The Diagnostic Potential of RNA Aptamers against the NS1 Protein of Dengue Virus Serotype 2
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Designing the Random ssDNA Library and Primers
2.2. PCR Amplification
2.3. In Vitro Transcription
2.4. SELEX
2.5. Nitrocellulose Filter Binding Assay
2.6. Cloning, Transformation and Sequence Analysis
2.7. Secondary Structure Determination and the Truncation of the Aptamers
2.8. Conversion of the RNA Sequences from 2D to 3D Structures
2.9. Rigid Molecular Docking
2.10. Molecular Dynamic (MD) Simulation of the RNA Aptamer–NS1 Protein Complex
2.11. Functionalization of the RNA Aptamer with Biotin at the 3′-End for ELASA
2.12. In Vitro Transcription of the Stabilized Truncated RNA Aptamers
2.13. Direct ELASA
2.13.1. Cross-Reactivity Analyses
2.13.2. Limit of Detection (LOD)
2.13.3. Direct ELASA of Stabilized TDENVs at Different Times of Incubation
2.14. Sandwich ELASA
2.14.1. Sandwich ELASA with Cumulative Incubations
2.14.2. Sandwich ELASA in Serum Diluents
2.15. Statistical Analysis
3. Results and Discussion
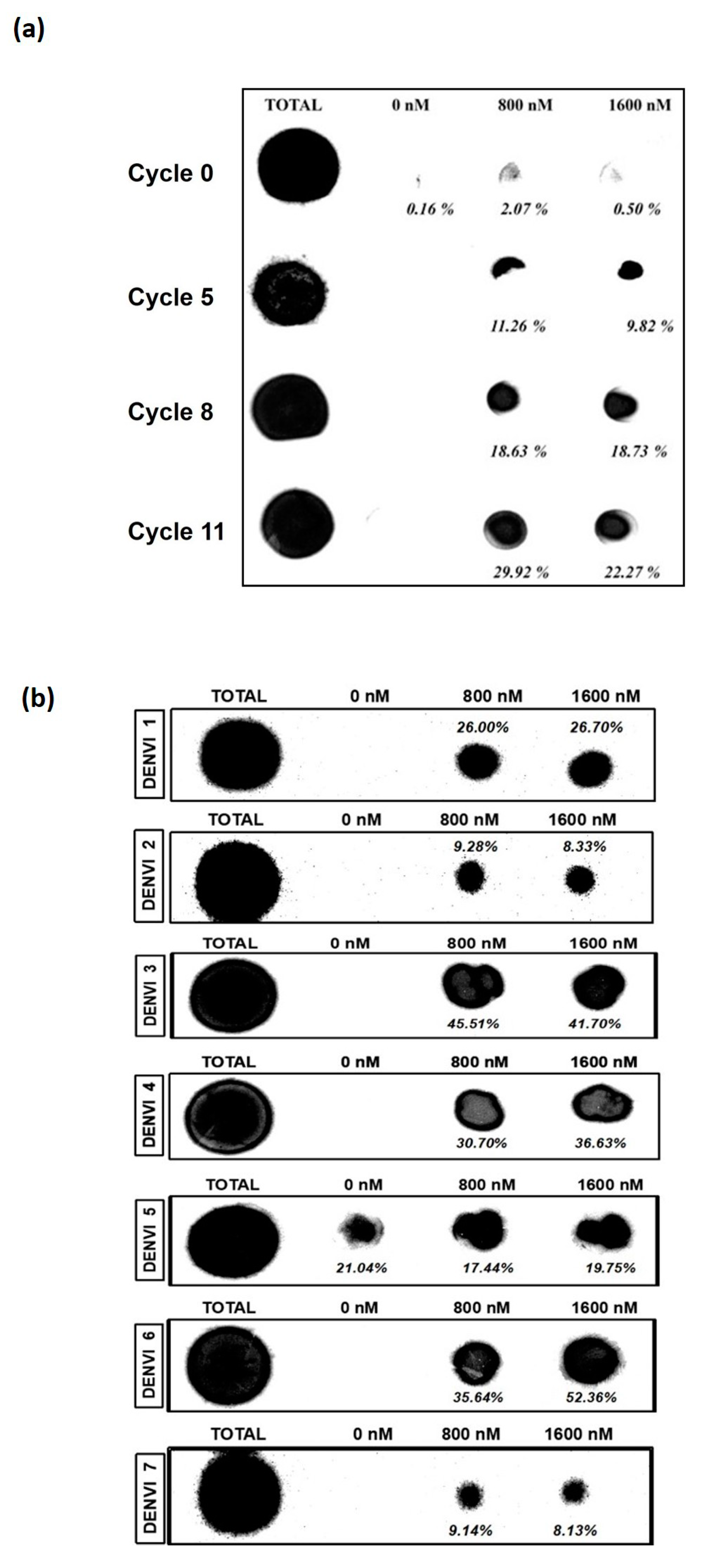
3.1. Nitrocellulose Filter Binding Assay Suggests Enrichment of the Nucleic Acid Pool from the 11th Cycle of SELEX
3.2. DENV-3 and -6 Are the Most Potent Aptamers
3.3. Estimation of the Equilibrium Dissociation Constant, Kd, of DENV-3 and DENV-6 RNA aptamers
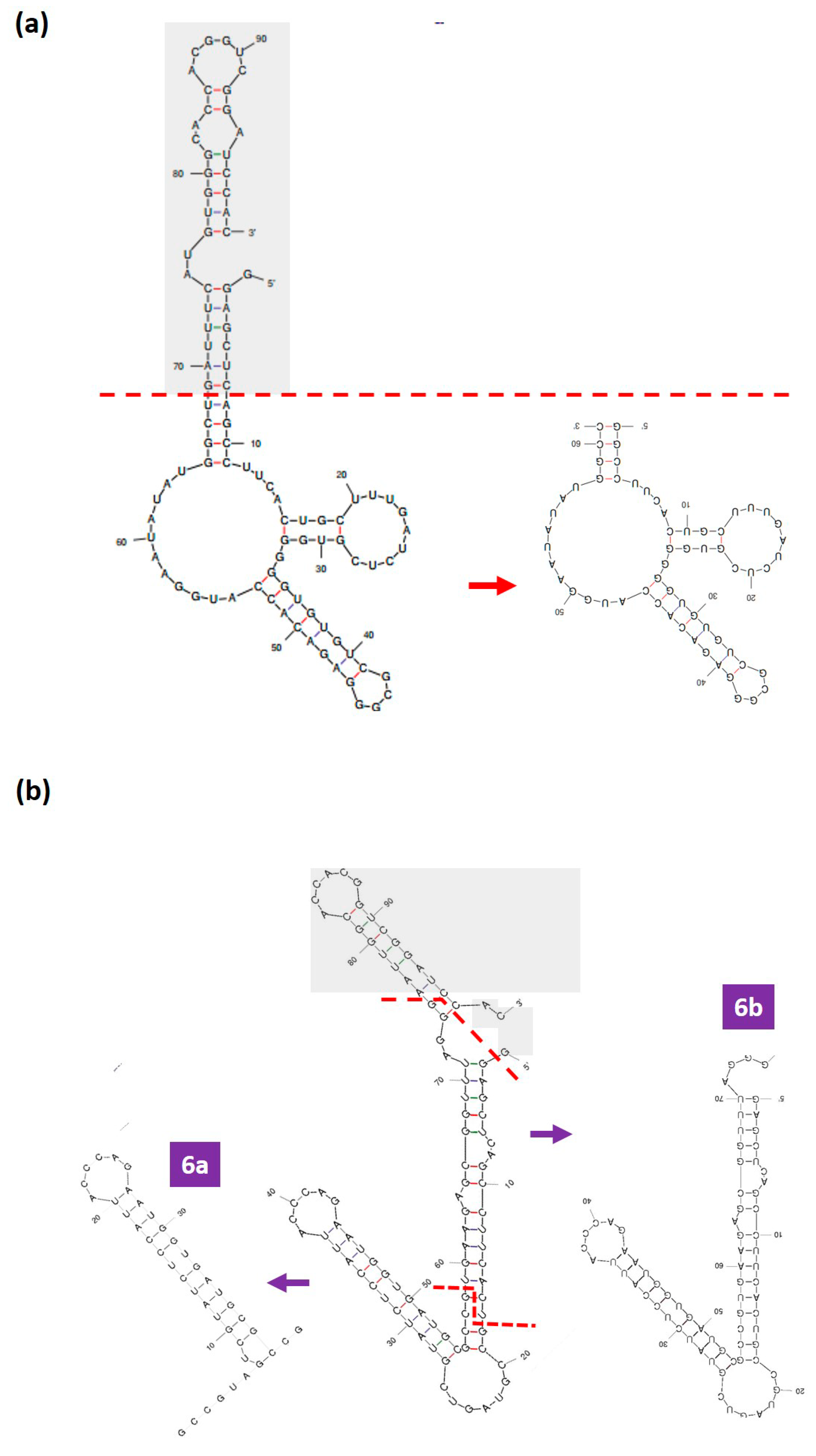
3.4. Rational Truncation of the Full-Length DENV Aptamers to Truncated Forms
3.5. Truncated Aptamers Display Higher Binding Scores Compared to Full-Length Aptamers, Suggesting an Improvement in their Binding Strength
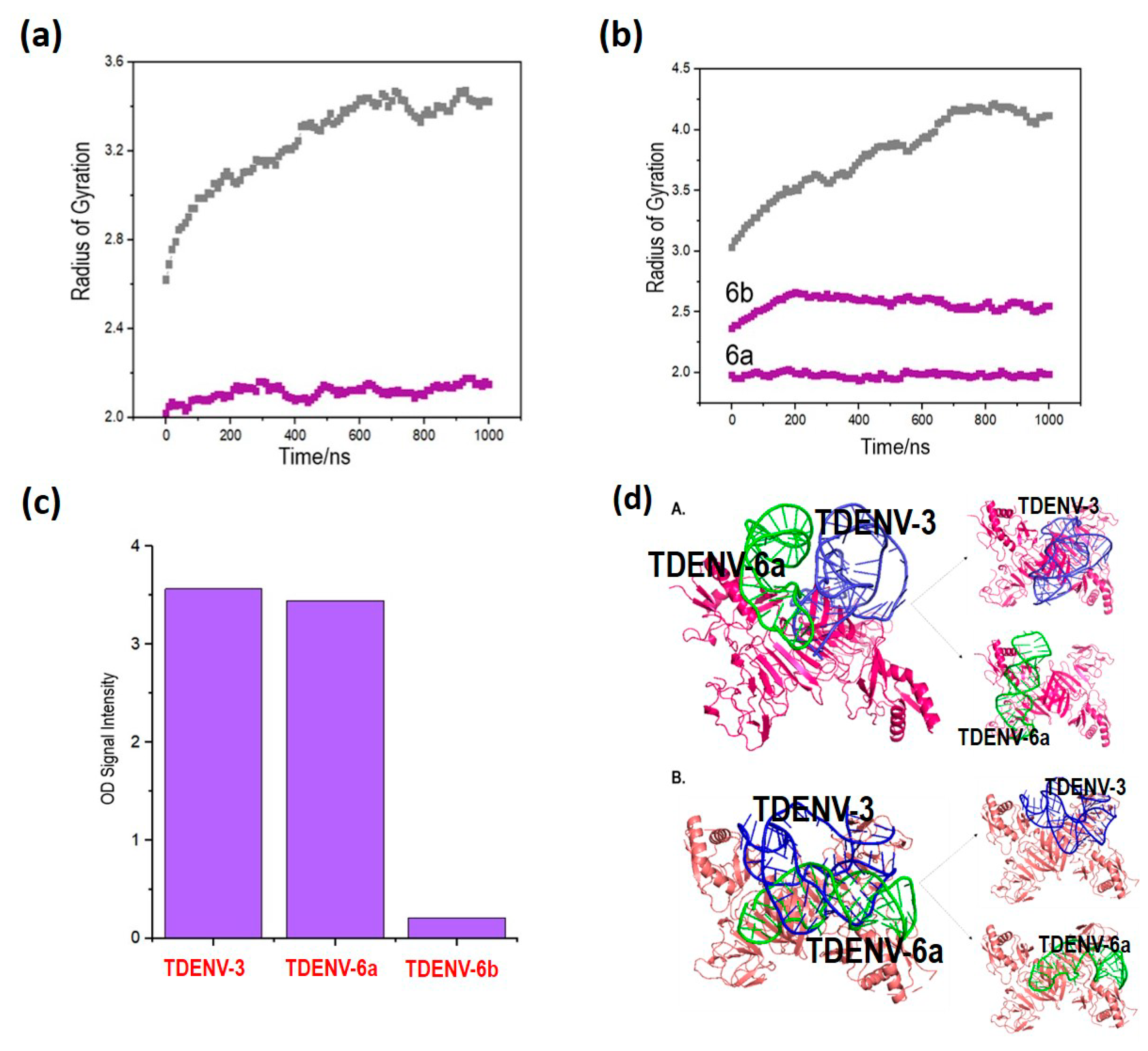
3.6. Lower Rg Values Corroborate Better Binding of the Truncated Aptamers Compared to Full-Length Aptamers
3.7. TDENV-3 and -6a as Potential Aptamers against Dengue NS1 as Corroborated by Direct ELASA
3.8. TDENV-3 and TDENV-6a Aptamers Occupy Distinct Epitopes of NS1, Suggesting Their Usage in Sandwich Assays
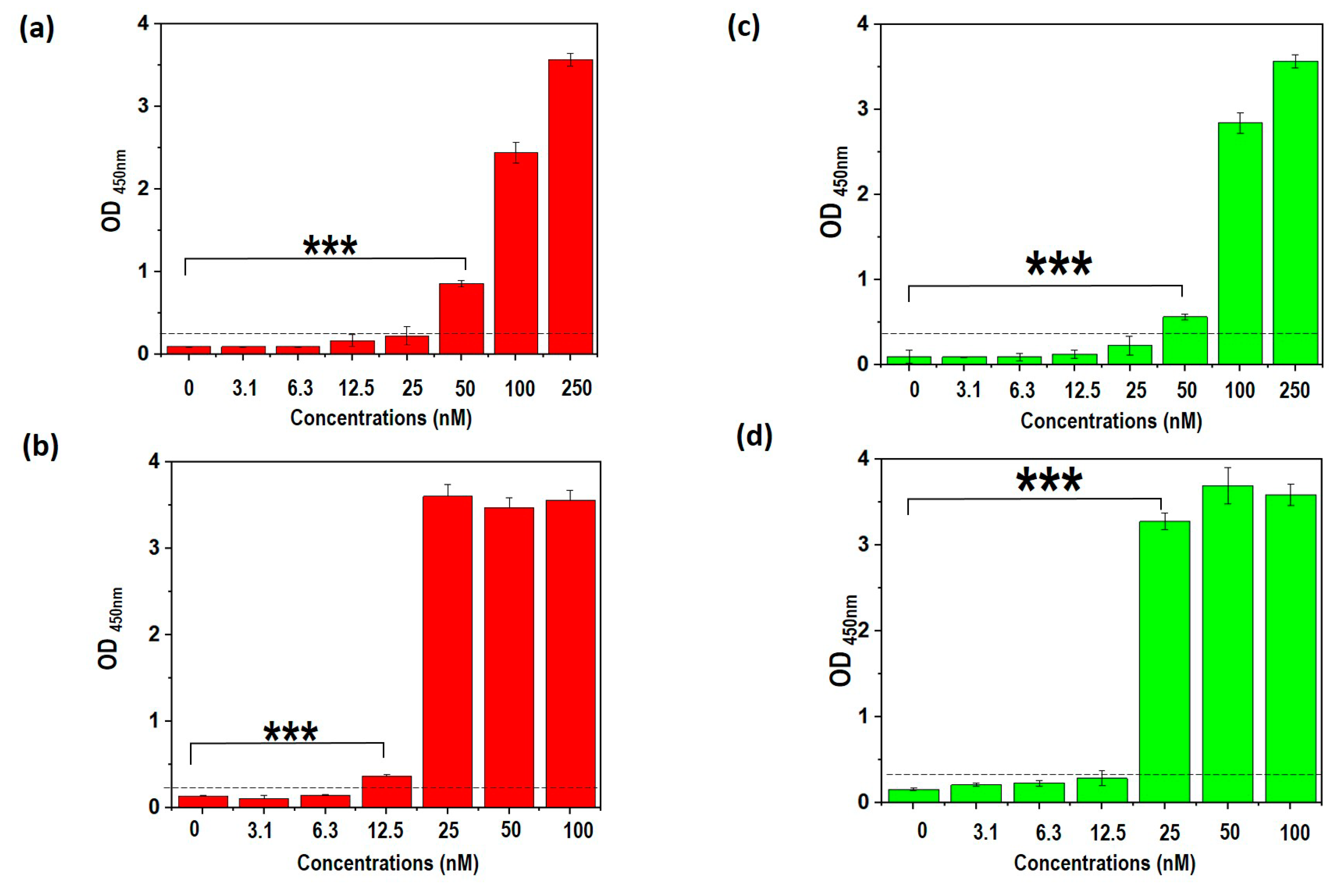
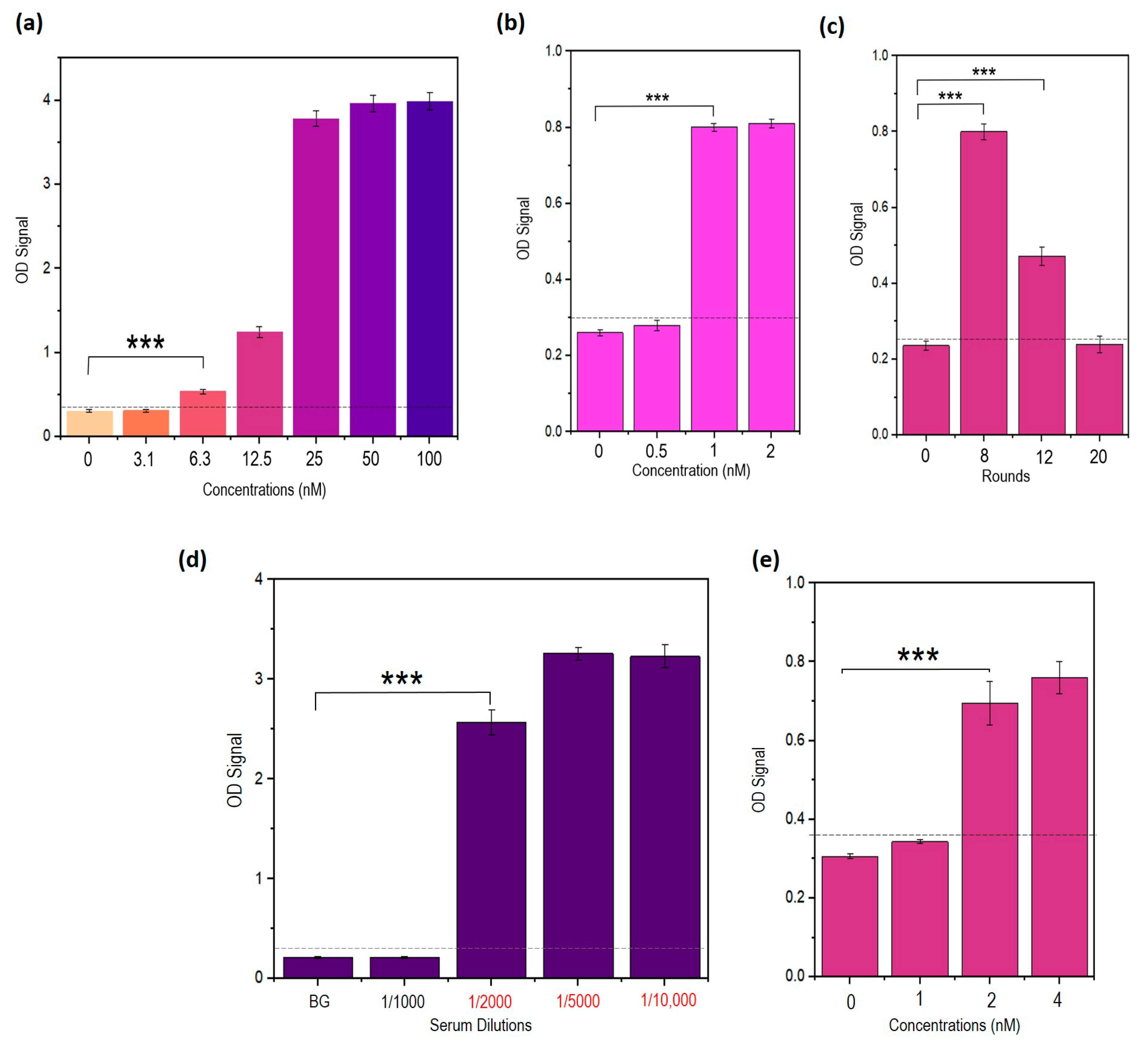
3.9. Truncated Aptamers Have a Better LOD Compared to Full-Length DENV
3.10. TDENV-3 and -6a Show High Specificity to Dengue NS1
3.11. TDENV-3 and -6a Selectively Target NS1 in Human Serum
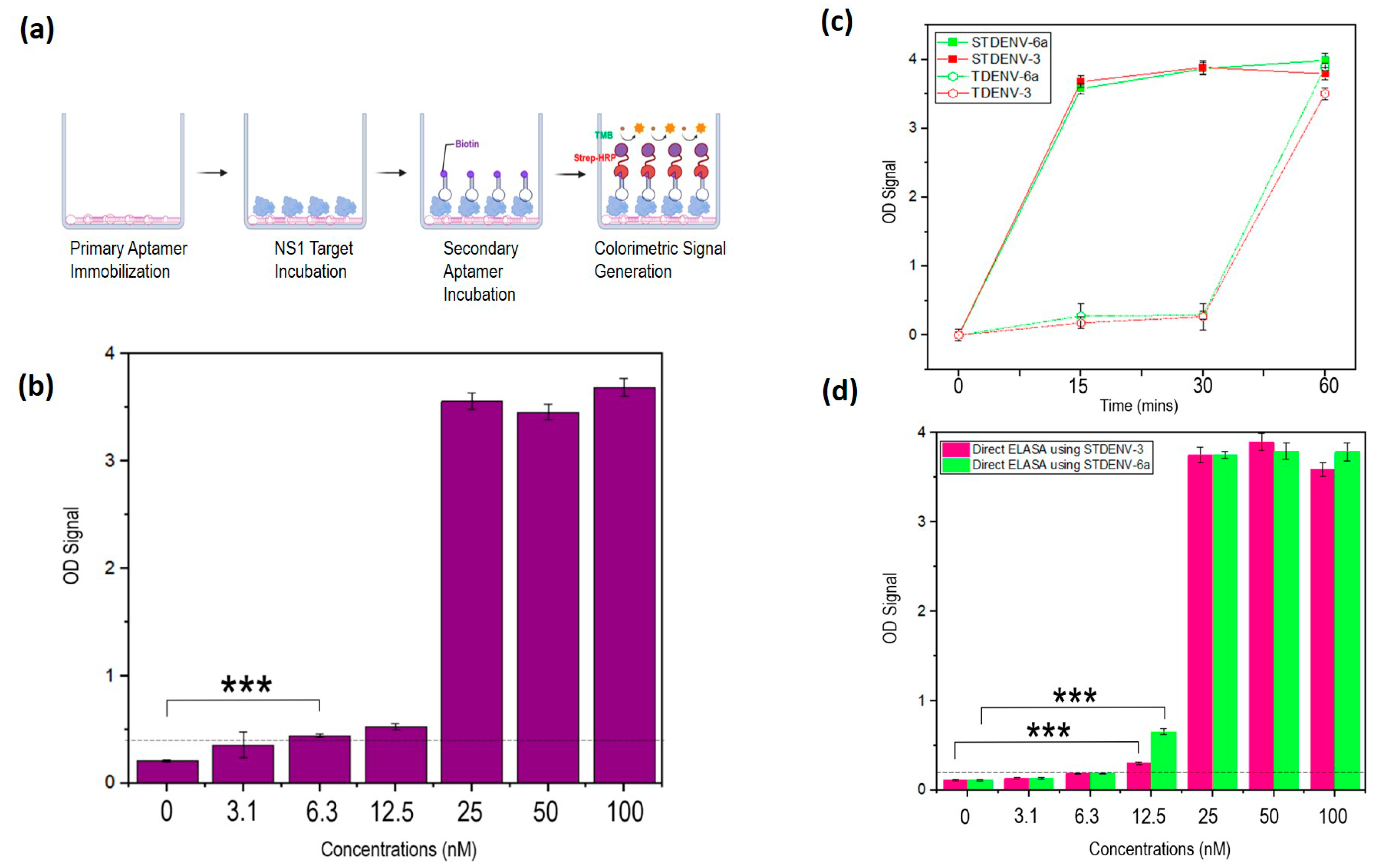
3.12. The Development of an Aptamer-Based Sandwich ELASA
3.13. Stabilized Aptamers Have Improved Binding Rates to the Target NS1
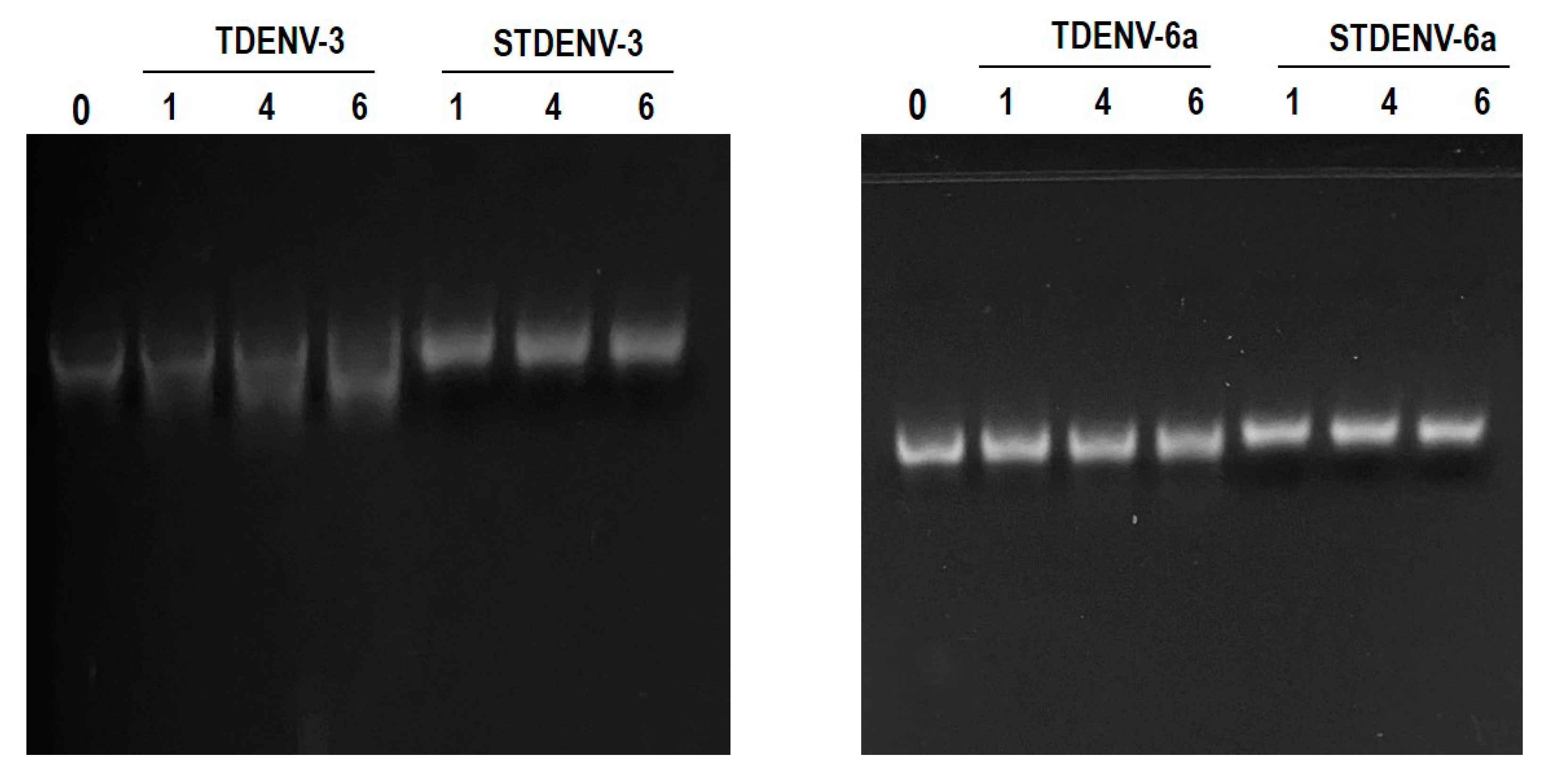
3.14. Both the Unmodified and Stabilized Aptamers Are Stable in Serum up to 6 h
3.15. Repeated Incubations Improved the Performance of the Sandwich ELASA
3.16. Developed Sandwich ELASA Detects Clinical NS1 Levels Indicative of Dengue Infection in Human Serum
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nani Mudin, R. Dengue Incidence and the Prevention and Control Program in Malaysia. Int. Med. J. Malaysia 2015, 14, 5–9. [Google Scholar] [CrossRef]
- Wiyono, L.; Rocha, I.C.N.; Cedeno, T.D.D.; Miranda, A.V.; Prisno, D.E.L. Dengue and COVID-19 infections in the ASEAN region: A concurrent outbreak of viral diseases. Epidemiol. Health 2021, 43, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Young, P.R.; Hilditch, P.A.; Bletchly, C.; Halloran, W. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 2000, 38, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Libraty, D.H.; Young, P.R.; Pickering, D.; Endy, T.P.; Kalayanarooj, S.; Green, S.; Vaughn, D.W.; Nisalak, A.; Ennis, F.A.; Rothman, A.L. High Circulating Levels of the Dengue Virus Nonstructural Protein NS1 Early in Dengue Illness Correlate with the Development of Dengue Hemorrhagic Fever. J. Infect. Dis. 2002, 186, 1165–1168. [Google Scholar] [CrossRef] [PubMed]
- Hermann, L.L.; Thaisomboonsuk, B.; Poolpanichupatam, Y.; Jarman, R.G.; Kalayanarooj, S.; Nisalak, A.; Yoon, I.K.; Fernandez, S. Evaluation of a Dengue NS1 Antigen Detection Assay Sensitivity and Specificity for the Diagnosis of Acute Dengue Virus Infection. PLoS Negl. Trop. Dis. 2014, 8, 1–8. [Google Scholar] [CrossRef]
- Citartan, M.; Tang, T.H. Recent developments of aptasensors expedient for point-of-care (POC) diagnostics. Talanta 2019, 199, 556–566. [Google Scholar] [CrossRef]
- Lee, K.H.; Zeng, H. Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal. Chem. 2017, 89, 12743–12748. [Google Scholar] [CrossRef]
- Zou, X.; Wu, J.; Gu, J.; Shen, L.; Mao, L. DNA aptamer against EV-A71 VP1 protein: Selection and application. Virol. J. 2021, 18, 1–10. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, Q.; Chen, J.; Ni, X.; Dai, J. A DNA Aptamer Based Method for Detection of SARS-CoV-2 Nucleocapsid Protein. Virol. Sin. 2020, 35, 351–354. [Google Scholar] [CrossRef]
- Citartan, M.; Tan, S.C.; Tang, T.H. A rapid and cost effective method in purifying small RNA. World J. Microbiol. Biotechnol. 2012, 28, 105–111. [Google Scholar] [CrossRef]
- Tsai, Y.T.; Liang, C.H.; Yu, J.H.; Huang, K.C.; Tung, C.H.; Wu, J.E.; Wu, Y.Y.; Chang, C.H.; Hong, T.M.; Chen, Y.L. A DNA Aptamer Targeting Galectin-1 as a Novel Immunotherapeutic Strategy for Lung Cancer. Mol. Ther. Nucleic Acids 2019, 18, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Murakami, K.; Summers, J.L.; Chen, C.H.B.; Bitan, G. RNA aptamers generated against oligomeric Aβ40 recognize common amyloid aptatopes with low specificity but high sensitivity. PLoS ONE 2009, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.; Arshad, S.; Seo, K.; Bowman, C.; Corley, M.; Jhaveri, S.D.; Ellington, A.D. In Vitro Selection of RNA Aptamers to a Protein Target by Filter Immobilization. Curr. Protoc. Mol. Biol. 2010, 89, 1–25. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Popenda, M.; Szachniuk, M.; Antczak, M.; Purzycka, K.J.; Lukasiak, P.; Bartol, N.; Blazewicz, J.; Adamiak, R.W. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012, 40, e112. [Google Scholar] [CrossRef]
- Scaturro, P.; Cortese, M.; Chatel-Chaix, L.; Fischl, W.; Bartenschlager, R. Dengue Virus Non-structural Protein 1 Modulates Infectious Particle Production via Interaction with the Structural Proteins. PLoS Pathog. 2015, 11, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Lebars, I.; Legrand, P.; Aimé, A.; Pinaud, N.; Fribourg, S.; Di Primo, C. Exploring TAR-RNA aptamer loop-loop interaction by X-ray crystallography, UV spectroscopy and surface plasmon resonance. Nucleic Acids Res. 2008, 36, 7146–7156. [Google Scholar] [CrossRef]
- Zimbres, F.M.; Tárnok, A.; Ulrich, H.; Wrenger, C. Aptamers: Novel molecules as diagnostic markers in bacterial and viral infections? Biomed Res. Int. 2013, 2013. [Google Scholar] [CrossRef]
- Sánchez-Luque, F.J.; Stich, M.; Manrubia, S.; Briones, C.; Berzal-Herranz, A. Efficient HIV-1 inhibition by a 16 nt-long RNA aptamer designed by combining in vitro selection and in silico optimisation strategies. Sci. Rep. 2014, 4, 1–10. [Google Scholar] [CrossRef]
- Rockey, W.M.; Hernandez, F.J.; Huang, S.Y.; Cao, S.; Howell, C.A.; Thomas, G.S.; Liu, X.Y.; Lapteva, N.; Spencer, D.M.; McNamara, J.O.; et al. Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther. 2011, 21, 299–314. [Google Scholar] [CrossRef]
- Navien, T.N.; Thevendran, R.; Hamdani, H.Y.; Tang, T.H.; Citartan, M. In silico molecular docking in DNA aptamer development. Biochimie 2021, 180, 54–67. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.J.; Van Dijk, M.; Bonvin, A.M.J.J. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010, 5, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Aguado, J.A.; Penner, G. Fluorescence polarization based displacement assay for the determination of small molecules with aptamers. Anal. Chem. 2008, 80, 8853–8855. [Google Scholar] [CrossRef]
- Torabi, R.; Bagherzadeh, K.; Ghourchian, H.; Amanlou, M. An investigation on the interaction modes of a single-strand DNA aptamer and RBP4 protein: A molecular dynamic simulations approach. Org. Biomol. Chem. 2016, 14, 8141–8153. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.J.; Chow, A.; Zheng, X.; Carrasco, L.R.; Cook, A.R.; Lye, D.C.; Ng, L.C.; Leo, Y.S. Simple Clinical and Laboratory Predictors of Chikungunya versus Dengue Infections in Adults. PLoS Negl. Trop. Dis. 2012, 6. [Google Scholar] [CrossRef]
- Fornazari, F.; Richini-Pereira, V.B.; Joaquim, S.F.; Nachtigall, P.G.; Langoni, H. Leptospirosis diagnosis among patients suspected of dengue fever in Brazil. J. Venom. Anim. Toxins Incl. Trop. Dis. 2021, 27, 1–7. [Google Scholar] [CrossRef]
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.B.; Tang, T.H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2014, 64, 392–403. [Google Scholar] [CrossRef]
- Citartan, M.; Gopinath, S.C.B.; Tang, T.H. Generation of RNA aptamer against rHuEPO-α by SELEX. Asian Pac. J. Trop Dis 2014, 4, 224. [Google Scholar] [CrossRef]
- Prabu, S.S.; Ch’ng, E.S.; Woon, P.Y.; Chen, J.H.; Tang, T.H.; Citartan, M. Unravelling the diagnostic and therapeutic potentialities of a novel RNA aptamer isolated against human pituitary tumour transforming gene 1 (PTTG1) protein. Anal. Chim. Acta 2020, 1138, 181–190. [Google Scholar] [CrossRef]
- Thirunavukarasu, D.; Chen, T.; Liu, Z.; Hongdilokkul, N.; Romesberg, F.E. Selection of 2′-Fluoro-Modified Aptamers with Optimized Properties. J. Am. Chem. Soc. 2017, 139, 2892–2895. [Google Scholar] [CrossRef] [PubMed]
- Gruenke, P.R.; Alam, K.K.; Singh, K.; Burke, D.H. 2’-fluoro-modified pyrimidines enhance affinity of RNA oligonucleotides to HIV-1 reverse transcriptase. RNA 2020, 26, 1667–1679. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Dauner, A.L.; Mitra, I.; Forshey, B.M.; Garcia, P.; Morrison, A.C.; Halsey, E.S.; Kochel, T.J.; Wu, S.J.L. Evaluation of dengue ns1 antigen rapid tests and elisa kits using clinical samples. PLoS ONE 2014, 9, e113411. [Google Scholar] [CrossRef] [PubMed]


| SELEX Cycle | RNA Pool (µM) | Competitor (µM) | NS1 Protein (µM) | PCR Amplification Cycles |
|---|---|---|---|---|
| 1 | 6.6 | 10.2 | 2.5 | 8 |
| 2 | 3.9 | 30.6 | 2.0 | 8 |
| 3 * | 2.1 | 50.9 | 1.5 | 8 |
| 4 | 1.6 | 81.6 | 1.0 | 8 |
| 5 * | 1.1 | 112.2 | 0.75 | 8 |
| 6 | 0.8 | 152.9 | 0.5 | 8 |
| 7 | 0.4 | 193.7 | 0.25 | 6 |
| 8 * | 0.3 | 203.9 | 0.125 | 6 |
| 9 | 0.3 | 203.9 | 0.125 | 6 |
| 10 | 0.2 | 203.9 | 0.1 | 5 |
| 11 * | 0.1 | 203.9 | 0.05 | 8 |
| Class | Sequence | Number of Identical Sequences | Percentage of Appearance (%) |
|---|---|---|---|
| DENV-1 | GGAGCUCAGCCUUCACUGCUUAGUCCCGUCAACGUCGGAGGUGACAGCCGUUGGCGGGCCGGCUUGGCGUUAACAUGAAGGCACCACGGUCGGAUCCAC | 5 | 17.86 |
| DENV-2 | GGAGCUCAGCCUUCACUGCUGUCAGAUAACAUGCAUGAUAGACUGAUGAUCGUCCAUGUUUGAAACUGAUCAGUAUCGAGGCACCACGGUCGGAUCCAC | 3 | 10.71 |
| DENV-3 | GGAGCUCAGCCUUCACUGCUUUGAUCUCGUGGGGGUGUGUCGCGGGAGACACCAUGGAAUAUAUGGCUGAUUUCAUGUGGGCACCACGGUCGGAUCCAC | 5 | 17.86 |
| DENV-4 | GGAGCUCAGCCUUCACUGCGUGAUGCGUAGCUCGAUACAAUGGUUGUCUUAAAAGUGACGCCUUUGUCGAAAACGAGAUGGCACCACGGUCGGAUCCAC | 5 | 17.86 |
| DENV-5 | GGAGCUCAGCCUUCACUGCUUUAGGUGCCCAUAGGGACGGGUUGGGAUCAUAGUUCGUGAAGGUGCUAUGAUACGGGAGGGCACCACGGUCGGAUCCAC | 5 | 17.86 |
| DENV-6 | GGAGCUCAGCCUUCACUGCCGUAGUCGUAUCUCCAUUACCCAGAAUGGUGAUGCGCCGUGAAGAGCGGUUUAGGGAAUUGGCACCACGGUCGGAUCCAC | 3 | 10.71 |
| DENV-7 | GGAGCUCAGCCUUCACUGCUUGCUUUUGGGUGCCGUACGCAUUGCGGCAGGGGGAAGAGGAGGGUAGCGACCAGUCGAAGGCACCACGGUCGGAUCCAC | 2 | 7.14 |
| TDENV | Haddock Z-Score | Autodock Z-Score | Patchdock Z-Score | Total Z-Score |
| 3 | −1.2 | −2.37 | −1.77 | −5.34 |
| 4 | −1.3 | −1.29 | −1.66 | −4.25 |
| 6a | −2.2 | −1.34 | −1.72 | −5.26 |
| 6b | −1.5 | −1.45 | −1.82 | −4.77 |
| DENV | Haddock Z-score | Autodock Z-score | Patchdock Z-score | Total Z-score |
| 3 | −1.3 | −1.40 | −1.54 | −4.24 |
| 4 | −1.3 | −2.10 | −2.04 | −5.44 |
| 6 | −1.6 | −1.38 | −1.85 | −4.83 |
| No. | Control Sequences | Haddock Z-Score | Autodock Z-Score | Patchdock Z-Score | Total Z-Score |
|---|---|---|---|---|---|
| C1 | AAUACAAAUUGUGUU | −2.3 | −2.00 | −0.30 | −4.60 |
| C2 | CAUAGCAGACAGCUAUC | −2.5 | −1.80 | −0.50 | −4.80 |
| C3 | AGUAAUACUCGCUGC | −1.6 | −1.90 | −1.00 | −4.50 |
| Docked Models | Haddock Z-Score | Patchdock Z-Score |
|---|---|---|
| TDENV-3/NS1—TDENV-6a | −2.3 | −1.76 |
| TDENV-6a/NS1—TDENV-3 | −1.6 | −1.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thevendran, R.; Rogini, S.; Leighton, G.; Mutombwera, A.; Shigdar, S.; Tang, T.-H.; Citartan, M. The Diagnostic Potential of RNA Aptamers against the NS1 Protein of Dengue Virus Serotype 2. Biology 2023, 12, 722. https://doi.org/10.3390/biology12050722
Thevendran R, Rogini S, Leighton G, Mutombwera A, Shigdar S, Tang T-H, Citartan M. The Diagnostic Potential of RNA Aptamers against the NS1 Protein of Dengue Virus Serotype 2. Biology. 2023; 12(5):722. https://doi.org/10.3390/biology12050722
Chicago/Turabian StyleThevendran, Ramesh, Sivalingam Rogini, Glenn Leighton, Atherton Mutombwera, Sarah Shigdar, Thean-Hock Tang, and Marimuthu Citartan. 2023. "The Diagnostic Potential of RNA Aptamers against the NS1 Protein of Dengue Virus Serotype 2" Biology 12, no. 5: 722. https://doi.org/10.3390/biology12050722
APA StyleThevendran, R., Rogini, S., Leighton, G., Mutombwera, A., Shigdar, S., Tang, T.-H., & Citartan, M. (2023). The Diagnostic Potential of RNA Aptamers against the NS1 Protein of Dengue Virus Serotype 2. Biology, 12(5), 722. https://doi.org/10.3390/biology12050722





