The Post-Antibiotic Era: A New Dawn for Bacteriophages
Abstract
Simple Summary
Abstract
1. Introduction
2. Understanding Phages
2.1. Phage Discovery
2.2. Phage Classification
2.3. Recognition and Infection of Phage
3. Application of Phages
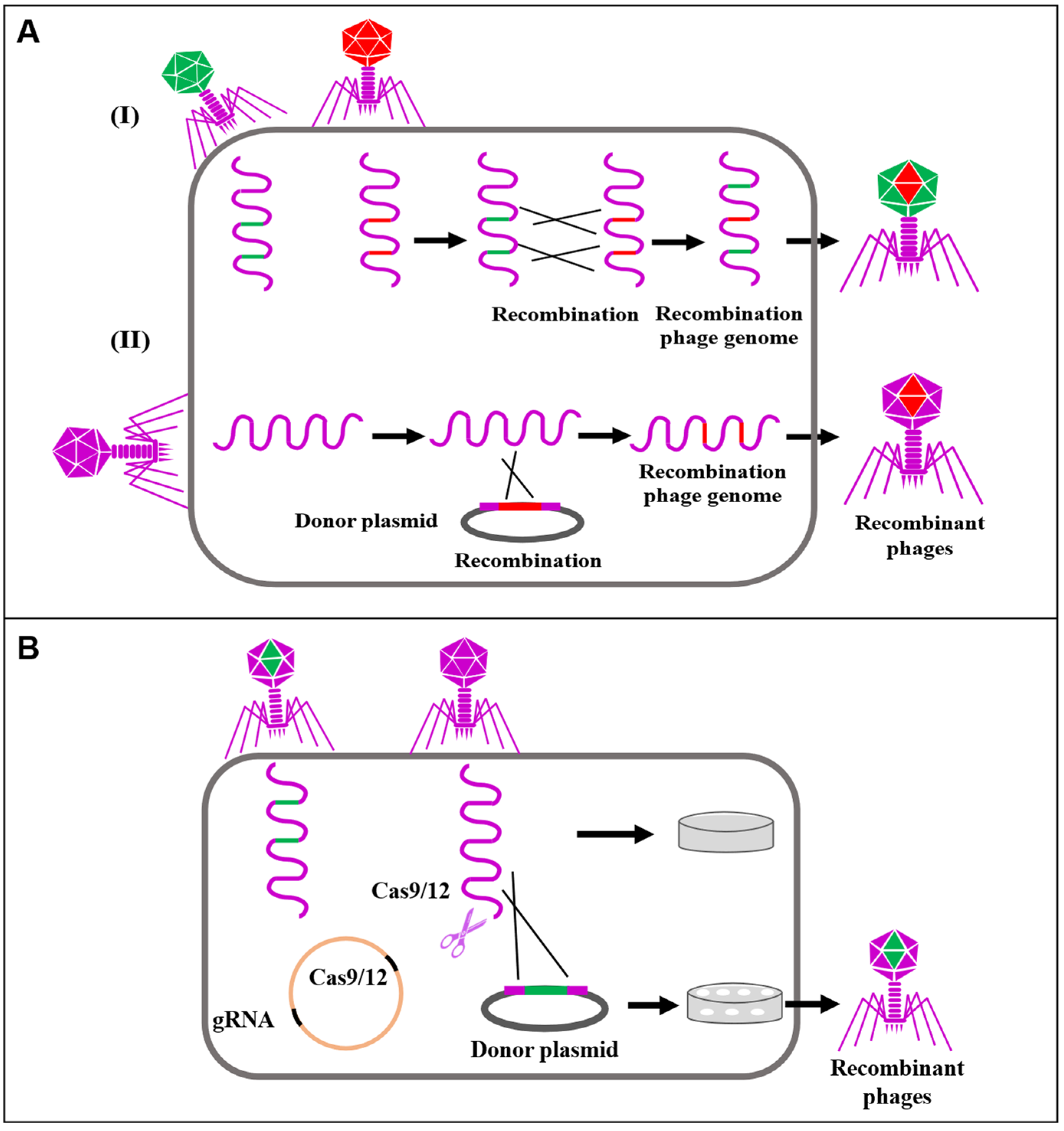
3.1. Gene Editing Using Bacteriophages
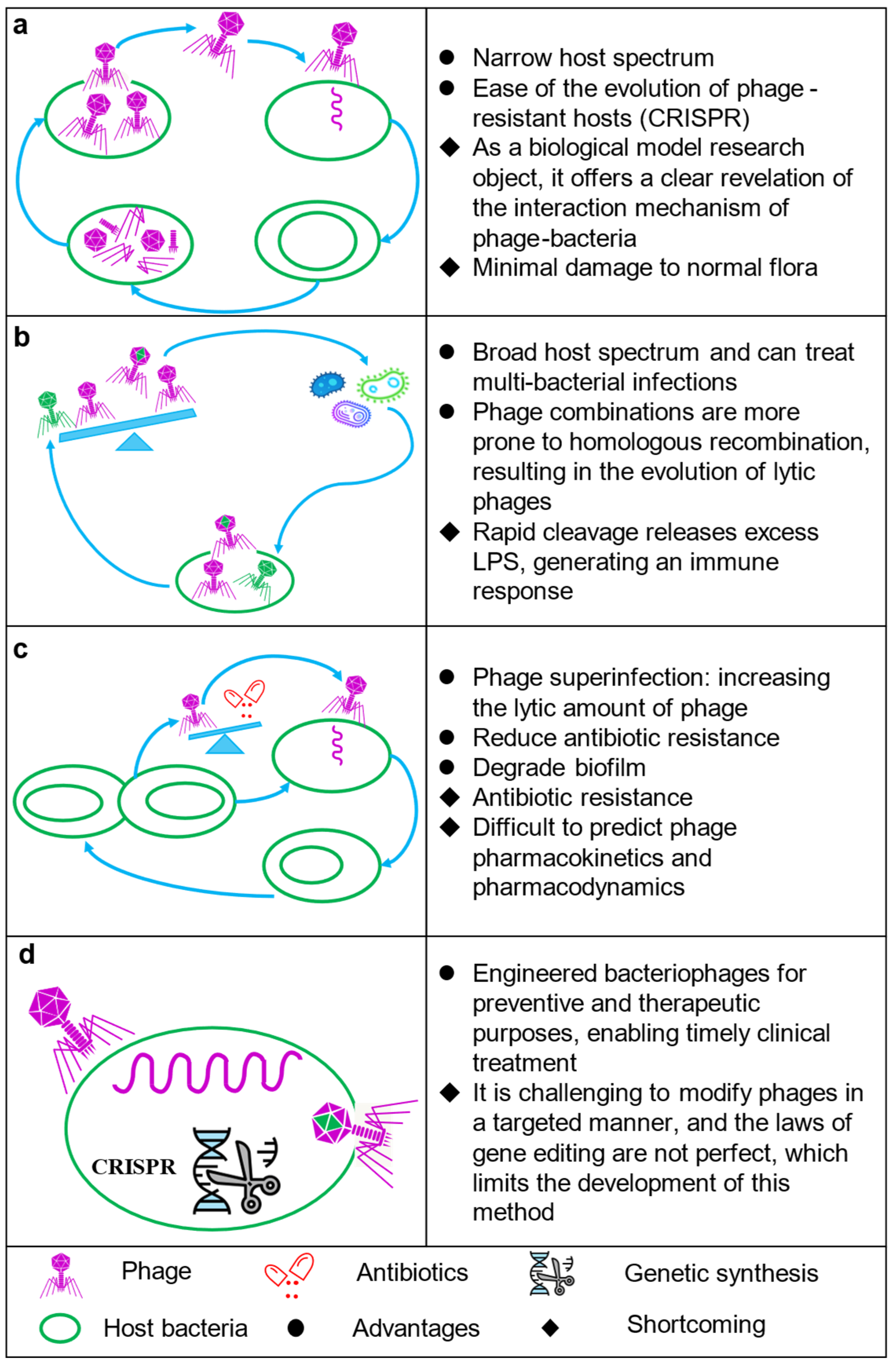
3.2. Phage Therapy
3.2.1. Phage Immune Mechanisms
3.2.2. Phages in the Gut
3.2.3. Biosafety of Phage Therapy
4. Conclusions and Future Directions
- Precise treatment: Synthetic omics can use phages as biological agents to treat MDR bacterial infections and antigen delivery to induce specific immune responses in the body.
- Regulating gut microbes: Gut phages can influence the host response by altering the composition of gut biota. However, theoretical models are still needed to support the phage-induced disturbance responses, including the mechanism of repairing and maintaining gut biota stability.
- Antibiotic alternatives: Phages can be used as antibiotic substitutes or supplements in animal husbandry, including daily healthcare and slaughter of livestock and poultry, to prevent food-borne bacteria from entering the food chain.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brum, J.R.; Sullivan, M.B. Rising to the challenge: Accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 2015, 13, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Makarova, K.S.; Gussow, A.B.; Krupovic, M.; Segall, A.; Edwards, R.A.; Koonin, E.V. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat. Microbiol. 2018, 3, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Altamirano, F.L.G.; Barr, J.J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef] [PubMed]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; Van Der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef]
- Adler, B.A.; Hessler, T.; Cress, B.F.; Lahiri, A.; Mutalik, V.K.; Barrangou, R.; Banfield, J.; Doudna, J.A. Broad-spectrum CRISPR-Cas13a enables efficient phage genome editing. Nat. Microbiol. 2022, 7, 1967–1979. [Google Scholar] [CrossRef]
- Meaden, S.; Koskella, B. Exploring the risks of phage application in the environment. Front. Microbiol. 2013, 4, 358. [Google Scholar] [CrossRef]
- Pirnay, J.-P.; Ferry, T.; Resch, G. Recent progress toward the implementation of phage therapy in Western medicine. FEMS Microbiol. Rev. 2022, 46, fuab040. [Google Scholar] [CrossRef]
- Stracy, M.; Snitser, O.; Yelin, I.; Amer, Y.; Parizade, M.; Katz, R.; Rimler, G.; Wolf, T.; Herzel, E.; Koren, G. Minimizing treatment-induced emergence of antibiotic resistance in bacterial infections. Science 2022, 375, 889–894. [Google Scholar] [CrossRef]
- Taylor, J.; Hafner, M.; Yerushalmi, E.; Smith, R.; Bellasio, J.; Vardavas, R.; Bienkowska-Gibbs, T.; Rubin, J. Estimating the Economic Costs of Antimicrobial Resistance. In Model and Results; RAND EUROPE, Ed.; report; RAND Corporation: Cambridge, UK, 2014; Available online: https://www.rand.org/pubs/research_reports/RR911.html (accessed on 16 March 2023).
- Ahmad, M.; Khan, A.U. Global economic impact of antibiotic resistance: A review. J. Glob. Antimicrob. Resist. 2019, 19, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Barış, E.; Go, D.S.; Lofgren, H.; Osorio-Rodarte, I.; Thierfelder, K. Assessing the global poverty effects of antimicrobial resistance. World Dev. 2018, 111, 148–160. [Google Scholar] [CrossRef]
- Qiao, M.; Ying, G.-G.; Singer, A.C.; Zhu, Y.-G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef]
- Ben, Y.; Fu, C.; Hu, M.; Liu, L.; Wong, M.H.; Zheng, C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: A review. Environ. Res. 2019, 169, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef]
- Le Page, G.; Gunnarsson, L.; Snape, J.; Tyler, C.R. Integrating human and environmental health in antibiotic risk assessment: A critical analysis of protection goals, species sensitivity and antimicrobial resistance. Environ. Int. 2017, 109, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Vikesland, P.J.; Pruden, A.; Alvarez, P.J.; Aga, D.; Bu, H.; Li, X.-D.; Manaia, C.M.; Nambi, I.; Wigginton, K.; Zhang, T. Toward a comprehensive strategy to mitigate dissemination of environmental sources of antibiotic resistance. Environ. Sci. Technol. 2017, 51, 13061–13069. [Google Scholar] [CrossRef]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Xuan, G.; Lin, H.; Tan, L.; Zhao, G.; Wang, J. Quorum sensing promotes phage infection in Pseudomonas aeruginosa PAO1. Mbio 2022, 13, e03174-21. [Google Scholar] [CrossRef]
- Twort, F.W. An investigation on the nature of ultra-microscopic viruses. Acta Kravsi. 1961, 186, 1241–1243. [Google Scholar] [CrossRef]
- D’Herelle, F. On an invisible microbe antagonistic toward dysenteric bacilli: Brief note by Mr. F. D’Herelle, presented by Mr. Roux. 1917. Res. Microbiol. 2007, 158, 553–554. [Google Scholar] [PubMed]
- Breitbart, M. Marine viruses: Truth or dare. Annu. Rev. Mar. Sci. 2012, 4, 425–448. [Google Scholar] [CrossRef]
- Williamson, K.E.; Fuhrmann, J.J.; Wommack, K.E.; Radosevich, M. Viruses in Soil Ecosystems: An Unknown Quantity Within an Unexplored Territory. Annu. Rev. Virol. 2017, 4, 201–219. [Google Scholar] [CrossRef]
- Salmond, G.P.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol. 2015, 13, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Nordmann, P.; Ortiz de la Rosa, J.M.; Kutateladze, M.; Gatermann, S.; Corbellino, M. A phage-based decolonisation strategy against pan-resistant enterobacterial strains. Lancet Infect. Dis. 2020, 20, 525–526. [Google Scholar] [CrossRef]
- Chng, K.R.; Li, C.; Bertrand, D.; Ng, A.H.Q.; Kwah, J.S.; Low, H.M.; Tong, C.; Natrajan, M.; Zhang, M.H.; Xu, L. Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nat. Med. 2020, 26, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Jofre, J.; Muniesa, M. Bacteriophage isolation and characterization: Phages of Escherichia coli. In Horizontal Gene Transfer; Springer: New York, NY, USA, 2020; pp. 61–79. [Google Scholar]
- Guerin, E.; Shkoporov, A.N.; Stockdale, S.R.; Comas, J.C.; Khokhlova, E.V.; Clooney, A.G.; Daly, K.M.; Draper, L.A.; Stephens, N.; Scholz, D. Isolation and characterisation of ΦcrAss002, a crAss-like phage from the human gut that infects Bacteroides xylanisolvens. Microbiome 2021, 9, 89. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Khokhlova, E.V.; Fitzgerald, C.B.; Stockdale, S.R.; Draper, L.A.; Ross, R.P.; Hill, C. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 2018, 9, 4781. [Google Scholar] [CrossRef]
- Federici, S.; Kviatcovsky, D.; Valdés-Mas, R.; Elinav, E. Microbiome-phage interactions in inflammatory bowel disease. Clin. Microbiol. Infect. 2022; in press. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Phage classification and characterization. Bacteriophages 2009, 501, 127–140. [Google Scholar]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity; genomics; phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef]
- Guttman, B.; Raya, R.; Kutter, E. Basic phage biology. Bacteriophages Biol. Appl. 2005, 4, 30–63. [Google Scholar]
- Chen, P.; Sun, H.; Ren, H.; Liu, W.; Li, G.; Zhang, C. LamB, OmpC, and the core lipopolysaccharide of Escherichia coli K-12 function as receptors of bacteriophage Bp7. J. Virol. 2020, 94, e00325-20. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.R.; Dobias, D.T.; Weitz, J.S.; Barrick, J.E.; Quick, R.T.; Lenski, R.E. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 2012, 335, 428–432. [Google Scholar] [CrossRef]
- Chowdhury, S.; Carter, J.; Rollins, M.F.; Golden, S.M.; Jackson, R.N.; Hoffmann, C.; Bondy-Denomy, J.; Maxwell, K.L.; Davidson, A.R.; Fischer, E.R. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell 2017, 169, 47–57.e11. [Google Scholar] [CrossRef]
- Guo, T.W.; Bartesaghi, A.; Yang, H.; Falconieri, V.; Rao, P.; Merk, A.; Eng, E.T.; Raczkowski, A.M.; Fox, T.; Earl, L.A. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell 2017, 171, 414–426.e12. [Google Scholar] [CrossRef]
- Lu, M.; Stierhof, Y.; Henning, U. Location and unusual membrane topology of the immunity protein of the Escherichia coli phage T4. J. Virol. 1993, 67, 4905–4913. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Vos, T.; Lim, S.S.; Abbafati, C.; Abbas, K.M.; Abbasi, M.; Abbasifard, M.; Abbasi-Kangevari, M.; Abbastabar, H.; Abd-Allah, F.; Abdelalim, A. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Ikuta, K.S.; Swetschinski, L.R.; Aguilar, G.R.; Sharara, F.; Mestrovic, T.; Gray, A.P.; Weaver, N.D.; Wool, E.E.; Han, C.; Hayoon, A.G. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef]
- White, R.A. The Future of Virology is Synthetic. Msystems 2021, 6, e00770-21. [Google Scholar] [CrossRef] [PubMed]
- Venter, J.C.; Glass, J.I.; Hutchison, C.A.; Vashee, S. Synthetic chromosomes, genomes, viruses, and cells. Cell 2022, 185, 2708–2724. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Batra, H.; Dong, J.; Chen, C.; Rao, V.B.; Tao, P. Genetic engineering of bacteriophages against infectious diseases. Front. Microbiol. 2019, 10, 954. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.D.; Parks, A.R.; Adhya, S.; Rattray, A.J.; Court, D.L. λ Recombineering used to engineer the genome of phage T7. Antibiotics 2020, 9, 805. [Google Scholar] [CrossRef]
- Hatoum-Aslan, A. Phage genetic engineering using CRISPR–Cas systems. Viruses 2018, 10, 335. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Box, A.M.; McGuffie, M.J.; O’Hara, B.J.; Seed, K.D. Functional analysis of bacteriophage immunity through a type IE CRISPR-Cas system in Vibrio cholerae and its application in bacteriophage genome engineering. J. Bacteriol. 2016, 198, 578–590. [Google Scholar] [CrossRef]
- Lemay, M.-L.; Tremblay, D.M.; Moineau, S. Genome engineering of virulent lactococcal phages using CRISPR-Cas9. ACS Synth. Biol. 2017, 6, 1351–1358. [Google Scholar] [CrossRef]
- Bari, S.N.; Walker, F.C.; Cater, K.; Aslan, B.; Hatoum-Aslan, A. Strategies for editing virulent staphylococcal phages using CRISPR-Cas10. ACS Synth. Biol. 2017, 6, 2316–2325. [Google Scholar] [CrossRef]
- Tao, P.; Wu, X.; Tang, W.-C.; Zhu, J.; Rao, V. Engineering of bacteriophage T4 genome using CRISPR-Cas9. ACS Synth. Biol. 2017, 6, 1952–1961. [Google Scholar] [CrossRef]
- Roach, D.R.; Debarbieux, L. Phage therapy: Awakening a sleeping giant. Emerg. Top. Life Sci. 2017, 1, 93–103. [Google Scholar] [PubMed]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Shuwen, H.; Kefeng, D. Intestinal phages interact with bacteria and are involved in human diseases. Gut Microbes 2022, 14, 2113717. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Zhu, T. Potential of therapeutic bacteriophages in nosocomial infection management. Front. Microbiol. 2021, 12, 638094. [Google Scholar] [CrossRef]
- Loganathan, A.; Manohar, P.; Eniyan, K.; VinodKumar, C.; Leptihn, S.; Nachimuthu, R. Phage therapy as a revolutionary medicine against Gram-positive bacterial infections. Beni-Suef Univ. J. Basic Appl. Sci. 2021, 10, 49. [Google Scholar] [CrossRef]
- Khatami, A.; Lin, R.C.; Petrovic-Fabijan, A.; Alkalay-Oren, S.; Almuzam, S.; Britton, P.N.; Brownstein, M.J.; Dao, Q.; Fackler, J.; Hazan, R. Bacterial lysis, autophagy and innate immune responses during adjunctive phage therapy in a child. EMBO Mol. Med. 2021, 13, e13936. [Google Scholar] [CrossRef]
- Rao, S.; Betancourt-Garcia, M.; Kare-Opaneye, Y.O.; Swierczewski, B.E.; Bennett, J.W.; Horne, B.A.; Fackler, J.; Hernandez, L.P.S.; Brownstein, M.J. Critically ill patient with multidrug-resistant Acinetobacter baumannii respiratory infection successfully treated with intravenous and nebulized bacteriophage therapy. Antimicrob. Agents Chemother. 2022, 66, e00824-21. [Google Scholar] [CrossRef]
- Roach, D.R.; Donovan, D.M. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 2015, 5, e1062590. [Google Scholar] [CrossRef]
- Gogokhia, L.; Buhrke, K.; Bell, R.; Hoffman, B.; Brown, D.G.; Hanke-Gogokhia, C.; Ajami, N.J.; Wong, M.C.; Ghazaryan, A.; Valentine, J.F. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe 2019, 25, 285–299. [Google Scholar] [CrossRef]
- Sweere, J.M.; Van Belleghem, J.D.; Ishak, H.; Bach, M.S.; Popescu, M.; Sunkari, V.; Kaber, G.; Manasherob, R.; Suh, G.A.; Cao, X. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 2019, 363, eaat9691. [Google Scholar] [CrossRef]
- Górski, A.; Borysowski, J.; Mi, R. Bacteriophage interactions with epithelial cells: Therapeutic implications. Front. Microbiol. 2021, 11, 631161. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.K.; Maurice, C.F. Ménage à trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol. 2017, 15, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Hodyra-Stefaniak, K.; Miernikiewicz, P.; Drapała, J.; Drab, M.; Jończyk-Matysiak, E.; Lecion, D.; Kaźmierczak, Z.; Beta, W.; Majewska, J.; Harhala, M. Mammalian Host-Versus-Phage immune response determines phage fate in vivo. Sci. Rep. 2015, 5, 14802. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, Z.; Zou, T.; Chen, J.; Li, G.; Zheng, L.; Li, S.; You, J. Bacteriophage as an alternative to antibiotics promotes growth performance by regulating intestinal inflammation, intestinal barrier function and gut microbiota in weaned piglets. Front. Vet. Sci. 2021, 8, 623899. [Google Scholar] [CrossRef] [PubMed]
- Dufour, N.; Delattre, R.; Ricard, J.-D.; Debarbieux, L. The lysis of pathogenic Escherichia coli by bacteriophages releases less endotoxin than by β-lactams. Clin. Infect. Dis. 2017, 64, 1582–1588. [Google Scholar] [CrossRef]
- Krut, O.; Bekeredjian-Ding, I. Contribution of the immune response to phage therapy. J. Immunol. 2018, 200, 3037–3044. [Google Scholar] [CrossRef]
- Huh, H.; Wong, S.; Jean, J.S.; Slavcev, R. Bacteriophage interactions with mammalian tissue: Therapeutic applications. Adv. Drug Deliv. Rev. 2019, 145, 4–17. [Google Scholar] [CrossRef]
- Gogokhia, L.; Round, J.L. Immune–bacteriophage interactions in inflammatory bowel diseases. Curr. Opin. Virol. 2021, 49, 30–35. [Google Scholar] [CrossRef]
- Majewska, J.; Beta, W.; Lecion, D.; Hodyra-Stefaniak, K.; Kłopot, A.; Kaźmierczak, Z.; Miernikiewicz, P.; Piotrowicz, A.; Ciekot, J.; Owczarek, B. Oral application of T4 phage induces weak antibody production in the gut and in the blood. Viruses 2015, 7, 4783–4799. [Google Scholar] [CrossRef]
- Reyes, A.; Haynes, M.; Hanson, N.; Angly, F.E.; Heath, A.C.; Rohwer, F.; Gordon, J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010, 466, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Maurice, C.F. Considering the other half of the gut microbiome: Bacteriophages. Msystems 2019, 4, e00102-19. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Park, E.-J.; Roh, S.W.; Bae, J.-W. Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 2011, 77, 8062–8070. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.; Semenkovich, N.P.; Whiteson, K.; Rohwer, F.; Gordon, J.I. Going viral: Next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 2012, 10, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.A.; Berger, B.; Deng, Y.; Kieser, S.; Foata, F.; Moine, D.; Descombes, P.; Sultana, S.; Huq, S.; Bardhan, P.K. Oral application of E scherichia coli bacteriophage: Safety tests in healthy and diarrheal children from B angladesh. Environ. Microbiol. 2017, 19, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.M.; Lin, H.C. A theoretical model of temperate phages as mediators of gut microbiome dysbiosis. F1000Research 2019, 8, 997. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.R.; Lee, H.H.; Spina, C.S.; Collins, J.J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 2013, 499, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Bearson, B.L.; Allen, H.K.; Brunelle, B.W.; Lee, I.S.; Casjens, S.R.; Stanton, T.B. The agricultural antibiotic carbadox induces phage-mediated gene transfer in Salmonella. Front. Microbiol. 2014, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Novick, R.P. Phage-mediated intergeneric transfer of toxin genes. Science 2009, 323, 139–141. [Google Scholar] [CrossRef]
- Cepko, L.C.; Garling, E.E.; Dinsdale, M.J.; Scott, W.P.; Bandy, L.; Nice, T.; Faber-Hammond, J.; Mellies, J.L. Myoviridae phage PDX kills enteroaggregative Escherichia coli without human microbiome dysbiosis. J. Med. Microbiol. 2020, 69, 309. [Google Scholar] [CrossRef]
- Hsu, B.B.; Gibson, T.E.; Yeliseyev, V.; Liu, Q.; Lyon, L.; Bry, L.; Silver, P.A.; Gerber, G.K. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe 2019, 25, 803–814.e5. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Guo, X.; Dong, K.; Zhang, Y.; Li, Q.; Zhu, Y.; Zeng, L.; Tang, R.; Li, L. Safety assessment of Staphylococcus phages of the family Myoviridae based on complete genome sequences. Sci. Rep. 2017, 7, 41259. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Loessner, M.J. Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 2014, 4, e28137. [Google Scholar] [CrossRef] [PubMed]
- Suh, G.A.; Lodise, T.P.; Tamma, P.D.; Knisely, J.M.; Alexander, J.; Aslam, S.; Barton, K.D.; Bizzell, E.; Totten, K.M.; Campbell, J.L. Considerations for the use of phage therapy in clinical practice. Antimicrob. Agents Chemother. 2022, 66, e02071-21. [Google Scholar] [CrossRef]
- Uyttebroek, S.; Chen, B.; Onsea, J.; Ruythooren, F.; Debaveye, Y.; Devolder, D.; Spriet, I.; Depypere, M.; Wagemans, J.; Lavigne, R. Safety and efficacy of phage therapy in difficult-to-treat infections: A systematic review. Lancet Infect. Dis. 2022, 22, e208–e220. [Google Scholar] [CrossRef] [PubMed]
- Glonti, T.; Pirnay, J.-P. In vitro techniques and measurements of phage characteristics that are important for phage therapy success. Viruses 2022, 14, 1490. [Google Scholar] [CrossRef]
- Teklemariam, A.D.; Al Hindi, R.; Qadri, I.; Alharbi, M.G.; Hashem, A.M.; Alrefaei, A.A.; Basamad, N.A.; Haque, S.; Alamri, T.; Harakeh, S. Phage cocktails–an emerging approach for the control of bacterial infection with major emphasis on foodborne pathogens. Biotechnol. Genet. Eng. Rev. 2023, 39, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Meneses, L.; Brandão, A.C.; Azeredo, J. An overview of the current state of phage therapy for the treatment of biofilm-related infections. Curr. Opin. Virol. 2022, 53, 101209. [Google Scholar] [CrossRef]
- Rastogi, V.; Yadav, P.; Verma, N.; Verma, A. Preparation and characterization of transdermal mediated microemulsion delivery of T4 bacteriophages against E. coli bacteria: A novel anti-microbial approach. J. Pharm. Investig. 2018, 48, 393–407. [Google Scholar] [CrossRef]
- Nayak, T.; Singh, R.K.; Jaiswal, L.K.; Gupta, A.; Singh, J. A Survey of Recent Nanotechnology-Related Patents for Treatment of Tuberculosis. In Intellectual Property Issues in Nanotechnology; CRC Press: Boca Raton, FL, USA, 2020; pp. 277–286. [Google Scholar]
- Dedrick, R.M.; Smith, B.E.; Cristinziano, M.; Freeman, K.G.; Jacobs-Sera, D.; Belessis, Y.; Brown, A.W.; Cohen, K.A.; Davidson, R.M.; van Duin, D. Phage Therapy of Mycobacterium Infections: Compassionate Use of Phages in 20 Patients With Drug-Resistant Mycobacterial Disease. Clin. Infect. Dis. 2023, 76, 103–112. [Google Scholar] [CrossRef]
- Ferry, T.; Kolenda, C.; Laurent, F.; Leboucher, G.; Merabischvilli, M.; Djebara, S.; Gustave, C.-A.; Perpoint, T.; Barrey, C.; Pirnay, J.-P. Personalized bacteriophage therapy to treat pandrug-resistant spinal Pseudomonas aeruginosa infection. Nat. Commun. 2022, 13, 4239. [Google Scholar] [CrossRef] [PubMed]
- Hoyle, N.; Zhvaniya, P.; Balarjishvili, N.; Bolkvadze, D.; Nadareishvili, L.; Nizharadze, D.; Wittmann, J.; Rohde, C.; Kutateladze, M. Phage therapy against Achromobacter xylosoxidans lung infection in a patient with cystic fibrosis: A case report. Res. Microbiol. 2018, 169, 540–542. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.M.; Woodworth, B.A.; Fackler, J.; Brownstein, M.J. Case Report: Successful use of phage therapy in refractory MRSA chronic rhinosinusitis. Int. J. Infect. Dis. 2022, 121, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Johri, A.V.; Johri, P.; Hoyle, N.; Pipia, L.; Nadareishvili, L.; Nizharadze, D. Case report: Chronic bacterial prostatitis treated with phage therapy after multiple failed antibiotic treatments. Front. Pharmacol. 2021, 12, 692614. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Chen, H.; Zhang, M.; Zhao, Y.; Jiang, Y.; Liu, X.; Huang, W.; Ma, Y. Clinical experience of personalized phage therapy against carbapenem-resistant Acinetobacter baumannii lung infection in a patient with chronic obstructive pulmonary disease. Front. Cell. Infect. Microbiol. 2021, 11, 631585. [Google Scholar] [CrossRef]

| Year | Country | Clinical Condition and Number of Patients Treated with Phages | Targeted Bacterial | Phages Cocktail (Yes or No) | Results | References |
|---|---|---|---|---|---|---|
| 2022 | United States of America | Two or more positive mycobacterial cultures in at least one organ (based on ATS/ERS/ESCMID) (n = 20) | Non-tuberculous Mycobacterium | Yes (n = 9) | 11 patients were assessed as responding well, five patients could not be assessed for treatment effect, and four patients showed no significant improvement | [93] |
| 2022 | France | Discitis with spinal abscess (n = 1) | Multidrug resistant Pseudomonas aeruginosa | Yes (n = 3) | The patient could walk without pain and has a good prognosis | [94] |
| 2018 | Georgia | The patient suffered from respiratory complications, including intermittent infections caused by Pseudomonas aeruginosa and Staphylococcus aureus (n = 1) | A. xylosoxidans | Yes (n = 2) | The patient’s self-consciousness significantly improved, dyspnea disappeared, and cough was relieved | [95] |
| 2022 | United States of America | Treatment of chronic sinusitis and recurrent ear infections in a woman with a history of diabetes and sarcoidosis (n = 1) | Methicillin-resistant S. aureus | No | The symptoms were alleviated without signs of relapsing chronic sinusitis or otomastoiditis | [96] |
| 2021 | India | Severe pain in the right testicle, radiating to the right buttock, right lower back, and left and right pelvic area. Perineal pain, accompanied by sweating, general weakness and physical discomfort (n = 1) | S. aureus and S. mitis | Yes | The patient is in full remission | [97] |
| 2021 | China | The patient had a long history of type 2 diabetes and had recurrent lung infections during the past two years of hospitalization due to repeated use of mechanical ventilation (n = 1) | Carbapenem-resistant A. baumannii (CRAB) | No | No re-emergence of CRAB was observed, and the patient remained stable | [98] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Y.; Li, W.; Zhang, H.; Ba, X.; Li, Z.; Zhou, J. The Post-Antibiotic Era: A New Dawn for Bacteriophages. Biology 2023, 12, 681. https://doi.org/10.3390/biology12050681
Jin Y, Li W, Zhang H, Ba X, Li Z, Zhou J. The Post-Antibiotic Era: A New Dawn for Bacteriophages. Biology. 2023; 12(5):681. https://doi.org/10.3390/biology12050681
Chicago/Turabian StyleJin, Youshun, Wei Li, Huaiyu Zhang, Xuli Ba, Zhaocai Li, and Jizhang Zhou. 2023. "The Post-Antibiotic Era: A New Dawn for Bacteriophages" Biology 12, no. 5: 681. https://doi.org/10.3390/biology12050681
APA StyleJin, Y., Li, W., Zhang, H., Ba, X., Li, Z., & Zhou, J. (2023). The Post-Antibiotic Era: A New Dawn for Bacteriophages. Biology, 12(5), 681. https://doi.org/10.3390/biology12050681




