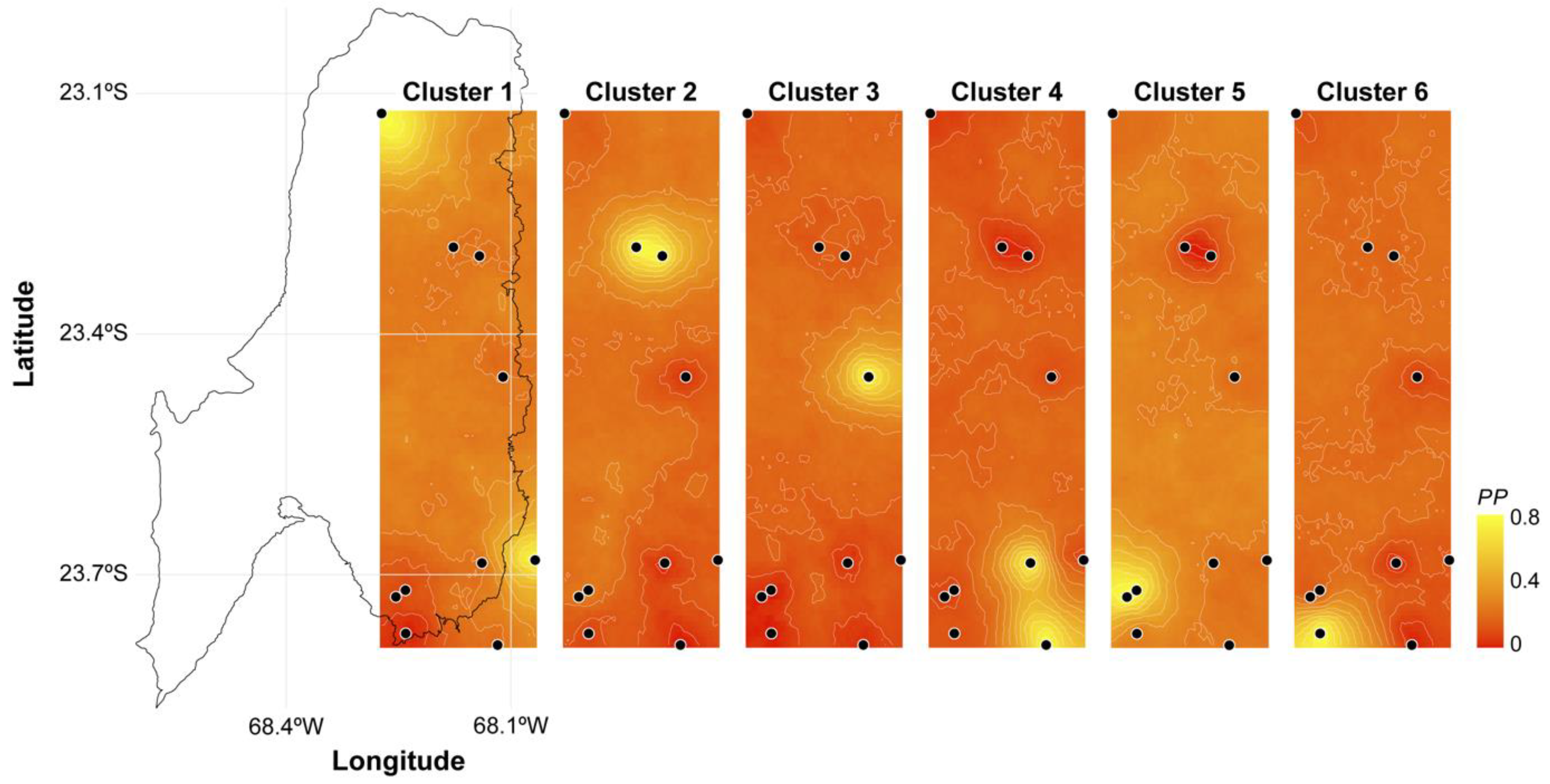
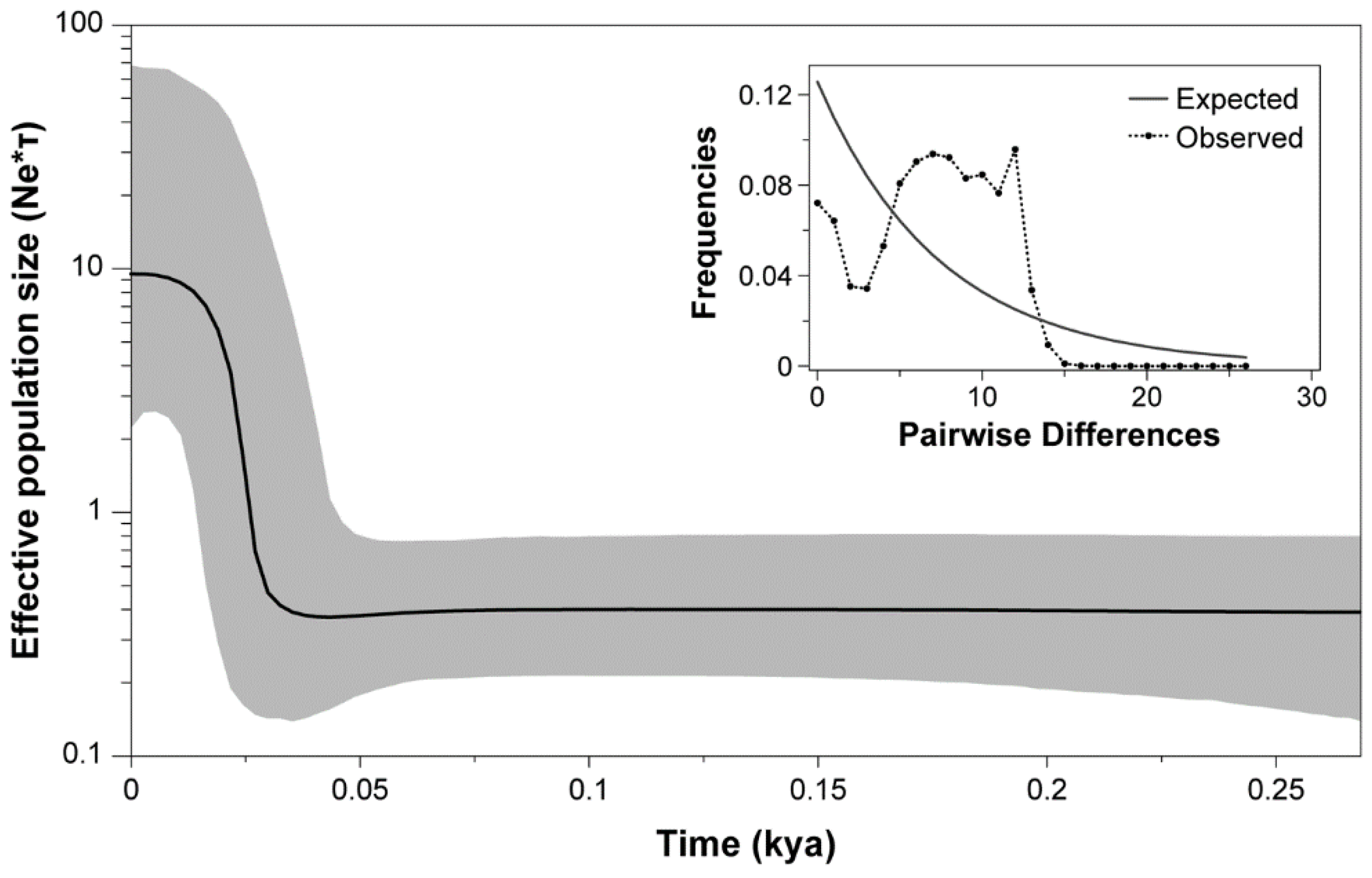
3.1. Molecular Analysis
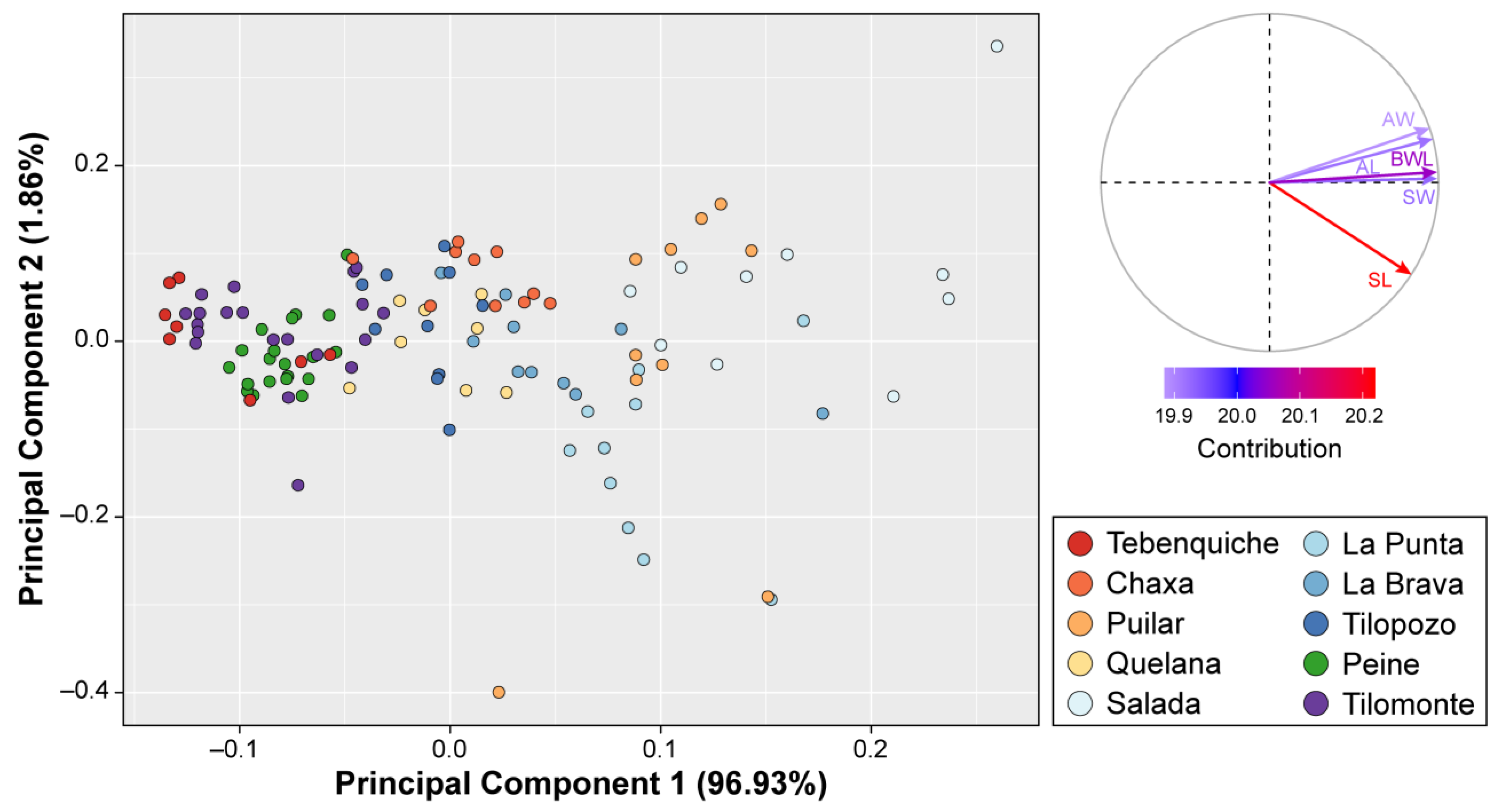
3.2. Morphological Analyses
3.3. Conservation Status
4. Systematic
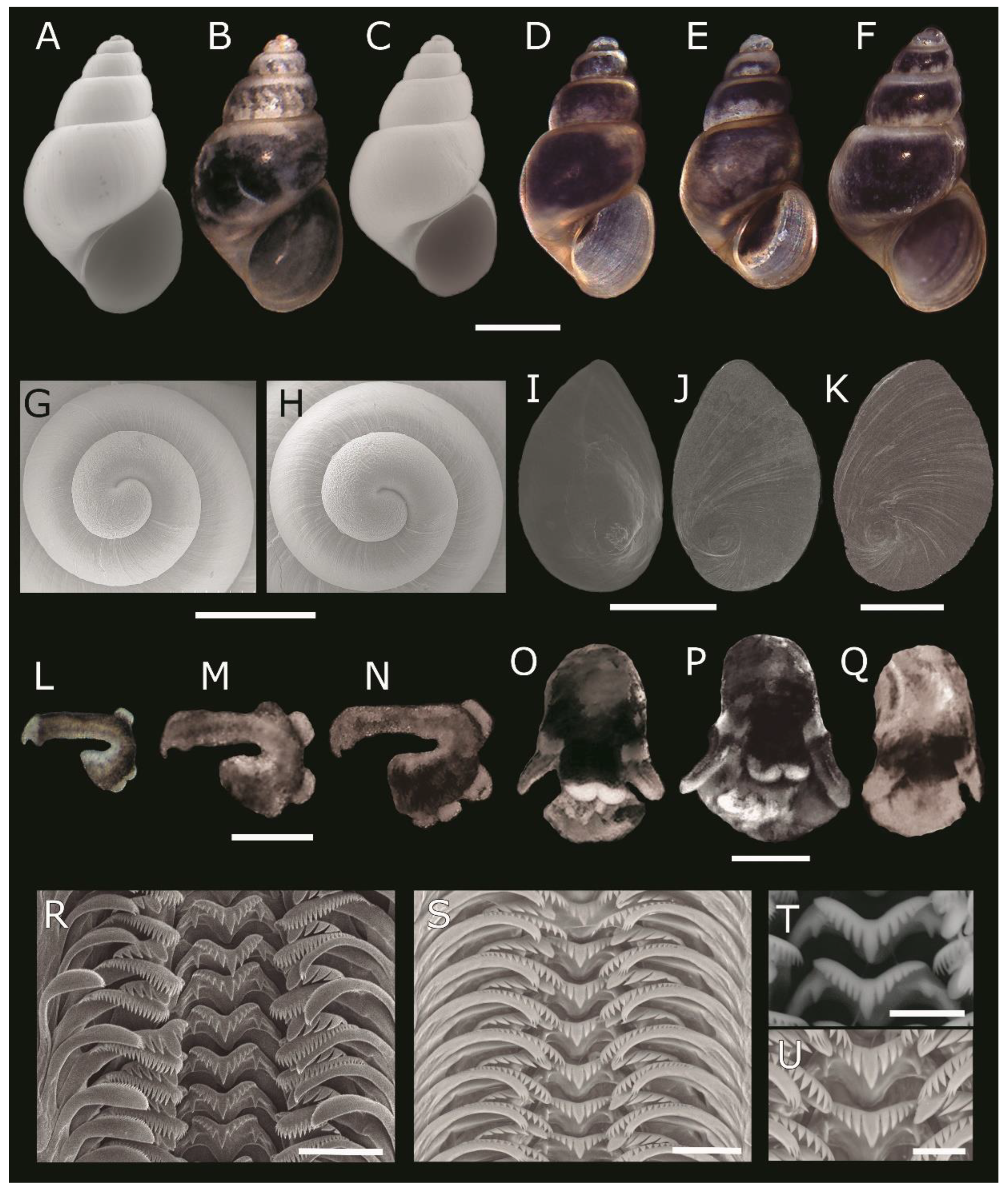
4.1. Description
4.2. Material Examined
4.3. Habitat
4.4. Distribution
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lowe, S.; Browne, M.; Boudlejas, S. 100 of the World’s Worst Invasive Alien Species; Invasive Species Specialist Group: Auckland, Australia, 2000. [Google Scholar]
- Barnosky, A.; Matzke, N.; Tomiya, S.; Wogan, G.O.U.; Swartz, B.; Quental, T.B.; Marshall, C.; McGuire, J.L.; Lindsey, E.L.; Maguire, K.C.; et al. Has the Earth’s sixth mass extinction already arrived? Nature 2011, 471, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, G.; Ehrlich, P.R.; Dirzo, R. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc. Natl. Acad. Sci. USA 2017, 114, E6089–E6096. [Google Scholar] [CrossRef] [PubMed]
- Slingenberg, A.; Braat, L.; van der Windt, H.; Eichler, L.; Turner, K. Study on Understanding the Causes of Biodiversity Loss and the Policy Assessment Framework; Directorate-General for Environment, European Commission: Rotterdam, The Netherlands, 2009. [Google Scholar]
- Sodhi, N.S.; Brook, B.W.; Bradshaw, C.J.A. Causes and Consequences of Species Extinctions. In The Princeton Guide to Ecology; Levin, S., Carpenter, S., Godfray, H., Kinzig, A., Loreau, M., Losos, J., Walker, B., Wilcove, D., Eds.; Princeton University Press: Princeton, NJ, USA, 2009; pp. 514–520. [Google Scholar]
- Swingland, I.R. Biodiversity, Definition of. In Encyclopedia of Biodiversity, 2nd ed.; Levin, S.A., Ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 399–410. [Google Scholar]
- Chapman, A.D. Number of Living Species in Australia and the World, 2nd ed.; Department of the Environment, Water, Heritage and the Arts, Australian Government: Canberra, Australian, 2009; pp. 1–80.
- Rosenberg, G. A new critical estimate of named species-level diversity of the recent Mollusca. Am. Malacol. Bull. 2014, 32, 308–322. [Google Scholar] [CrossRef]
- Ponder, W.; Hutchings, P.; Chapman, R. Overview of the Conservation of Australian Marine Invertebrates; Report for Environment Australia, Australian Museum: Sydney, Australia, 2002; pp. 1–588. [Google Scholar]
- IUCN 2021. The IUCN Red List of Threatened Species. Version 2021-2. Available online: https://www.iucnredlist.org (accessed on 13 January 2023).
- MolluscaBase. 2021. Available online: http://www.molluscabase.org (accessed on 18 October 2021).
- Downing, J.A.; Van Meter, P.; Woolnough, D.A. Suspects and evidence: A review of the causes of extirpation and decline of freshwater mussels. Anim. Biodivers. Conserv. 2010, 33, 151–185. [Google Scholar] [CrossRef]
- Strayer, D.E.; Dudgeon, D. Freshwater biodiversity conservation: Recent progress and future challenges. J. N. Am. Benthol. Soc. 2010, 29, 344–358. [Google Scholar] [CrossRef]
- Johnson, P.D.; Bogan, A.E.; Brown, K.M.; Burkhead, N.M.; Cordeiro, J.R.; Garner, J.T.; Hartfield, P.D.; Lepitzki, D.A.W.; Mackie, G.L.; Pip, E.; et al. Conservation status of freshwater gastropods of Canada and the United States. Fisheries 2013, 38, 247–282. [Google Scholar] [CrossRef]
- Philippi, R.A. Reise Durch Die Wueste Atacama auf Befehl der Chilenischen Regierung im Sommer 1853–54; E Anton: Halle, Germany, 1860; pp. 1–62. [Google Scholar]
- Courty, G. Explorations géologiques dans l’Amerique du Sud. In Mission Scientifique de G de Crèqui Montefort et E Sénéchal de la Grange, 1st ed.; Impremiere Nationale: Paris, France, 1907; pp. 1–258. [Google Scholar]
- Biese, W.A. Revisión de los moluscos terrestres y de agua dulce provistos de concha de Chile. Parte 4: Planorbidae. Bol. Mus. Nac. Hist. Nat. 1951, 25, 115–137. [Google Scholar]
- Collado, G.A.; Valladares, M.A.; Méndez, M.A. Unraveling cryptic species of freshwater snails (Caenogastropoda, Truncatelloidea) in the Loa River basin, Atacama Desert. Syst. Biodivers. 2016, 14, 417–429. [Google Scholar] [CrossRef]
- Collado, G.A.; Valladares, M.A.; Méndez, M.A. A new species of Heleobia (Caenogastropoda: Cochliopidae) from the Chilean Altiplano. Zootaxa 2016, 4137, 277–280. [Google Scholar] [CrossRef]
- Ministerio del Medio Ambiente, Chile (MMA). Inventario Nacional de Especies de Chile: Biomphalaria costata (Biese, 1951). 10° Proceso de Clasificación de Especies. 2014. Available online: http://especies.mma.gob.cl/CNMWeb/Web/WebCiudadana/ficha_indepen.aspx?EspecieId=5395&Version=1 (accessed on 30 December 2022).
- Ministerio del Medio Ambiente, Chile (MMA). Inventario Nacional de Especies de Chile: Heleobia atacamensis (Philippi, 1860). 10° Proceso de Clasificación de Especies. 2014. Available online: http://especies.mma.gob.cl/CNMWeb/Web/WebCiudadana/ficha_indepen.aspx?EspecieId=5418&Version=1 (accessed on 30 December 2022).
- Ministerio del Medio Ambiente, Chile (MMA). Inventario Nacional de Especies de Chile: Heleobia transitoria (Biese, 1947). 11° Proceso de Clasificación de Especies. 2015. Available online: http://especies.mma.gob.cl/CNMWeb/Web/WebCiudadana/ficha_indepen.aspx?EspecieId=5419&Version=1 (accessed on 30 December 2022).
- Pastorino, G.; Darrigran, G. Heleobia atacamensis. The IUCN Red List of Threatened Species 2011: E.T189150A8693243. 2011. Available online: https://dx.doi.org/10.2305/IUCN.UK.2011-2.RLTS.T189150A8693243.en (accessed on 30 December 2022).
- Cabello, J. Reservas, recursos y exploración de litio en salares del norte de Chile. Andean Geol. 2022, 49, 297–306. [Google Scholar] [CrossRef]
- Liu, W.; Agusdinata, D.B.; Myint, S.W. Spatiotemporal patterns of lithium mining and environmental degradation in the Atacama Salt Flat, Chile. Int. J. Appl. Earth Obs. Geoinf. 2019, 80, 145–156. [Google Scholar] [CrossRef]
- Liu, W.; Agusdinata, D.B. Interdependencies of lithium mining and communities sustainability in Salar de Atacama, Chile. J. Clean. Prod. 2020, 260, 120838. [Google Scholar] [CrossRef]
- Marazuela, M.A.; Vázquez-Suñé, E.; Ayora, C.; García-Gil. A. Towards more sustainable brine extraction in salt flats: Learning from the Salar de Atacama. Sci. Total Environ. 2020, 703, 135605. [Google Scholar] [CrossRef]
- Collado, G.A.; Valladares, M.A.; Méndez, M.A. Hidden diversity in spring snails from the Andean Altiplano, the second highest plateau on Earth, and the Atacama Desert, the driest place in the world. Zool. Stud. 2013, 52, 50. [Google Scholar] [CrossRef]
- Valladares, M.A.; Fabres, A.A.; Collado, G.A.; Sáez, P.A.; Méndez, M.A. Coping With Dynamism: Phylogenetics and Phylogeographic Analyses Reveal Cryptic Diversity in Heleobia Snails of Atacama Saltpan, Chile. Front. Ecol. Evol. 2022, 10, 869626. [Google Scholar] [CrossRef]
- Chiesa, S.; Scalici, M.; Negrini, R.; Gibertini, G.; Marzano, F.N. Fine-scale genetic structure, phylogeny and systematics of threatened crayfish species complex. Mol. Phylogenet. Evol. 2011, 61, 1–11. [Google Scholar] [CrossRef]
- Eizaguirre, C.; Baltazar-Soares, M. Evolutionary conservation—Evaluating the adaptive potential of species. Evol. Appl. 2014, 7, 963–967. [Google Scholar] [CrossRef]
- Darwall, W.; Bremerich, V.; De Wever, A.; Dell, A.I.; Freyhof, J.; Gessner, M.O.; Grossart, H.P.; Harrison, I.; Irvine, K.; Jähnig, S.C.; et al. The alliance for freshwater life: A global call to unite efforts for freshwater biodiversity science and conservation. Aquat. Conserv. 2018, 28, 1015–1022. [Google Scholar] [CrossRef]
- Lovrenčić, L.; Bonassin, L.; Boštjančić, L.L.; Podnar, M.; Jelić, M.; Klobučar, G.; Jaklič, M.; Slavevska-Stamenković, V.; Hinić, J.; Maguire, I. New insights into the genetic diversity of the stone crayfish: Taxonomic and conservation implications. BMC Evol. Biol. 2020, 20, 146. [Google Scholar] [CrossRef]
- Lan, D.; Ji, W.; Xiong, X.; Liang, Q.; Yao, W.; Mipam, T.D.; Zhong, J.; Li, J. Population genome of the newly discovered Jinchuan yak to understand its adaptive evolution in extreme environments and generation mechanism of the multirib trait. Integr. Zool. 2021, 16, 685–695. [Google Scholar] [CrossRef]
- Bláha, M.; Patoka, J.; Japoshvili, B.; Let, M.; Buřič, M.; Kouba, A.; Mumladze, L. Genetic diversity, phylogenetic position and morphometric analysis of Astacus colchicus (Decapoda, Astacidae): A new insight into Eastern European crayfish fauna. Integr. Zool. 2021, 16, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proc. Gatew. Comput. Environ. Work. 2010, 1, 1–8. [Google Scholar]
- Collado, G.A.; Fuentealba, C.G.; Cazzaniga, N.J.; Valladares, M.A. Assessing biodiversity within the range of Heleobia chimbaensis (Caenogastropoda: Cochliopidae) on the Atacama Desert coast. Syst. Biodivers. 2020, 18, 708–719. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Guillot, G.; Mortier, F.; Estoup, A. Geneland: A computer package for landscape genetics. Mol. Ecol. Notes 2005, 5, 712–715. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef]
- Wilke, T.; Schultheiß, R.; Albrecht, C. As Time Goes by: A simple fool’s guide to molecular clock approaches in invertebrates. Am. Malacol. Bull. 2009, 27, 25–45. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing 3.6.1; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- IUCN 2022. Guidelines for Using the IUCN Red List Categories and Criteria. Version 14. Prepared by the Standards and Petitions Committee. Available online: https://nrl.iucnredlist.org/resources/redlistguidelines (accessed on 13 January 2023).
- Collado, G.A.; Chihuailaf, E.; Muñoz, N.; Novoa, F.; Valladares, M.A. Reproductive aspects of the poorly known and critically endangered freshwater snail Heleobia atacamensis (Gastropoda: Truncatelloidea). PeerJ 2021, 9, e11550. [Google Scholar] [CrossRef]
- Bachman, S.; Moat, J.; Hill, A.W.; Scott, B. Supporting Red List threat assessments with GeoCat: Geospatial conservation assessment tool. ZooKeys 2011, 150, 117–126. [Google Scholar] [CrossRef]
- Frauenfeld, G. Verzeichniss der Namen der fossilen und lebenden Arten der Gattung Paludina LAM. nebst jenen der nächststehenden und Einreihung derselben in die verschiedenen neueren Gattungen. Verh. Zool.-Bot. Ges. Wien 1864, 14, 561–672. [Google Scholar]
- Biese, W.A. Revisión de los moluscos terrestres y de agua dulce provistos de concha de Chile. Parte I, Familia Amnicolidae. Bol. Mus. Nac. Hist. Nat. 1944, 22, 169–190. [Google Scholar]
- Biese, W.A. Revisión de los moluscos terrestres y de agua dulce provistos de concha de Chile. Parte II, Familia Amnicolidae. Bol. Mus. Nac. Hist. Nat. 1947, 23, 63–77. [Google Scholar]
- Stuardo, J. Contribución a un catálogo de los moluscos chilenos de agua dulce. Gayana 1961, 1, 7–32. [Google Scholar]
- Valdovinos Zarges, C. Biodiversidad de moluscos chilenos: Base de datos taxonómica y distribucional. Gayana 1999, 63, 111–164. [Google Scholar]
- Sielfeld, W. Phylum Mollusca. Guías de Identificación y Biodiversidad Fauna Chilena; Universidad Arturo Prat: Iquique, Chile, 2001; pp. 1–115. [Google Scholar]
- Valdovinos Zarges, C. Estado de conocimiento de los gastrópodos dulceacuícolas de Chile. Gayana 2006, 70, 88–95. [Google Scholar] [CrossRef]
- Hershler, R.; Thompson, F.G. A review of the aquatic gastropod subfamily Cochliopinae (Prosobranchia: Hydrobiidae). Malacol. Rev. (Suppl.) 1992, 5, 1–140. [Google Scholar]
- Collado, G.A.; Méndez, M.A.; Letelier, S.; Veliz, D.; Sabando, M.C. Morfología peniana y taxonomía del género Heleobia Stimpson, 1865 en Chile junto a una revisión de los ejemplares tipo del Museo Nacional de Historia Natural de Chile. Amici Molluscarum (Número Especial) 2011, 49–58. [Google Scholar]
- Collado, G.A.; Vila, I.; Méndez, M.A. Monophyly, candidate species and vicariance in Biomphalaria snails (Mollusca: Planorbidae) from the Southern Andean Altiplano. Zool. Scr. 2011, 40, 613–622. [Google Scholar] [CrossRef]
- Vila, I.; Morales, P.; Scott, S.; Poulin, E.; Veliz, D.; Harrod, C.; Mendez, M.A. Phylogenetic and phylogeographic analysis of the genus Orestias (Teleostei: Cyprinodontidae) in the southern Chilean Altiplano: The relevance of ancient and recent divergence processes in speciation. J. Fish Biol. 2013, 82, 927–943. [Google Scholar] [CrossRef]
- Sáez, P.A.; Zúñiga-Reinoso, Á.; Fibla, P.; Cruz-Jofré, F.; Aguilar, C.; Aparicio, J.; Cusi, J.C.; Otálora, K.; Méndez, M.A. Phylogeny of Telmatobius marmoratus complex (Anura, Telmatobiidae) reveals high cryptic diversity in the Andean Altiplano. Mol. Phylogenet. Evol. 2022, 176, 107594. [Google Scholar] [CrossRef]
- Seidel, R.A.; Lang, B.K.; Berg, D.J. Phylogeographic analysis reveals multiple cryptic species of amphipods (Crustacea: Amphipoda) in Chihuahuan Desert springs. Biol. Conserv. 2009, 142, 2303–2313. [Google Scholar] [CrossRef]
- Murphy, N.P.; Breed, M.F.; Guzik, M.T.; Cooper, S.J.B.; Austin, A.D. Trapped in desert springs: Phylogeography of Australian desert spring snails. J. Biogeogr. 2012, 39, 1573–1582. [Google Scholar] [CrossRef]
- Collado, G.A.; Méndez, M.A. Microgeographic differentiation among closely related species of Biomphalaria (Gastropoda: Planorbidae) from the Andean Altiplano. Zool. J. Linn. Soc. 2013, 169, 640–652. [Google Scholar] [CrossRef]
- Valladares, M.A.; Méndez, M.A.; Collado, G.A. Influenced but not determined by historical events: Genetic, demographic and morphological differentiation in Heleobia ascotanensis from the Chilean Altiplano. PeerJ 2018, 6, e5802. [Google Scholar] [CrossRef]
- Bobst, A.L.; Lowenstein, T.K.; Jordan, T.E.; Godfrey, L.V.; Ku, T.L.; Luo, S. A 106 ka paleoclimate record from drill core of the Salar de Atacama, northern Chile. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2001, 173, 21–42. [Google Scholar] [CrossRef]
- Sáez, A.; Cabrera, L.; Garcés, M.; van den Bogaard, P.; Jensen, A.; Gimeno, D. The stratigraphic record of changing hyperaridity in the Atacama desert over the last 10Ma. Earth Planet. Sci. Lett. 2012, 355–356, 32–38. [Google Scholar] [CrossRef]
- Ritter, B.; Binnie, S.A.; Stuart, F.M.; Wennrich, V.; Dunai, T.J. Evidence for multiple Plio-Pleistocene lake episodes in the hyperarid Atacama Desert. Quat. Geochronol. 2018, 44, 1–12. [Google Scholar] [CrossRef]
- Gajardo, G.; Redón, S. Andean hypersaline lakes in the Atacama Desert, northern Chile: Between lithium exploitation and unique biodiversity conservation. Conserv. Sci. Pract. 2019, e94. [Google Scholar]
- Hubendick, B. XVIII. The anatomy of the gastropoda. The Percy Sladen Trust Expedition to Lake Titicaca in 1937. Trans. Linn. Soc. London 1955, 1, 309–327. [Google Scholar]
- Gaillard, M.C.; de Castellanos, Z.A. Mollusca, Gasteropoda, Hydrobiidae. In Fauna de Agua Dulce de la República Argentina; Ringuelet, R.A. (Director) Fundación para la Educación, la Ciencia y la Cultura (FECIC): Buenos Aires, Argentina, 1976; Volume 15, pp. 1–40. [Google Scholar]
- Cazzaniga, N.J. Nota sobre los hidrobidos Argentinos. I (Gastropoda: Rissoidea), Acerca de Littoridina occidentalis (Doering, 1884). Neotropica 1980, 26, 187–191. [Google Scholar]
- Cazzaniga, N.J. Nota sobre los hidrobidos Argentinos. 5. Conquiliometría de Littoridina parchappii (d’Orbigny, 1835) (Gastropoda Rissoidea) referida a su ciclo de vida en poblaciones Australes. Iheringia Ser. Zool. 1982, 61, 97–118. [Google Scholar]
- Cazzaniga, N.J. Nota sobre los hidróbidos Argentinos. II (Gastropoda: Rissoidea), Una Littoridina del “grupo parchappii” en Península Valdés (Chubut). Rev. Mus. La Plata 1982, 129, 11–16. [Google Scholar]
- Pons da Silva, M.C.P. Dados morfológicos de Heleobia parchappei (Orbigny, 1835) (Prosobranchia, Hydrobiidae, Littoridininae). Iheringia Ser. Zool. 1993, 75, 81–87. [Google Scholar]
- Pons da Silva, M.C.; Veitenheimer-Mendes, I.L. Nova espécie de Heleobia (Rissooidea, Hydrobiidae) da planicie costeira do sul do Brasil. Iheringia Ser. Zool. 2004, 94, 89–94. [Google Scholar] [CrossRef]
- Collado, G.A. A new freshwater snail (Caenogastropoda: Cochliopidae) from the Atacama Desert, northern Chile. Zootaxa 2015, 3925, 445–449. [Google Scholar] [CrossRef]
- Merlo, M.J.; Parietti, M.; Etchegoin, J.A. Stunting of the penis in Heleobia parchappii (Mollusca: Cochliopidae) and its relationship with parasitism. Dis. Aquat. Org. 2017, 123, 81–85. [Google Scholar] [CrossRef]
- Laguna, E.; Fos, S.; Jiménez, J.; Volis, S. Role of micro-reserves in conservation of endemic, rare and endangered plants of the Valencian region (Eastern Spain). Isr. J. Plant Sci. 2016, 63, 320–332. [Google Scholar] [CrossRef]
- Tonge, S.; Bloxam, Q. A review of the captive-breeding programme for Polynesian tree snails Partula spp. Int. Zoo Yearb. 1991, 30, 51–59. [Google Scholar] [CrossRef]
- Pearce-Kelly, P.; Clarke, D.; Walker, C.; Atkin, P. A conservation programme for the partulid tree snails of the Pacific region. Mem. Mus. Vic. 1997, 56, 431–433. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Collado, G.A.; Torres-Díaz, C.; Vidal, M.A.; Valladares, M.A. Genetic Diversity, Morphometric Characterization, and Conservation Reassessment of the Critically Endangered Freshwater Snail, Heleobia atacamensis, in the Atacama Saltpan, Northern Chile. Biology 2023, 12, 791. https://doi.org/10.3390/biology12060791
Collado GA, Torres-Díaz C, Vidal MA, Valladares MA. Genetic Diversity, Morphometric Characterization, and Conservation Reassessment of the Critically Endangered Freshwater Snail, Heleobia atacamensis, in the Atacama Saltpan, Northern Chile. Biology. 2023; 12(6):791. https://doi.org/10.3390/biology12060791
Chicago/Turabian StyleCollado, Gonzalo A., Cristian Torres-Díaz, Marcela A. Vidal, and Moisés A. Valladares. 2023. "Genetic Diversity, Morphometric Characterization, and Conservation Reassessment of the Critically Endangered Freshwater Snail, Heleobia atacamensis, in the Atacama Saltpan, Northern Chile" Biology 12, no. 6: 791. https://doi.org/10.3390/biology12060791
APA StyleCollado, G. A., Torres-Díaz, C., Vidal, M. A., & Valladares, M. A. (2023). Genetic Diversity, Morphometric Characterization, and Conservation Reassessment of the Critically Endangered Freshwater Snail, Heleobia atacamensis, in the Atacama Saltpan, Northern Chile. Biology, 12(6), 791. https://doi.org/10.3390/biology12060791





