Simple Summary
Holstein is the most popular dairy cattle breed worldwide due to its milk yield. When these cows are exposed to heat stress, they reduce feed intake and milk production in order to minimize body heat production. The large variability associated with this response appears to be genetically regulated. Therefore, we combined genomic and marker-assisted technologies with the objective to validate genetic markers associated with milk production and thermotolerance. A genome-wide association study detected six candidate single nucleotide polymorphisms (SNPs) as predictors for milk production in heat-stressed Holstein cows. Only three of these SNPs were further validated as markers for milk production and thermotolerance traits (i.e., rectal temperature and respiratory rate) in two independent Holstein cow populations. Such markers belong to genes that regulate metabolic functions needed to accomplish energy demands and minimal heat production. The results of this study revealed that heat-stressed Holstein cows with favorable markers were able to reduce rectal temperature and respiratory rate, which allowed them to maintain adequate milk production levels. In conclusion, we validated three genetic markers in heat-stressed Holstein dairy cows, which are useful to be included in selection programs to improve milk yield and tolerance to heat stress.
Abstract
Dairy production in Holstein cows in a semiarid environment is challenging due to heat stress. Under such conditions, genetic selection for heat tolerance appears to be a useful strategy. The objective was to validate molecular markers associated with milk production and thermotolerance traits in Holstein cows managed in a hot and humid environment. Lactating cows (n = 300) exposed to a heat stress environment were genotyped using a medium-density array including 53,218 SNPs. A genome-wide association study (GWAS) detected six SNPs associated with total milk yield (MY305) that surpassed multiple testing (p < 1.14 × 10−6). These SNPs were further validated in 216 Holstein cows from two independent populations that were genotyped using the TaqMan bi-allelic discrimination method and qPCR. In these cows, only the SNPs rs8193046, rs43410971, and rs382039214, within the genes TLR4, GRM8, and SMAD3, respectively, were associated (p < 0.05) with MY305, rectal temperature (RT), and respiratory rate. Interestingly, these variables improved as the number of favorable genotypes of the SNPs increased from 0 to 3. In addition, a regression analysis detected RT as a significant predictor (R2 = 0.362) for MY305 in cows with >1 favorable genotype, suggesting this close relationship was influenced by genetic markers. In conclusion, SNPs in the genes TLR4, GRM8, and SMAD3 appear to be involved in the molecular mechanism that regulates milk production in cows under heat-stressed conditions. These SNPs are proposed as thermotolerance genetic markers for a selection program to improve the milk performance of lactating Holstein cows managed in a semiarid environment.
1. Introduction
Milk production is a very important livestock activity because it generates a basic food for humans. Cattle account for 81% of the total milk produced worldwide [1]. The dairy industries located in semiarid regions face the challenge of producing quality milk in a warm climate, which is characterized by a high ambient temperature and humidity leading to heat stress. Installation of cooling systems to dissipate heat during summer seasons, automated feed machinery, and herd improvement using genomic-based technologies are strategies that dairy producers are preparing for the future [2].
Heat stress is an imbalance between the ratio of heat acquired from different sources such as body metabolism and environmental conditions versus the heat dissipation system, which causes an increase in the animal’s body temperature [3]. The Holstein breed of dairy cattle is the most popular worldwide due to its outstanding milk yield and ease of management. During summer, dairy cows’ ability to dissipate heat through skin evaporation is limited by their low live weight to body surface area ratio, the presence of underdeveloped sweat glands, and their short, dense body surface hair, all of which affect their milk production [4].
Fermentation of forage consumed in the rumen produces considerable metabolic heat, which also generates additional heat load for the cow’s body [5]. Additionally, enhanced warming of the climate with the consequences of the intensification of hot periods has caused dairy cows to be subjected to long periods of intense heat stress [6], which threatens to severely reduce productive performance, especially milk production [7]. Therefore, the selection of livestock adapted to semiarid regions with a superior ability to tolerate drought and extreme temperatures will be essential to ensure profitable production and cope with the challenging climate scenario expected in the coming years [8,9].
With the progress of bovine genome sequencing and high-density SNP genotyping technologies, genome-wide association studies (GWASs) have become an important tool for locating QTL or chromosomal regions associated with traits of economic importance [10]. In various dairy breeds of cattle, GWASs have been successfully employed to identify regions in DNA involved in heat tolerance [11], lipid movement [12], milk production and fertility [13], production persistency [14], and lactation curve characteristics [15]. These traits and QTLs are summarized in the Animal QTL database: milk production and yield (9121), milk composition—fat (45,625), milk composition—protein (25,916), reproduction (45,715), production traits (22,722), meat and carcass (22,695), exterior (10,388), and health (including heat tolerance; 8242) (QTLdb; http://www.animalgenome.org/QTLdb. Date accessed: 23 April 2023).
GWASs examine thousand of small variants across the genome called single-nucleotide polymorphisms (SNPs), which are very helpful to identify functional genes likely involved in phenotypic traits [16]. For instance, the SNP K2323A from the gene DGAT1 has been associated with genetic variation in milk yield and composition, as this SNP decreased milk production and increased milk fat, then showed a genetic marker effect of the SNP K232A on milk traits [17,18].
Recently, GWASs have been performed in Holstein cows subjected to heat stress conditions. Potential candidate genes involving the heat stress response have been discovered as associated with milk production traits [19]; physiological traits such as rectal temperature, respiratory rate, drooling score [20,21]; and milk fatty acid composition [22]. The identification of specific genes or SNP markers inferring thermotolerance in Holstein cows could be an important strategy to ensure dairy cows' productivity [23]. However, these markers must be validated in independent cattle populations to be available for incorporation into genetic selection programs focused on improving complex and economically important traits such as milk production and thermotolerance.
Therefore, our objective was to validate candidate genomic SNPs as molecular markers associated with milk production and physiological traits indicative of thermotolerance in Holstein cows managed in a hot and humid environment.
2. Materials and Methods
The Institutional Animal Care and Use Committee of the Instituto Tecnologico de Sonora approved all procedures performed on animals (approval code 2017-02).
2.1. Experimental Locations and Animals
The study was conducted at three neighboring dairy farms located in the Yaqui Valley, Sonora, Mexico (27°21′ N 109°54′ W). These farms were managed under an intensive system for milk production. Climatic conditions throughout the year were characterized by significant variations in temperature (−2.5 to 45.9 °C), relative humidity (25 to 90%), and solar radiation (0.05 to 761.7 W/m2). Annual precipitation was ~341 mm/year ranging from 3 to 90 mm on rainy days.
Three hundred spring-calved Holstein cows, about 4 to 6 years old, average body weight of 645.1 ± 32.5 kg, and average body condition of 3.8 ± 0.01, were used in this study. Selection criteria included age, body weight, body condition, and calving season because spring-calving cows experience heat stress during their peak milk production period.
Similar management was provided to all cows, which were housed in shaded barns with free access to a water source and a commercial supplement of trace minerals. The floor and shade spaces were provided as per the need of Holstein cows. In order to supply the nutritional requirements for dairy cattle in lactation, cows received a mixed ration twice a day that was formulated considering an average weight of 650 kg and milk production of ~30 kg/day, with an average composition of 3.5% fat and 3.2% true protein [24].
2.2. Milk Yield Records and Climatic Data
Cows were milked twice per day (0700 and 1700) and milk production (MY; kg) was recorded daily using an electronic system (Metraton 21TM, Westfalia-Surge Farm Technologies, Siemensstraße, Bönen, Germany). A total of 183,000 milk records were collected from 300 complete lactations. Data were filtered by the exclusion of wrong records (e.g., 5 L, 80 L) and then used to calculate the adjusted 305-day milk yield (MY305) per cow in kg. The MY305 was calculated by multiplying milk production levels by an adjustment factor provided by the Dairy Herd Improvement Association, which included days in milk and the age of the cow [25]. These Holstein cows were daughters of 93 purebred bulls and 265 dams.
Ambient temperature (AT; °C) and relative humidity (RH; %) data were collected by a nearby meteorological station (Network of Automatic Meteorological Stations of Sonora) and extracted from the REMAS website (http://www.siafeson.com/remas; accessed on 8 April 2022). Daily records of AT and RH were used to calculate the temperature–humidity index (THI) using the formula THI = (0.8 × AT) + [(RH/100) × (AT − 14.4)] + 46.4 [26].
2.3. Genotyping and Quality Control
Disposable sterile syringes were used to collect from each cow a blood sample (3 mL) through venipuncture of the coccygeal vein. Five drops of whole blood were spotted on Fast Technology for Analysis of Nucleic Acids cards (FTA®), which were sent to Neogen AgriGenomics (Lincoln, NE, USA) for DNA extraction. DNA was genotyped using the SNP panel BovineSNP50 containing 53,218 highly informative SNPs uniformly distributed across the entire bovine genome (Illumina Inc., San Diego, CA, USA) [27].
Quality control was performed using PLINK software to remove all SNPs that did not comply with the following criteria: (1) minor allele frequency of an SNP greater than 10% (MAF > 0.10), (2) no deviation from Hardy–Weinberg equilibrium (p-value of chi-square goodness-of-fit test greater than 0.05, X2 > 0.05), (3) call rate of an individual genotype higher than 95%, and (4) call rate of a single SNP genotype higher than 90%. Additionally, the SNPs located on the sex chromosome or those with unknown positions were removed from the current study. After quality control measures, 43,691 SNP markers were retained for further analyses.
2.4. Genome-Wide Association Analysis (GWAS)
The principal component analysis (PCA) option was used to correct batch effects/stratification of the test input data. The GWAS was performed using a single-locus mixed model to study associations between genotypes of each SNP marker with the trait MY305 (i.e., single-marker SNP GWAS), by fitting all SNPs simultaneously. The additive, single-locus, mixed model used was: y = Xβ + Za + e, where y was the vector of phenotypic observations (MY305), X was the design matrix of fixed effects, Z was the design matrix of random additive genetic effects, β was the vector of fixed effects, a was the vector of random additive genetic effects, and e was the vector of residual effects. It was assumed that a~N (0, Gσ2a) and e~N (0, Iσ2e), where σ2a is the additive genetic variance, σ2e is the residual variance component, G is the genomic relationship matrix, and I is the identity matrix.
The software SNP & Variation Suite v8 (SVSv8; Golden Helix, Inc., Bozeman, MT, USA, www.goldenhelix.com; accessed on 12 June 2022) was used to perform single-marker GWAS analysis. The statistical model included genotype, herd, and lactation number as fixed effects, days in milk as the covariate, and sire as the random term to account for family effects. A total of 93 Holstein bulls were included as sires in the current study.
2.5. Multiple-Testing Correction
The Bonferroni correction test (b = α/n), which assumed independence between SNPs, was used to adjust and set for multiple comparisons the p-values for the SNPs obtained from GWAS analyses [28]. The experiment-wise error was α = 0.05, and the number of tests (n) was taken to be the number of the useful SNPs (n = 43,691). Therefore, the corrected p-value threshold for the genome-wide significant level was 1.18 × 10−6 (i.e., 0.05/43,691), which corresponded to 5.93 on a −log10(p-value) scale.
2.6. Candidate Genes and Pathway Analyses
The functional genes that contained or were located less than 0.1 MB of distance from the significant SNPs associated with MY305 were selected as candidate genes. The genome assembly Bos_taurus_UMD_3.1.1. was used as a reference genome to search for candidate genes according to the chromosomal locations of each significant SNP.
Functional pathway enrichment analysis was performed using candidate genes that were significant at a P less than 0.01 (n = 36) and the bioinformatics online database KOBAS (http://kobas.cbi.pku.edu.cn/; accessed on 12 August 2022). Ensembl (https://useast.ensembl.org/index.html; accessed on 23 July 2022) and GeneCards (http://www.genecards.org; accessed on 15 August 2022) were used for gene descriptions (i.e., registration code, name, and chromosomal location).
2.7. SNP Validation Study
Two independent cattle populations including 216 Holstein cows (n = 112, n = 104) from neighboring dairy farms were used to validate, as molecular markers for milk yield and thermotolerance, three SNPs previously detected as associated (p < 0.05) with milk yield in Holstein cows exposed to heat stress. These SNPs were rs8193046, rs43410971, and rs382039214, which were within the genes TLR4 (toll-like receptor 4), GRM8 (glutamate metabotropic receptor 8), and SMAD3 (SMAD family member 3), respectively.
Cows included in the validation study were spring-calved, 3 to 6 years old, and body conditions ranged from 2.5 to 4.0. Milk records were collected daily from each cow through the entire lactation using an electronic system and were adjusted to obtain the individual MY305. Physiological traits of rectal temperature (RT) and respiratory rate (RR) were measured biweekly (06:00 and 16:00 h) as indicators of thermotolerance. A digital thermometer (TES-1310R) with a contact sounding line (Type K; -com large) that touched the rectal mucus was used to record RT, whereas RR was evaluated by visual counting of the flank intercostal movements for 60 s (breaths/min). Both RT and RR were evaluated in the waiting parlor before milking, once the cow remained quiet to facilitate collection of physiological data by the same person all the time.
Vacutainer tubes that contained the anticoagulant disodium ethylenediamine tetraacetic acid (EDTA-Na2; Venoject®, Terumo, Lakewood, CA, USA) were used to collect a blood sample from each cow by puncturing the jugular vein. After placing in a thermal cooler, the samples were transported to the Instituto Tecnologico de Sonora and centrifuged at 3500 RMP for 15 min. A pipet was used to collect 200 μL of leukoplatelet layer from each sample, which was refrigerated at −20 °C before proceeding with DNA extraction. A commercial kit was used for DNA extraction (DNeasy Blood & Tissue Kits; Cat. 69504, QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. A NanoDrop automatic spectrophotometer (NanoDrop, Thermo Fisher Scientific, Waltham, MA, USA) was used to measure the concentration (260 nm absorbance) and purity (260/280 ratio) of DNA obtained. Then, electrophoresis in 1% agarose gel stained with 1.5 μL ethidium bromide was used to confirm the integrity of the DNA.
The TaqMan method for allelic discrimination and RT-qPCR (StepOneTM, Applied Biosystems, Foster City, CA, USA) were used to genotype the four significant SNPs reported in our study, according to the procedures described by Castillo-Salas et al. [29]. The StepOne Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) was used to perform the PCR. Finally, the StepOne software (version 2.3, Life Technologies Corporation, Carlsbad, CA, USA) was used for PCR data analysis and genotyping.
2.8. Statistical Analyses
The procedure MEANS was used to calculate descriptive statistical measures for the traits MY305, RT, and RR. Similarly, the procedures UNIVARIATE and GLM (Levene’s test) were performed to evaluate the assumption of normality of data distribution and equality of variances, respectively. Then, PROC ALLELE was used to calculate both allele and genotype frequencies. A chi-square (X2) test was performed in PROC FREQ to analyze Hardy–Weinberg equilibrium (HWE). The SAS software (Version 9.4; SAS Inst. Inc., Cary, NC, USA) was used to perform all statistical procedures.
An associative study between genotype and phenotype was conducted to validate three genomic SNPs as molecular markers for milk production and thermotolerance in two independent Holstein cattle populations under heat stress. This study was conducted after verifying that the data had a normal distribution within each validation dairy population. Statistical mixed procedures were used to test the candidate SNPs as predictors for the traits MY305, RT, and RR. The statistical model included the response variable; the SNP genotype, herd, and lactation number as fixed effects; days in milk as the covariate; and sire as the random effect.
Preplanned pairwise comparisons of least-squares means were generated using the PDIFF option when the associative analyses detected the genotype term as a significant source of variation (p < 0.05). The option LSMEANS with Bonferroni adjustment was used to process these means separation tests [30]. The MIXED procedure in SAS was used to calculate the estimated effects of the average allele substitution, which were generated by regressing the phenotype on the number of copies of one SNP’s allele as a covariate (e.g., the effect of substituting 1 allele with another allele) [31]. Additive and dominance (e.g., non-additive) genetic effects were calculated in SAS following the procedures described by Falconer and Mackay [32].
The data from the validation population were represented in tables with the average values ± standard errors according to SNP genotypes. Data were also represented in figures that included average values ± standard errors according to the number of SNPs with favorable genotypes.
2.9. Gene Marker Effects on Milk Production and Thermotolerance Traits
One-way ANOVA was used to compare MY305, RT, and RR in cows according to the number of favorable SNP genotypes. Data processed for this analysis belonged to a normal and homoscedastic population. The Tukey HSD test was performed for pairwise comparisons. For these analyses, statistical significance was declared at p < 0.05. In order to confirm the thermotolerance effect of the SNP markers identified in this study, a correlation between MY305 and physiological traits was performed using PROC CORR within cow groups with different numbers of favorable SNP genotypes. Finally, a linear regression analysis including RT or RR as predictors for MY305 was performed using PROC REG in cow’s groups with 0, 1, and >1 favorable SNP genotypes.
3. Results
3.1. Climatic Conditions
According to ambient temperature and relative humidity data, the THI through the study averaged 68.5 units in late spring, ranged from 70 to 82 in the summer, and averaged 72.6 units in fall. These THI values suggested environmental conditions leading to heat stress in dairy cows involved in the current study.
3.2. Whole-Genome Association Study
The single-marker GWAS identified 18 SNPs associated with the trait MY305 at a significant p-value less than 0.001. Only six of these SNPs surpassed Bonferroni multiple-testing corrections because they showed a p-value less than the threshold 1.14 × 10−6, as presented in Figure 1. The SNPs rs8193046, rs43410971, and rs382039214 were intronic variants located within the genes TLR4, GRM8, and SMAD3, respectively. The SNP rs109479519 was near to GLRX5 gene (0.032 Mb), whereas the SNPs rs29015299 and rs108988401 were intergenic but more than 0.1 MB from the nearest gene (Table 1).
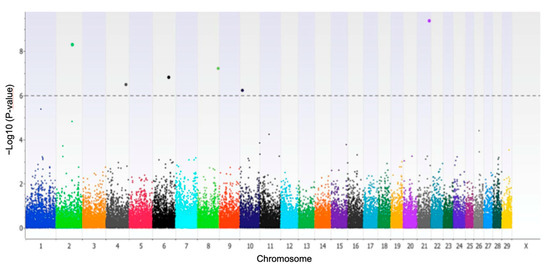
Figure 1.
Manhattan plot displaying single-marker GWAS for total milk yield at 305 days (MY305) in dairy Holstein cows exposed to climatic heat stress.

Table 1.
Summary of significant SNPs (p < 1.18 × 10−6) from single-marker genome-wide association study (GWAS) in Holstein cows managed under heat-stressed environmental conditions.
3.3. Functional Enrichment Analyses
Functional pathways for the candidate genes associated with MY305 at p < 0.01 are presented in Table 2. Only pathways that were significant (p < 0.05) after Benjamini–Hochberg correction were retained for the validation study. Genes involved in such pathways were TLR4, SMAD3, GRM8, CARD11, TCL1A, NFKB1, SMAD6, and TRPC1.

Table 2.
Enriched pathways for SNP in protein-coding genes associated with milk production in cows affected by heat stress.
3.4. SNP Marker Association Study
Of the six SNPs that were associated with MY305, only four of them were retained for further study because they were within or near a candidate gene. However, only three out of these four SNPs were accomplished with the criteria for minor allele frequency higher than 10% (MAF > 0.10) and no deviation from the Hardy–Weinberg equilibrium (HWE, X2 > 0.05). These 3 SNPs were rs8193046, rs43410971, and rs382039214 within the genes TLR4, GRM8, and SMAD3, respectively (Table 3), and they were designated as suitable to be analyzed in additional genotype to phenotype association and validation studies.

Table 3.
Identification, gene name, favorable SNP allele, allele frequencies, and Hardy–Weinberg equilibrium analysis for genomic SNPs associated with milk and thermotolerance phenotypes.
The least-square means for milk yield at 305 d and physiological traits are reported in Table 4. The SNPs rs8193046 and rs43410971 were associated with MY305, RT, and RR (p < 0.001), whereas the SNP rs382039214 was associated with MY305 and RT (p < 0.001).

Table 4.
Least-square means ± SE according to SNP’s marker genotypes for milk yield and physiological traits in Holstein cattle validation populations.
The most favorable genotypes for the SNPs rs8193046, rs43410971, and rs382039214 were AA, GG, and TT, respectively, because they were associated with higher milk production and more beneficial measurements of RT and RR in cows. Combined with genotype average values, these results appeared to confirm the favorable effect of the genes TLR4, GRM8, and SMAD3 on milk production and physiological traits indicative of thermotolerance in Holstein cows exposed to a heat-stressed environment.
3.5. Allele and SNP Genotype Effects on Phenotypic Traits
Effects of allele substitution and fixed estimates are presented in Table 5. The SNPs rs8193046 and rs43410971 had the highest allele contribution (p < 0.01) for MY305 and RR, whereas the SNP rs382039214 was the most beneficial contributor for RT. In addition, an additive fixed effect was confirmed (p < 0.01) for these SNP markers, suggesting that the sum of their individual effects was equal to their combined allele effects.

Table 5.
Allele substitution effects and fixed estimates for additive and dominance effects of the favorable allele for MY305, RT, and RR in the validation Holstein cattle populations.
Further genotype analysis confirmed a significant improvement (p < 0.05) in MY305 (Figure 2a) as the number of favorable genotypes of the three significant SNP markers increased from 0 to 3 in Holstein cows. Similarly, RT and RR also improved (p < 0.05) as the number of favorable genotypes increased (Figure 2b,c).
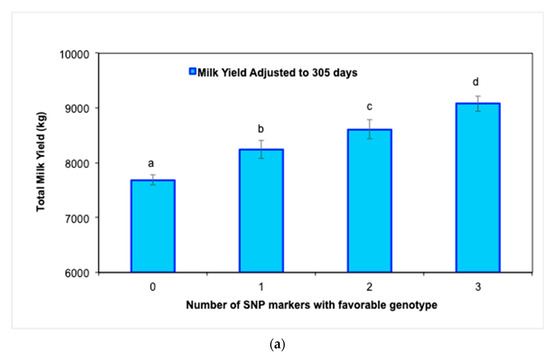
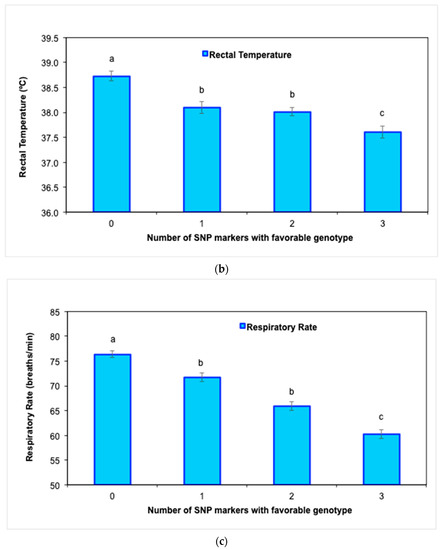
Figure 2.
Average values (±SE) for milk production and physiological traits in Holstein cows during the experimental period according to the number of favorable genotypes (i.e., 0, 1, 2, or 3) of the three SNP markers: (a) milk yield adjusted to 305 days (MY305), (b) rectal temperature (RT), and (c) respiratory rate (RR) (abcd p < 0.05).
3.6. SNP Markers Effects on Relationship between Milk Yield and Thermotolerance
Pearson correlation analysis showed a moderate association between MY305 and RT in cows carrying 0 and 1 favorable genotype (r = −0.4587 and r = −0.5345, respectively; p < 0.01). Interestingly, this relationship increased in cows carrying more than one favorable genotype (r = −0.6021; p < 0.01). However, a low relationship was detected between MY305 and RR in cows from all favorable genotype groups (p < 0.05; Table 6).

Table 6.
Pearson correlations between physiological variables and milk yield at 305 days (MY305) according to the number of favorable genotypes from the 3 significant SNP markers.
Linear regression analysis identified RT as a significant predictor for MY305 (p < 0.01). The regression coefficient (β1) improved as the number of SNPs with favorable genotypes increased, as well as the coefficient of determination (R2). These results suggested a change of −140.35, −277.51, and −470.39 L in MY305 per unit of change in RT in cows carrying 0, 1, and >1 favorable SNP genotypes, respectively (p < 0.01; Figure 3a–c).
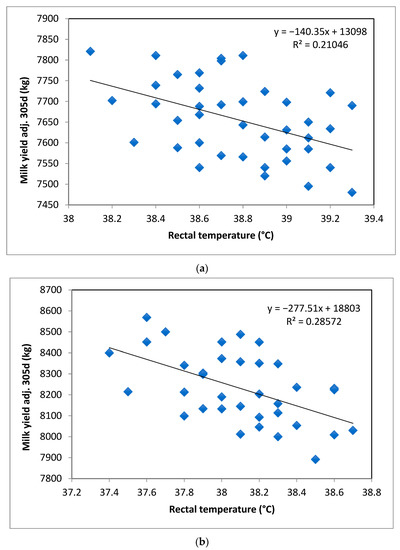
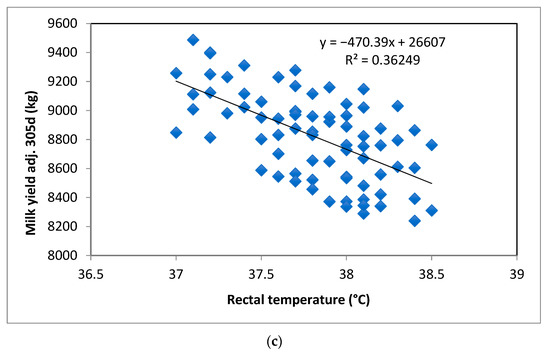
Figure 3.
Rectal temperature (RT) as a predictor for milk yield at 305 days (MY305) according to the number of favorable genotypes of the three SNP markers: (a) no favorable genotypes, (b) one favorable genotype, and (c) more than one favorable genotype.
4. Discussion
An environmental factor that exerts a substantial negative effect on dairy performance is heat stress, which results when the internal heat production is higher than the cow’s body’s ability to dissipate it [4]. In recent times, lactating dairy cows have been exposed to even more severe heat stress due to the steadily warming global climate, the lengthening of drought periods, and the intensification of the warm season throughout the year [6]. Moreover, the intensive selection for high milk-yielding Holstein cows that are subjected to a tremendous metabolic heat load has impaired these cow’s ability to maintain homeothermy, especially when they are exposed to severe heat stress conditions, making these cows more sensitive or susceptible to environmental changes [33,34].
The Yaqui Valley in Sonora Mexico is characterized by extended periods of high ambient temperature and high relative humidity throughout the year, which elevates the THI value above the threshold of 68 units in the middle spring. As the warm environmental conditions become more intense, the THI begins to increase steadily in late spring, peaking in the summer and then declining in fall. This very hot weather is unique in northwest Mexico and creates adverse heat stress conditions for cattle. This location provides an opportunity to study the genetic basis of thermotolerance in lactating Holstein cattle. However, only a few dairy farmers are willing to collaborate in this type of research because of the decrease in milk production of the cows under study.
Genomic technologies and marker-assisted selection programs have been used as successful strategies to study candidate genes and genetic markers associated with heat stress response in Holstein cows [35]. In the current study, we performed both technologies sequentially in order to identify and validate SNPs as potential molecular markers to be used in genetic selection and herd improvement programs. After executing GWAS, we identified 18 SNPs as predictors of milk production performance in heat-stressed Holstein cows. Four of these SNPs were found to meet with the selection criteria and were genotyped in independent Holstein populations using the TaqMan molecular assay to obtain the SNP genotypes. From these SNPs, only three were tested through a genotype-to-phenotype association study and validated as molecular markers for milk production and thermotolerance traits. Further analyses confirmed the favorable effect of these markers on milk production and physiological traits indicative of heat stress tolerance (i.e., rectal temperature and respiratory frequency), as well as the positive influence of the markers on the predictive relationship between rectal temperature and milk production.
A genetic component has been reported to influence milk production in dairy cows exposed to heat stress, suggesting the presence of thermotolerance-related genes [19,36]. The response or sensitivity to heat stress in milking cows appears to be associated with pronounced chromosome-wide genetic variances, indicating that genomic regions and major candidate genes should underlie the heat stress response [37]. Identification of molecular markers or SNPs associated with tolerance to heat stress has been proposed as an important strategy to ensure productivity in cattle exposed to thermal stress [38]. Therefore, the study of the genetic basis associated with thermotolerance is potentially helpful for the selection of cattle adaptable to warm semiarid conditions [39].
In the current study, single-marker GWAS analysis detected 18 SNPs associated with milk production in heat-stressed cows. Of these SNPs, only six of them surpassed multiple-testing selection criteria and were proposed as genomic candidate SNPs for milk production in Holstein cows, which were managed in a warm semiarid environment from northwest Mexico (i.e., Yaqui Valley). This low number of significant SNPs may have been influenced by the small sample size. As GWASs involve thousands of SNP markers, a large sample size is usually required to reach appropriate statistical power. However, when a small sample is available, GWASs may be limited to detecting true associations between SNPs and phenotypes leading to increase false negative rates [16].
Genome-wide association studies (GWASs) have become a widely used approach to identify genomic regions and genetic variants associated with phenotypes of interest [10]. A GWAS performed for milk yield in heat-stressed Holstein cows identified three genomic regions harboring candidate genes involved in the cellular response to heat stress [19]. Chromosomal regions and candidate genes associated with milk fatty acid composition were reported in Holstein cattle also subjected to a GWAS for heat stress response [33]. Similar GWAS analyses in Holstein cows identified non-overlapping genomic regions and candidate genes associated with physiological heat stress indicators and confirmed the polygenic nature of heat tolerance, as well as complementary mechanisms associated with heat stress response [21,40].
A QTL with major effects on milk production traits and milk fatty acid profiles has been detected in the centromeric region of cattle chromosome 14 (BTA14) [22]. Although several genes with major effects on milk yield and milk fat have been reported, the major contribution to the genetic variance of BTA14 associated with these milk traits is attributed to the DGAT1 gene [41]. According to Grisart et al. [42], a nonconservative lysine to alanine (K232A) mutation in the DGAT1 gene explained daughter deviations for milk yield (18%), fat yield (15%), and protein yield (8%).
Research involving DGAT1 mutation K232A (Lys232 → Ala) has focused on the study of milk fat production because it encodes the enzyme diglyceride O-acyltransferase 1 (DGAT1), which catalyzes the synthesis of triglycerides in the mammary gland. However, this SNP is also involved in the structure of mammary gland epithelial cells suggesting its influence on milk synthesis [43]. The DGAT1 K232A mutation has been reported to be associated with an increase in milk fat production and a reduction in milk yield and milk protein in New Zealand [44] and German Holstein dairy cattle [17].
From the six genomic SNPs detected in our study, only three were within a candidate gene and surpassed selection criteria (i.e., MAF > 0.10 and HWE > 0.05), which were tested in a marker SNP association study. These SNPs were associated with MY305, RT, and RR, and they were proposed as molecular markers for milk production and heat stress tolerance. The results supported prior reports from GWAS about the existence of a genetic component underlying heat stress tolerance in Holstein dairy cows. However, to our knowledge, this is the first report of genomic SNPs from candidate genes associated with milk production in heat-stressed Holstein cows, which were further validated as molecular markers for milk performance and thermotolerance in independent Holstein cattle populations managed under extremely warm environmental conditions.
Genomic analyses using GWASs are able to detect markers distributed across the whole genome; it is likely to be more successful compared to the candidate-marker approach, which is generally based on testing a small number of SNP markers [45]. However, the combination of both technologies has been proposed for the identification of genomic candidate SNPs and their further validation as molecular markers in independent cattle populations [46]. This approach was also suggested to validate candidate genes identified from gene expression studies [47].
Statistical models of GWAS are able to detect individual significant associations between thousands of SNPs and a specific phenotype. In order to avoid false positives due to multiple SNP testing, a correction procedure must be applied and it results in a very high significance threshold lacking to detect polygenic effects [48]. Controlling the false discovery rate (FDR), as well as similar methods, were developed as an alternative to control the rate of the experiment-wide error in GWAS results [49,50]. However, the validation of the SNP effects in independent animal populations appears to be the most reliable procedure to test the importance of SNPs [51], because of the small probability of an SNP being significant in two different populations [52]. This information reinforces our rationale for the validation of SNPs at different dairies.
Pryce et al. [53] identified eleven genomic SNPs in three regions on chromosomes 6 and 26 associated with lactation persistency in Australian dairy cattle; these SNPs were further validated as potential genetic markers in two dairy cow populations from different breeds (i.e., Holstein and Jersey). In a similar study, Chamberlain et al. [46] identified 43 genomic SNPs on chromosome 20 associated with milk production traits, also in Australian dairy cattle, after executing three GWAS studies. These SNPs were further evaluated in a validation population that revealed molecular markers for milk protein composition.
In the current study, three genomic SNPs were validated as marker predictors for milk production and thermotolerance. These SNPs were in the genes of TLR4, GRM8, and SMAD3. Although these SNPs were located within intronic regions, they may be able to influence the expression of their corresponding genes [54]. Such ability is attributed to the presence of cis-elements or long non-coding RNAs within the intronic sequences that interact with transcription factors to regulate gene expression [55].
Toll-like receptor 4 (TLR4) is expressed by cells of the innate immune system in mammals, and it appears to be involved in lipid release pathways [56]. A dramatic increase in TLR4 was reported in pathogen-induced inflammatory processes related to milk production [57]. Conversely, downregulation of TLR4 cell signaling has been detected in heat-stressed dairy cows, making them more susceptible to disease [58]. Such a decrease in TLR4 induced by thermal heat was also associated with a significant reduction in lipolytic activity and lipid milk content [59]. Adipose tissue, an endocrine organ highly sensitive to climate changes, had reduced adipocyte mobilization in heat-stressed dairy cows [60]. Lactating dairy cattle under heat stress experienced a reduced responsiveness to the lipolytic effect of norepinephrine [61]. As reported by Gupta et al. [62], a close relationship has been observed between endocrine responses to heat stress and immune system activation, which appeared to involve the TLR4 signaling pathway.
In our study, results suggested that the SNP rs8193046 in the gene TLR4 was a predictor for milk production and thermotolerance in Holstein cows because it was able to induce an immune response against heat stress, as well as modifications in lipid metabolism to minimize internal heat load. Therefore, the TLR4 gene appears to coordinate adaptive endocrine and metabolic functions in heat-stressed dairy cows, allowing them to maintain milk production.
The glutamate metabotropic receptor 8 (GRM8) gene encodes a presynaptic receptor that modulates glutamate release at the axon neuro-terminals. Glutamate is involved as a chemical messenger in most of the excitatory synapses and participates in the modulation of the adenylate cyclase and stimulation of phospholipase C in the cell membrane [63]. The maintenance of these functions during exposure to heat stress appears to be critical to prevent structural damaging changes and alterations in membrane fluidity [64]. Moreover, heat stress-induced glutamate activation appears to be essential for carbohydrate and amino acid metabolism; this adaptive response increases glucose synthesis, which is required to supplement the energy demands [65]. This glucose supply is the main energy source because heat stress limits lipolysis and fat mobilization to minimize the generation of heat [66].
In our study, we assumed that the SNP rs43410971 within the gene GRM8 was validated as a genomic marker for milk production because it activated metabolic pathways allowing the heat-stressed cows to comply with their energy demands. In a similar genomic study, Cheruyoit et al. [67] reported the GRM8 gene as a heat tolerance candidate gene that was enriched in two metabolic pathways, the neuroactive ligand–receptor interaction pathway and the glutamatergic synapse pathway. Biological pathways related to the nervous system were associated with complex adaptations in animals exposed to heat stress because these pathways allowed maintenance of a stable core body temperature by connecting the internal and external environments [68].
The SMAD family member 3 (SMAD3) gene encodes for an intracellular protein that is a key mediator of the TGFβ (Transforming growth factor β) signaling pathway, regulating signal transmission and transcription of their target genes [69]. In cattle, SMAD3 mediates the inhibition of adipogenesis caused by TGFBβ through the transcriptional suppression of the PPARγ promoter [70]. In addition, SMAD3 acts as a transcriptional regulator on the central promoter region of the PLIN1 gene. Phosphorylation of PLIN1 is essential for fat mobilization in adipose tissue and plays an important role in the regulation of lipid storage in adipocytes [71]. Lipogenesis in bovine primary adipocytes exposed to heat stress was reduced, which was attributed to low responsiveness to the lipogenic signals of insulin, as well as a decrease in the insulin-stimulated activation of the rate-limiting enzyme acetyl-CoA carboxylase [60].
There is strong reason to propose the SNP rs382039214 in the gene SMAD3 as a genomic marker for milk production and thermotolerance based on its ability to regulate other genes, as well as molecular pathways highly involved in adipose tissue metabolism, specifically adipogenesis. The SMAD3 gene appeared to be a negative regulator of adipocyte synthesis during heat stress, probably as a strategy of the body to minimize heat production. For milk production, Zhang et al. [72] constructed a miR-143 and SMAD3 regulatory network that revealed its role to increase milk fat synthesis through the formation of lipid droplets and the synthesis of triglycerides in bovine mammary epithelial cells. Such results suggested that the SMAD3 gene reduces adipogenesis but ensures an adequate fat supply to the mammary gland in milking cows exposed to heat stress.
The allele substitution effects observed in the current study indicated that the 3 SNPs validated as genomic markers had an important contribution of the favorable allele on the traits MY305, RT, and RR. The negative estimated effects observed in the physiological variables could suggest a key role of the corresponding genes to maintain heat-stressed cows within a thermoneutral zone. These adaptive adjustments appear to favor milk production as suggested by the estimated effect of the favorable allele.
Interestingly, MY305, RT, and RR showed an improvement as the number of SNPs with favorable genotypes increased, which highlighted the positive effect of the genes TLR4, GRM8, and SMAD3 on milk production and thermotolerance. A similar study identified one SNP from four genomic SNPs previously associated with milk yield in heat-stressed cows, as a thermotolerant marker associated with an increase in sweating rate [73]. An SNP in the HSP90AA1 gene was reported to be associated with thermoresistance in Holstein cows, and it was proposed as a genetic marker for heat stress tolerance [74]. A recent study combining GWAS and RNAseq analysis reported the genes PMAIP1, SBK1, TMEM33, GATB, CHORDC1, RTN4IP1, and BTBD7 as candidate markers associated with RT, RR, and drooling score in Holstein cows [21].
In our study, as the number of favorable SNP genotypes increased, Holstein cows appeared to improve their ability to maintain RT and RR at a physiological level that allowed adequate milk production. Conversely, dairy cows with no favorable genotypes appeared to be heat-sensitive, as they showed a reduction in their milk performance that was associated with an elevation in RT. Jensen et al. [75] reported that RT was 0.12 °C higher in heat-sensitive dairy cows compared to heat-tolerant cows.
Rectal temperature (RT) is a useful phenotype indicative of thermal equilibrium in animals. Cattle are able to maintain their physiological and productive functions when RT is within a thermoneutral zone. Once the RT increases above this threshold due to heat stress exposure, normal physiology is altered, leading to an increase in respiration rate (RR) and a decrease in feed intake [76]. Similarly, RR is considered a predictive marker for heat stress in dairy cattle; this physiological function allows the body to maintain temperature through evaporative cooling, acting as a powerful thermoregulatory mechanism [77].
In the current study, a negative correlation was observed between MY305 and the physiological traits RT and RR, which increased as more favorable SNP genotypes were inherited. This inverse relationship between milk yield and thermotolerance was confirmed when RT resulted as a significant predictor of MY305. Interestingly, the predictive model supporting this negative association became more powerful as the number of favorable genotypes of the genes TLR4, GRM8, and SMAD3 increased. These results suggested that the greater the number of favorable genotypes present in the cow, the more dramatic the change in milk production will be due to variations in the RT.
Nguyen et al. [78] also reported a strong negative correlation between milk production and thermotolerance, and they suggested that selection for heat tolerance should also be included in a selection program to improve milk yield. Similarly, Jensen et al. [75] observed a reduction in milk yield and milk quality traits in dairy cows showing a high estimated breeding value for heat tolerance.
Results reported in the current study demonstrated a close association of the tested SNPs with milk yield at 305 days and thermotolerance indicators such as RT and RR. In addition, these SNPs showed a favorable contribution to improving MY305, RT, and RR. Moreover, these SNPs were in the genes TLR4, GRM8, and SMAD3, which appeared to be implicated in several physiological mechanisms required to maintain milk performance in dairy cows exposed to heat stress. Together, these results provided strong evidence to propose the genes TLR4, GRM8, and SMAD as potential molecular markers for milk production and thermotolerance in heat-stressed Holstein cows.
5. Conclusions
Validation of genomic markers is considered a beneficial strategy to improve our understanding of adaptive mechanisms in Holstein dairy cows, which are exposed to a heat stress environment common in semiarid or subtropical regions. In the current study, we validated three genomic SNPs in the genes TLR4, GRM8, and SMAD3 as molecular markers for milk production and thermotolerance in heat-stressed Holstein cows. These three genes appeared to regulate metabolic processes needed to comply with energy demands and minimal heat production, which will favor the productive performance in cattle managed under extremely warm environmental conditions. Therefore, our findings may assist marker selection programs focused on the genetic improvement of milk production and thermotolerance traits in Holstein dairy cows.
Author Contributions
Conceptualization, J.F.M., M.G.T., R.M.E., S.E.S. and P.L.-N.; methodology, R.Z.-A.; software, R.Z.-A., M.A.S.-C. and G.L.-N.; validation, R.Z.-A. and J.C.L.-C.; formal analysis, R.Z.-A., G.L.-N. and P.L.-N.; investigation, R.Z.-A. and P.L.-N.; resources, J.F.M., M.G.T. and P.L.-N.; data curation, R.Z.-A., J.C.L.-C. and M.A.S.-C.; writing—original draft preparation, M.G.T. and P.L.-N.; writing—review and editing, J.F.M., M.G.T., R.M.E. and S.E.S.; visualization, R.Z.-A. and P.L.-N.; supervision, J.F.M., M.G.T. and P.L.-N.; project administration, J.F.M. and P.L.-N.; funding acquisition, J.F.M., M.G.T. and P.L.-N. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by UCMEXUS-CONACYT Grant Program 2016 through the project titled “Genomic analyses of thermotolerance in Holstein dairy cattle managed during summer in southern Sonora, Mexico” (Project Number CN-16-123). This project was also funded by PROFAPI-ITSON Grant Program 2017 through the project titled “Validation of genetic markers associated with thermotolerance and milk production in Holstein dairy cattle managed under heat-stressed environmental conditions” (Project Number 2017-0079).
Institutional Review Board Statement
The Institutional Animal Care and Use Committee of the Instituto Tecnologico de Sonora approved all procedures performed on animals (Protocol code 2017-02 approved on 1 March 2017).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, P.L.-N., upon reasonable request.
Acknowledgments
We thank the respective staff of the cooperative dairy herds, dairy producers, and students involved during the project.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAO. Dairy and Dairy Products. OECD-FAO Agricultural Outlook 2020–2029; OECD Publishing/FAO: Paris, France, 2020; pp. 174–183. [Google Scholar]
- Becker, C.A.; Collier, R.J.; Stone, A.E. Invited review: Physiological and behavioral effects of heat stress in dairy cows. J. Dairy Sci. 2020, 103, 6751–6770. [Google Scholar] [CrossRef]
- Collier, R.J.; Baumgard, L.H.; Zimbelman, R.B.; Xiao, Y. Heat stress: Physiology of acclimation and adaptation. Anim. Front. 2018, 29, 12–19. [Google Scholar] [CrossRef]
- Liu, J.; Li, L.; Chen, X.; Lu, Y.; Wang, D. Effects of heat stress on body temperature, milk production, and reproduction in dairy cows: A novel idea for monitoring and evaluation of heat stress—A review. Asian-Australas. J. Anim. Sci. 2019, 32, 1332–1339. [Google Scholar] [CrossRef]
- Cheruiyot, E.K.; Haile-Mariam, M.; Cocks, B.G.; Pryce, J.E. Improving genomic selection for heat tolerance in dairy cattle: Current opportunities and future directions. Front. Genet. 2022, 13, 894067. [Google Scholar] [CrossRef]
- Fournel, S.; Ouellet, V.; Charbonneau, É. Practices for alleviating heat stress of dairy cows in humid continental climates: A literature review. Animals 2017, 7, 37. [Google Scholar] [CrossRef]
- Wen, Y.L. Effects of Heat Stress on Performance and Physiological Functions in Dairy Cows; Inner Mongolia Agricultural University: Huhehot, China, 2011. [Google Scholar]
- Joy, A.; Pragna, P.; Archana, P.R.; Sejian, P.R.; Bagath, M. Significance of metabolic response in livestock for adapting to heat stress challenges. Asian J. Anim. Sci. 2016, 10, 224–234. [Google Scholar]
- Joy, A.; Dunshea, F.R.; Leury, B.J.; Clarke, I.J.; DiGiacomo, K.; Chauhan, S.S. Resilience of small ruminants to climate change and increased environmental temperature: A review. Animal 2020, 10, 867. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, L.; Prakapenka, D.; VanRaden, P.M.; Cole, J.B.; Da, Y. A large-scale genome-wide association study in U.S. Holstein cattle. Front. Genet. 2019, 14, 412. [Google Scholar] [CrossRef]
- Dikmen, S.; Cole, J.B.; Null, D.J.; Hansen, P.J. Genome-wide association mapping for identification of quantitative trait loci for rectal temperature during heat stress in Holstein cattle. PLoS ONE 2013, 8, e69202. [Google Scholar] [CrossRef]
- Hammami, H.; Vandenplas, J.; Vanrobays, M.L.; Rekik, B.; Bastin, C.; Gengler, N. Genetic analysis of heat stress effects on yield traits, udder health, and fatty acids of Walloon Holstein cows. J. Dairy Sci. 2015, 98, 4956–4968. [Google Scholar] [CrossRef]
- Nayeri, S.; Sargolzaei, M.; Abo-Ismail, M.K.; May, N.; Miller., S.P.; Schenkel, F.; Moore, S.S.; Stothard, P. Genome-wide association for milk production and female fertility traits in Canadian dairy Holstein cattle. BMC Genet. 2016, 17, 75. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Bissonnette, N.; Lacasse, P.; Miglior, F.; Sargolzaei, M.; Zhao, X.; Ibeagha-Awemu, E.M. Genome-wide association analysis and pathways enrichment for lactation persistency in Canadian Holstein cattle. J. Dairy Sci. 2017, 100, 1955–1970. [Google Scholar] [CrossRef] [PubMed]
- Yodklaew, P.; Koonawootrittriron, S.; Elzo, M.A.; Suwanasopee, T.; Laodim, T. Genome-wide association study for lactation characteristics, milk yield and age at first calving in a Thai multibreed dairy cattle population. Agric. Nat. Resour. 2017, 51, 223–230. [Google Scholar] [CrossRef]
- Hong, E.P.; Park, J.W. Sample size and statistical power calculation in genetic association studies. Genom. Inform. 2012, 10, 117–122. [Google Scholar] [CrossRef]
- Streit, M.; Neugebauer, N.; Meuwissen, T.H.; Bennewitz, J. Short communication: Evidence for a major gene by polygene interaction for milk production traits in German Holstein dairy cattle. J. Dairy Sci. 2011, 94, 1597–1600. [Google Scholar] [CrossRef]
- Molee, A.; Duanghaklang, N.; Na-Lampang, P. Effects of acyl-CoA:diacylglycerol acyl transferase 1 (DGAT1) gene on milk production traits in crossbred Holstein dairy cattle. Trop. Anim. Health Prod. 2012, 44, 751–755. [Google Scholar] [CrossRef]
- Sigdel, A.; Abdollahi-Arpanahi, R.; Aguilar, I.; Peñagaricano, F. Whole genome mapping reveals novel genes and pathways involved in milk production under heat stress in US Holstein cows. Front. Genet. 2019, 4, 928. [Google Scholar] [CrossRef] [PubMed]
- Otto, P.I.; Guimarães, S.E.F.; Verardo, L.L.; Azevedo, A.L.S.; Vandenplas, J.; Soares, A.C.C.; Sevillano, C.A.; Veroneze, R.; de Fatima, A. Genome-wide association studies for tick resistance in Bos taurus × Bos indicus crossbred cattle: A deeper look into this intricate mechanism. J. Dairy Sci. 2018, 101, 11020–11032. [Google Scholar] [CrossRef]
- Luo, H.; Hu, L.; Brito, L.F.; Dou, J.; Sammad, A.; Chang, Y.; Ma, L.; Guo, G.; Liu, L.; Zhai, L.; et al. Weighted single-step GWAS and RNA sequencing reveals key candidate genes associated with physiological indicators of heat stress in Holstein cattle. J. Anim. Sci. Biotechnol. 2022, 13, 108. [Google Scholar] [CrossRef]
- Bohlouli, M.; Halli, K.; Yin, T.; Gengler, N.; König, S. Genome-wide associations for heat stress response suggest potential candidate genes underlying milk fatty acid composition in dairy cattle. J. Dairy Sci. 2022, 105, 3323–3340. [Google Scholar] [CrossRef]
- Hassan, F.; Nawaz, A.; Rehman, M.S.; Ali, M.A.; Dilshad, S.; Yan, C. Prospects of HSP70 as a genetic marker for thermo-tolerance and immuno-modulation in animals under climate change scenario. Anim. Nutr. 2019, 5, 340–350. [Google Scholar] [CrossRef] [PubMed]
- NRC. Nutrient Requirements of Dairy Cattle, 7th ed.; National Academy Press: Washington, DC, USA, 2001. [Google Scholar]
- Norman, H.D.; Miller, P.D.; McDaniel, B.T.; Dickinson, F.N. USDA-DHIA Factors for Standardizing 305 Day Lactation Records for Age and Moth to Calving; Forgotten Books: London, UK, 1974. [Google Scholar]
- Mader, T.L.; Johnson, L.J.; Gaughan, J.B. A comprehensive index for assessing environmental stress in animals. J. Anim Sci. 2010, 88, 2153–2165. [Google Scholar] [CrossRef]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’Connell, J.; Moore, S.S.; Smith, T.P.; Sonstegard, T.S.; et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Multiple significance tests: The Bonferroni method. BMJ 1995, 21, 170. [Google Scholar] [CrossRef]
- Castillo-Salas, C.A.; Luna-Nevárez, G.; Reyna-Granados, J.R.; Luna-Ramirez, R.I.; Limesand, S.W.; Luna-Nevárez, P. Molecular markers for thermo-tolerance are associated with reproductive and physiological traits in Pelibuey ewes raised in a semiarid environment. J. Therm. Biol. 2023, 112, 103475. [Google Scholar] [CrossRef]
- Weir, B.S. Forensics: Handbook of Statistical Genetics; John Wiley and Sons: Hoboken, NJ, USA, 2001. [Google Scholar]
- Sherman, E.L.; Nkrumah, J.D.; Murdoch, B.M.; Li, C.; Wang, Z.; Fu, A.; Moore, S.S. Polymorphisms and haplotypes in the bovine neuropeptide Y, growth hormone receptor, ghrelin, insulin-like growth factor 2, and uncoupling proteins 2 and 3 genes and their associations with measures of growth, performance, feed efficiency, and carcass merit in beef cattle. J. Anim. Sci. 2008, 86, 11–16. [Google Scholar]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Longman Scientific and Technical: New York, NY, USA, 1996. [Google Scholar]
- Purwanto, B.P.; Abo, Y.; Sakamoto, R.; Furumoto, F.; Yamamoto, S. Diurnal patterns of heat production and heart rate under thermoneutral conditions in Holstein Friesian cows differing in milk production. J. Agric. Sci. 1990, 114, 139–142. [Google Scholar] [CrossRef]
- Garner, J.B.; Douglas, M.L.; Williams, S.R.; Wales, W.J.; Marett, L.C.; Nguyen, T.T.; Reich, C.M.; Hayes, B.J. Genomic selection improves heat tolerance in dairy cattle. Sci. Rep. 2016, 29, 34114. [Google Scholar] [CrossRef] [PubMed]
- Macciotta, N.P.P.; Biffani, S.; Bernabucci, U.; Lacetera, N.; Vitali, A.; Ajmone-Marsan, P.; Nardone, A. Derivation and genome-wide association study of a principal component-based measure of heat tolerance in dairy cattle. J. Dairy Sci. 2017, 100, 4683–4697. [Google Scholar] [CrossRef]
- Tiezzi, F.; de Los Campos, G.; Parker-Gaddis, K.L.; Maltecca, C. Genotype by environment (climate) interaction improves genomic prediction for production traits in US Holstein cattle. J. Dairy Sci. 2017, 100, 2042–2056. [Google Scholar] [CrossRef]
- Bohlouli, M.; Yin, T.; Hammami, H.; Gengler, N.; König, S. Climate sensitivity of milk production traits and milk fatty acids in genotyped Holstein dairy cows. J. Dairy Sci. 2021, 104, 6847–6860. [Google Scholar] [CrossRef]
- Hariyono, D.N.H.; Prihandini, P.W. Association of selected gene polymorphisms with thermotolerance traits in cattle—A review. Anim. Biosci. 2022, 35, 1635–1648. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.; Eckard, R.; Beauchemin, K. Adaptation of ruminant livestock production systems to climate changes. Animal 2018, 12, s445–s456. [Google Scholar] [CrossRef]
- Luo, H.; Li, X.; Hu, L.; Xu, W.; Chu, Q.; Liu, A.; Guo, G.; Liu, L.; Brito, L.F.; Wang, Y. Genomic analyses and biological validation of candidate genes for rectal temperature as an indicator of heat stress in Holstein cattle. J. Dairy Sci. 2021, 104, 4441–4451. [Google Scholar] [CrossRef] [PubMed]
- Gebreyesus, G.; Buitenhuis, A.J.; Poulsen, N.A.; Visker, M.H.P.W.; Zhang, Q.; van Valenberg, H.J.F.; Sun, D.; Bovenhuis, H. Multi-population GWAS and enrichment analyses reveal novel genomic regions and promising candidate genes underlying bovine milk fatty acid composition. BMC Genom. 2019, 20, 178. [Google Scholar] [CrossRef]
- Grisart, B.; Coppieters, W.; Farnir, F.; Karim, L.; Ford, C.; Berzi, P.; Cambisano, N.; Mni, M.; Reid, S.; Simon, P.; et al. Positional candidate cloning of a QTL in dairy cattle: Identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002, 12, 222–231. [Google Scholar] [CrossRef]
- Lu, J.; Boeren, S.; van Hooijdonk, T.; Vervoort, J.; Hettinga, K. Effect of the DGAT1 K232A genotype of dairy cows on the milk metabolome and proteome. J. Dairy Sci. 2015, 98, 3460–3469. [Google Scholar] [CrossRef] [PubMed]
- Spelman, R.J.; Ford, C.A.; McElhinney, P.; Gregory, G.C.; Snell, R.G. Characterization of the DGAT1 gene in the New Zealand dairy population. J. Dairy Sci. 2002, 85, 3514–3517. [Google Scholar] [CrossRef]
- Hayes, B.J.; Bowman, P.J.; Chamberlain, A.J.; Goddard, M.E. Invited review: Genomic selection in dairy cattle: Progress and challenges. J. Dairy Sci. 2009, 92, 433–443. [Google Scholar] [CrossRef]
- Chamberlain, A.J.; Hayes, B.J.; Savin, K.; Bolormaa, S.; McPartlan, H.C.; Bowman, P.J.; Van der Jagt, C.; MacEachern, S.; Goddard, M.E. Validation of single nucleotide polymorphisms associated with milk production traits in dairy cattle. J. Dairy Sci. 2012, 95, 864–875. [Google Scholar] [CrossRef]
- Naukkarinen, J.; Surakka, I.; Pietiläinen, K.H.; Rissanen, A.; Salomaa, V.; Ripatti, S.; Yki-Järvinen, H.; van Duijn, C.M.; Wichmann, H.E.; Kaprio, J.; et al. ENGAGE Consortium. Use of genome-wide expression data to mine the “Gray Zone” of GWA studies leads to novel candidate obesity genes. PLoS Genet. 2010, 6, e1000976. [Google Scholar] [CrossRef]
- Weller, J.I.; Song, J.Z.; Heyen, D.W.; Lewin, H.A.; Ron, M. A new approach to the problem of multiple comparisons in the genetic dissection of complex traits. Genetics 1998, 150, 1699–1706. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Mosig, M.O.; Lipkin, E.; Khutoreskaya, G.; Tchourzyna, E.; Soller, M.; Friedmann, A. A whole genome scan for quantitative trait loci affecting milk protein percentage in Israeli-Holstein cattle, by means of selective milk DNA pooling in a daughter design, using an adjusted false discovery rate criterion. Genetics 2001, 157, 1683–1698. [Google Scholar] [CrossRef]
- Visscher, P.M. Sizing up human height variation. Nat. Genet. 2008, 40, 489–490. [Google Scholar] [CrossRef]
- Karlsson, E.K.; Baranowska, I.; Wade, C.M.; Salmon-Hillbertz, N.H.; Zody, M.C.; Anderson, N.; Biagi, T.M.; Patterson, N.; Pielberg, G.R.; Kulbokas, E.J.; et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 2007, 39, 1321–1328. [Google Scholar] [CrossRef]
- Pryce, J.E.; Haile-Mariam, M.; Verbyla, K.; Bowman, P.J.; Goddard, M.E.; Hayes, B.J. Genetic markers for lactation persistency in primiparous Australian dairy cows. J. Dairy Sci. 2010, 93, 2202–2214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bailey, S.D.; Lupien, M. Laying a solid foundation for Manhattan—‘setting the functional basis for the post-GWAS era’. Trends Genet. 2014, 30, 140–149. [Google Scholar] [CrossRef]
- Deng, N.; Zhou, H.; Fan, H.; Yuan, Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget 2017, 8, 110635–110649. Available online: https://www.oncotarget.com/article/22372/text/ (accessed on 19 November 2022).
- Salcedo-Tacuma, D.; Parales-Giron, J.; Prom, C.; Chirivi, M.; Laguna, J.; Locj, A.L.; Contreras, G.A. Transcriptomic profiling of adipose tissue inflammation, remodeling, and lipid metabolism in periparturient dairy cows (Bos taurus). BMC Genom. 2020, 21, 824. [Google Scholar] [CrossRef]
- Wang, M.; Song, H.; Zhu, X.; Xing, S.; Zhang, M.; Zhang, H.; Wang, X.; Yang, Z.; Ding, X.; Karrow, N.A.; et al. Toll-like receptor 4 gene polymorphisms influence milk production traits in Chinese Holstein cows. J. Dairy Res. 2018, 85, 407–411. [Google Scholar] [CrossRef]
- Rajamanickam, K.; Leela, V.; Suganya, G.; Basha, S.H.; Parthiban, M.; Visha, P.; Elango, A. Thermal cum lipopolysaccharide-induced stress challenge downregulates functional response of bovine monocyte-derived macrophages. J. Dairy Sci. 2018, 101, 11020–11032. [Google Scholar]
- Liu, Z.; Ezernieks, V.; Wang, J.; Arachchillage, N.W.; Garner, J.B.; Wales, W.J.; Cocks, B.G.; Rochfort, S. Heat stress in dairy cattle alters lipid composition of milk. Sci. Rep. 2017, 7, 961. [Google Scholar] [CrossRef]
- Faylon, M.P.; Baumgard, L.H.; Rhoads, R.P.; Spurlock, D.M. Effects of acute heat stress on lipid metabolism of bovine primary adipocytes. J. Dairy Sci. 2015, 98, 8732–8740. [Google Scholar] [CrossRef]
- Baumgard, L.H.; Rhoads, R.P., Jr. Effects of heat stress on postabsorptive metabolism and energetics. Annu. Rev. Anim. Biosci. 2013, 1, 311–337. [Google Scholar] [CrossRef]
- Gupta, S.; Sharma, A.; Joy, A.; Dunshea, F.R.; Chauhan, S.S. The impact of heat stress on immune status of dairy cattle and strategies to ameliorate the negative effects. Animals 2022, 13, 107. [Google Scholar] [CrossRef]
- Scherer, S.W.; Soder, S.; Duvoisin, R.M.; Huizenga, J.J.; Tsui, L.C. The human metabotropic glutamate receptor 8 (GRM8) gene: A disproportionately large gene located at 7q31.3-q32.1. Genomics 1997, 44, 232–236. [Google Scholar] [CrossRef]
- Belhadj, S.I.; Najar, T.; Ghram, A.; Abdrrabba, M. Heat stress effects on livestock: Molecular, cellular and metabolic aspects, a review. J. Anim. Physiol. Anim. Nutr. 2016, 100, 401–412. [Google Scholar] [CrossRef]
- Yue, S.; Ding, S.; Zhou, J.; Yang, C.; Hu, X.; Zhao, X.; Wang, Z.; Wang, L.; Peng, Q.; Xue, B. Metabolomics Approach Explore Diagnostic Biomarkers and Metabolic Changes in Heat-Stressed Dairy Cows. Animals 2020, 10, 1741. [Google Scholar] [CrossRef]
- Qu, M.; Wei, S.; Chen, Z.; Wang, G.; Zheng, Y.; Yan, P. Differences of hormones involved in adipose metabolism and lactation between high and low producing Holstein cows during heat stress. Anim. Nutr. 2015, 1, 339–343. [Google Scholar] [CrossRef]
- Cheruiyot, E.K.; Haile-Mariam, M.; Cocks, B.G.; MacLeod, I.M.; Xiang, R.; Pryce, J.E. New loci and neuronal pathways for resilience to heat stress in cattle. Sci. Rep. 2021, 11, 16619. [Google Scholar] [CrossRef]
- Nakamura, K.; Morrison, S.F. A thermosensory pathway that controls body temperature. Nat. Neurosci. 2008, 11, 62–71. [Google Scholar] [CrossRef]
- Zou, M.L.; Chen, Z.H.; Teng, Y.Y.; Liu, S.Y.; Jia, Y.; Zhang, K.W.; Sun, Z.L.; Wu, J.J.; Yuan, Z.D.; Feng, Y.; et al. The Smad dependent TGF-β and BMP signaling pathway in bone remodeling and therapies. Front. Mol. Biosci. 2021, 8, 593310. [Google Scholar] [CrossRef]
- Marchildon, F.; St-Louis, C.; Akter, R.; Roodman, V.; Wiper-Bergeron, N.L. Transcription factor Smad3 is required for the inhibition of adipogenesis by retinoic acid. J. Biol. Chem. 2010, 285, 13274–13284. [Google Scholar] [CrossRef]
- Shijun, L.; Khan, R.; Raza, S.H.A.; Jieyun, H.; Chugang, M.; Kaster, N.; Gong, C.; Chunping, Z.; Schreurs, N.M.; Linsen, Z. Function and characterization of the promoter region of perilipin 1 (PLIN1): Roles of E2F1, PLAG1, C/EBPβ, and SMAD3 in bovine adipocytes. Genomics 2020, 112, 2400–2409. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, Z.Q.; Wang, Y.J.; Wang, M.; Yang, W.C. MiR-143 Regulates milk fat synthesis by targeting Smad3 in bovine mammary epithelial cells. Animals 2020, 10, 1453. [Google Scholar] [CrossRef]
- Dikmen, S.; Wang, X.Z.; Ortega, M.S.; Cole, J.B.; Null, D.J.; Hansen, P.J. Single nucleotide polymorphisms associated with thermoregulation in lactating dairy cows exposed to heat stress. J. Anim. Breed. Genet. 2015, 132, 409–419. [Google Scholar] [CrossRef]
- Badri, T.M.; Chen, K.L.; Alsiddig, M.A.; Li, L.; Cai, Y.; Wang, G.L. Genetic polymorphism in Hsp90AA1 gene is associated with the thermotolerance in Chinese Holstein cows. Cell Stress Chaperones 2018, 23, 639–651. [Google Scholar] [CrossRef]
- Jensen, L.M.; Jannaman, E.A.; Pryce, J.E.; De Vries, A.; Hansen, P.J. Effectiveness of the Australian breeding value for heat tolerance at discriminating responses of lactating Holstein cows to heat stress. J. Dairy Sci. 2022, 105, 7820–7828. [Google Scholar] [CrossRef]
- Afsal, A.; Sejian, V.; Bagath, M.; Krishnan, G.; Devaraj, C.; Bhatta. Heat stress and livestock adaptation: Neuro-endocrine regulation. Int. J. Vet. Anim. Med. 2018, 1, 2. [Google Scholar]
- Otto, P.I.; Guimarães, S.E.F.; Verardo, L.L.; Azevedo, A.L.S.; Vandenplas, J.; Sevillano, C.A.; Marques, D.B.D.; Pires, M.F.A.; de Freitas, C.; Verneque, R.S.; et al. Genome-wide association studies for heat stress response in Bos taurus × Bos indicus crossbred cattle. J. Dairy Sci. 2019, 102, 8148–8158. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Bowman, P.J.; Haile-Mariam, M.; Pryce, J.E.; Hayes, B.J. Genomic selection for tolerance to heat stress in Australian dairy cattle. J. Dairy Sci. 2016, 99, 2849–2862. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).