In Vitro Cytotoxic Effect of Aqueous Extracts from Leaves and Rhizomes of the Seagrass Posidonia oceanica (L.) Delile on HepG2 Liver Cancer Cells: Focus on Autophagy and Apoptosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Extracts from Green Leaves (GLE), Brown Leaves (BLE), and Rhizomes (RE) of P. oceanica
2.2. Chromatographic Analysis
2.3. Proteomic Analysis
2.4. Cell Culture and Viability Assay
2.5. Clonogenic Assay
2.6. Wound-Healing Assay
2.7. Flow Cytometry Assays
2.7.1. Cell-Cycle Analysis
2.7.2. Apoptosis Assay
2.7.3. Analysis of Mitochondrial Transmembrane Potential (MMP)
2.7.4. Reactive Oxygen Species (ROS) Detection
2.7.5. Acidic Vesicular Organelles’ (AVO) Accumulation Analysis
2.8. Caspase Activity Assay
2.9. Preparation of Cell Lysates and Western Blot
2.10. RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR (qRT-PCR)
2.11. Statistics
3. Results
3.1. Characterization of GLE, BLE, and RE
3.2. Short-Term Viability and Long-Term Proliferative Potential of HepG2 Cells Exposed to P. oceanica Extracts
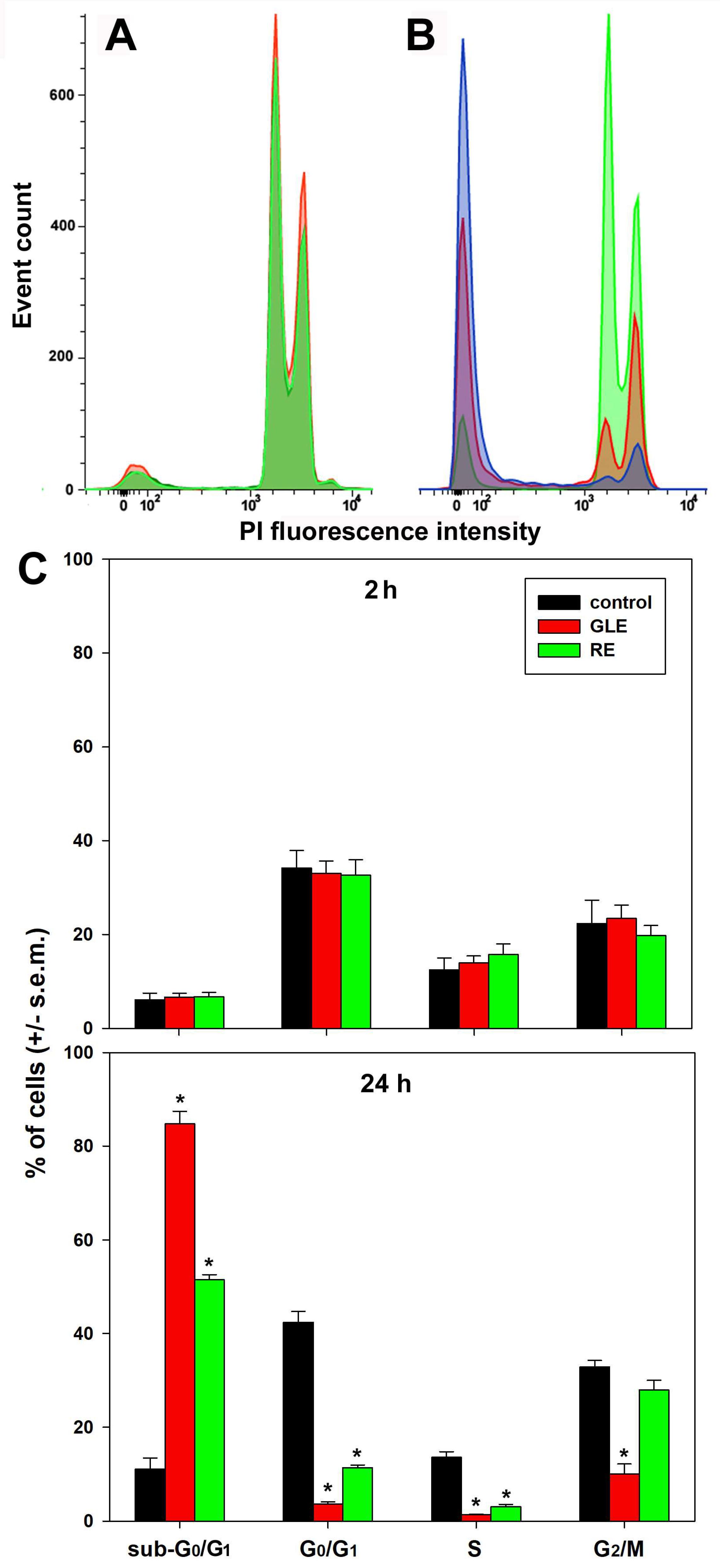
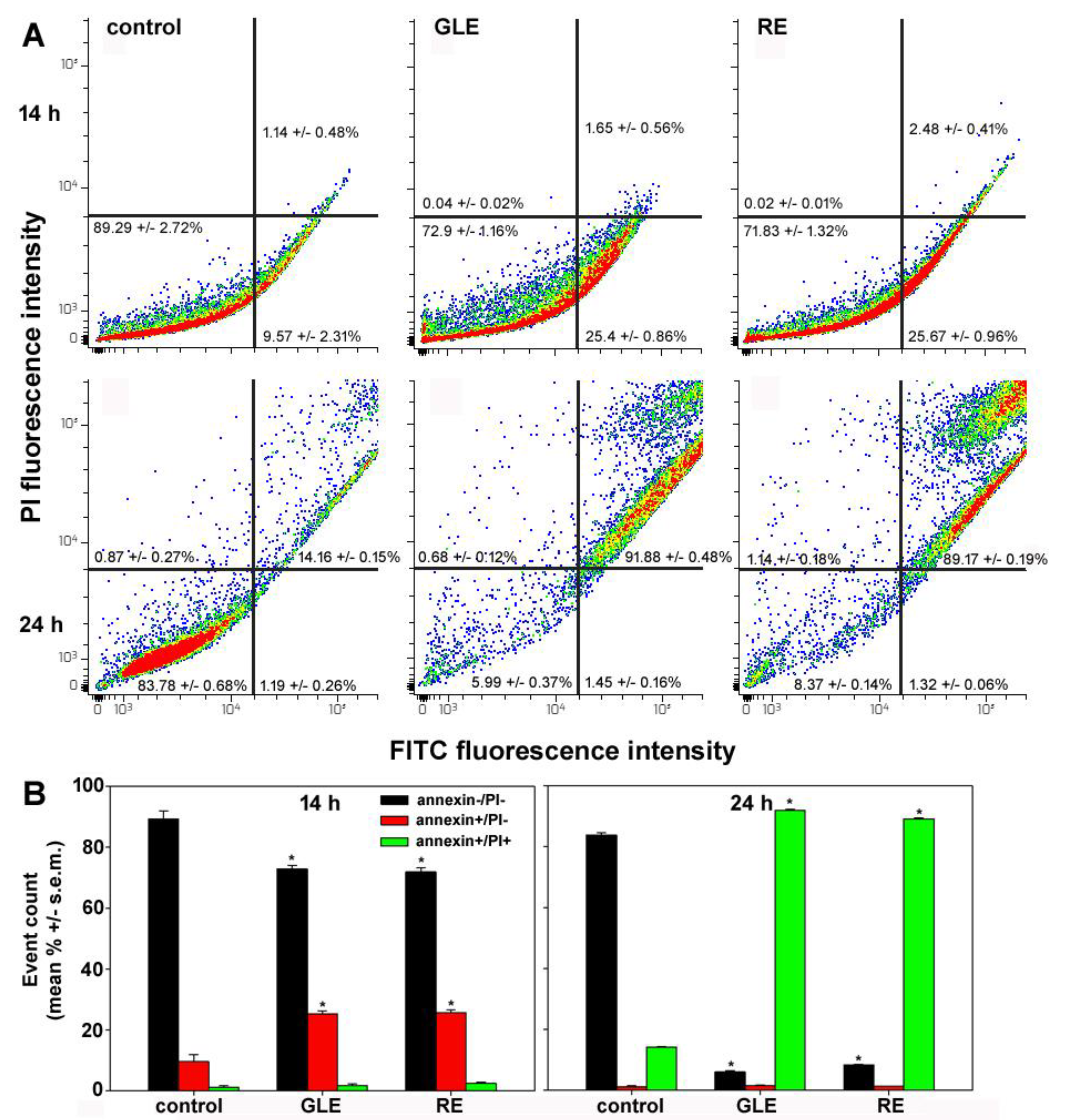
3.3. Cell-Cycle Status and Apoptosis Induction in HepG2 Cells Exposed to P. oceanica Extracts
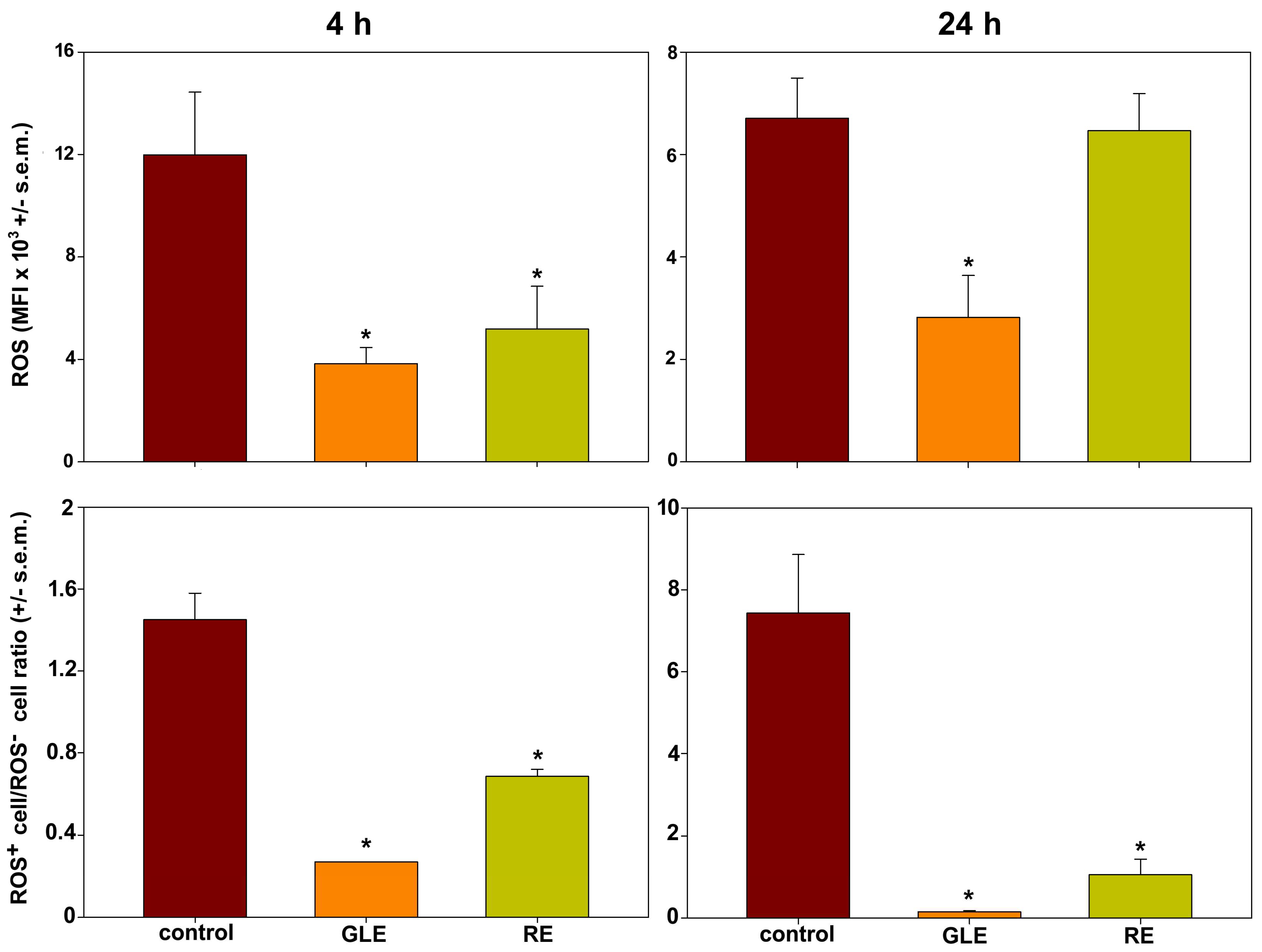
3.4. Mitochondrial Membrane Polarization and Redox Status in HepG2 Cells Exposed to P. oceanica Extracts
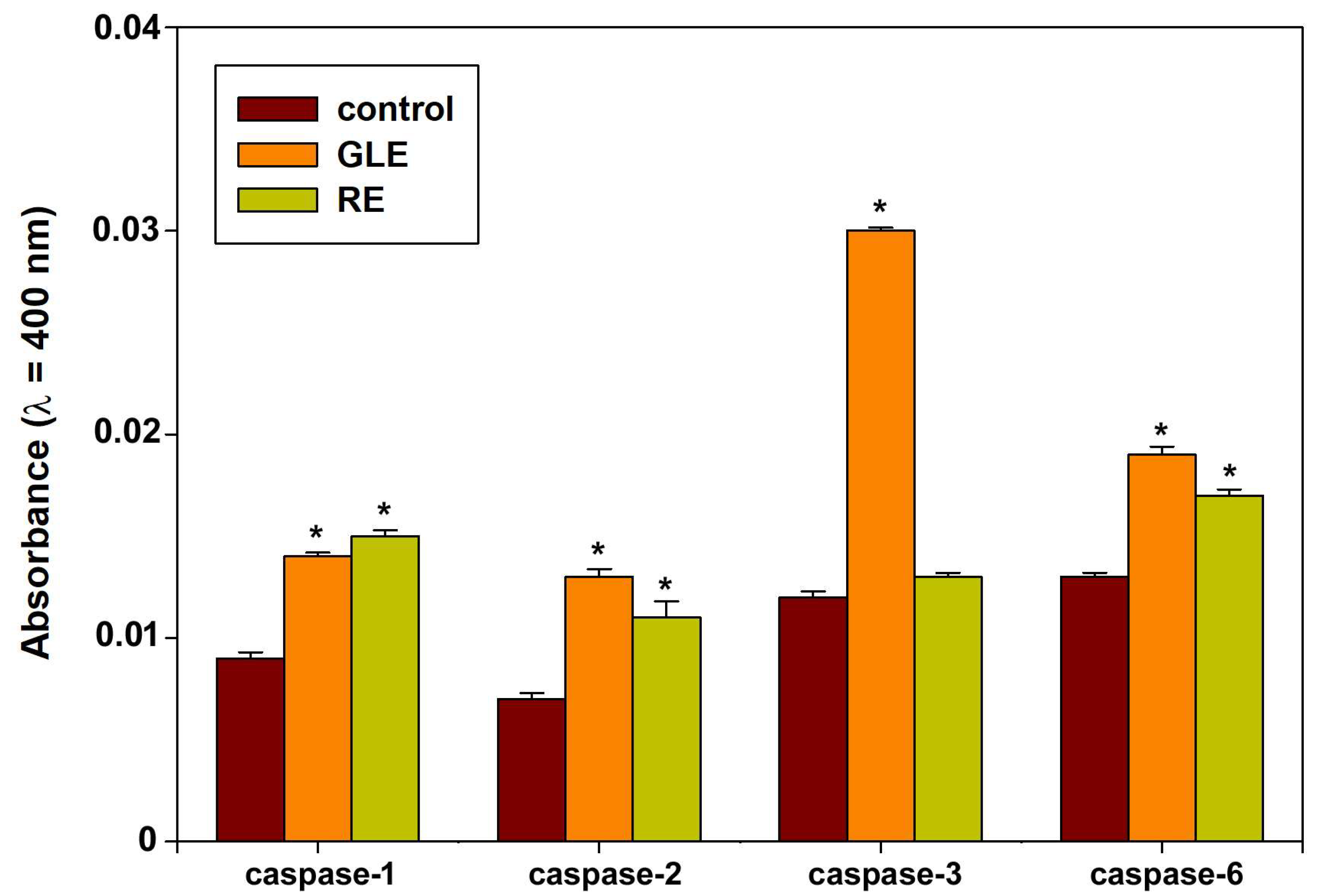
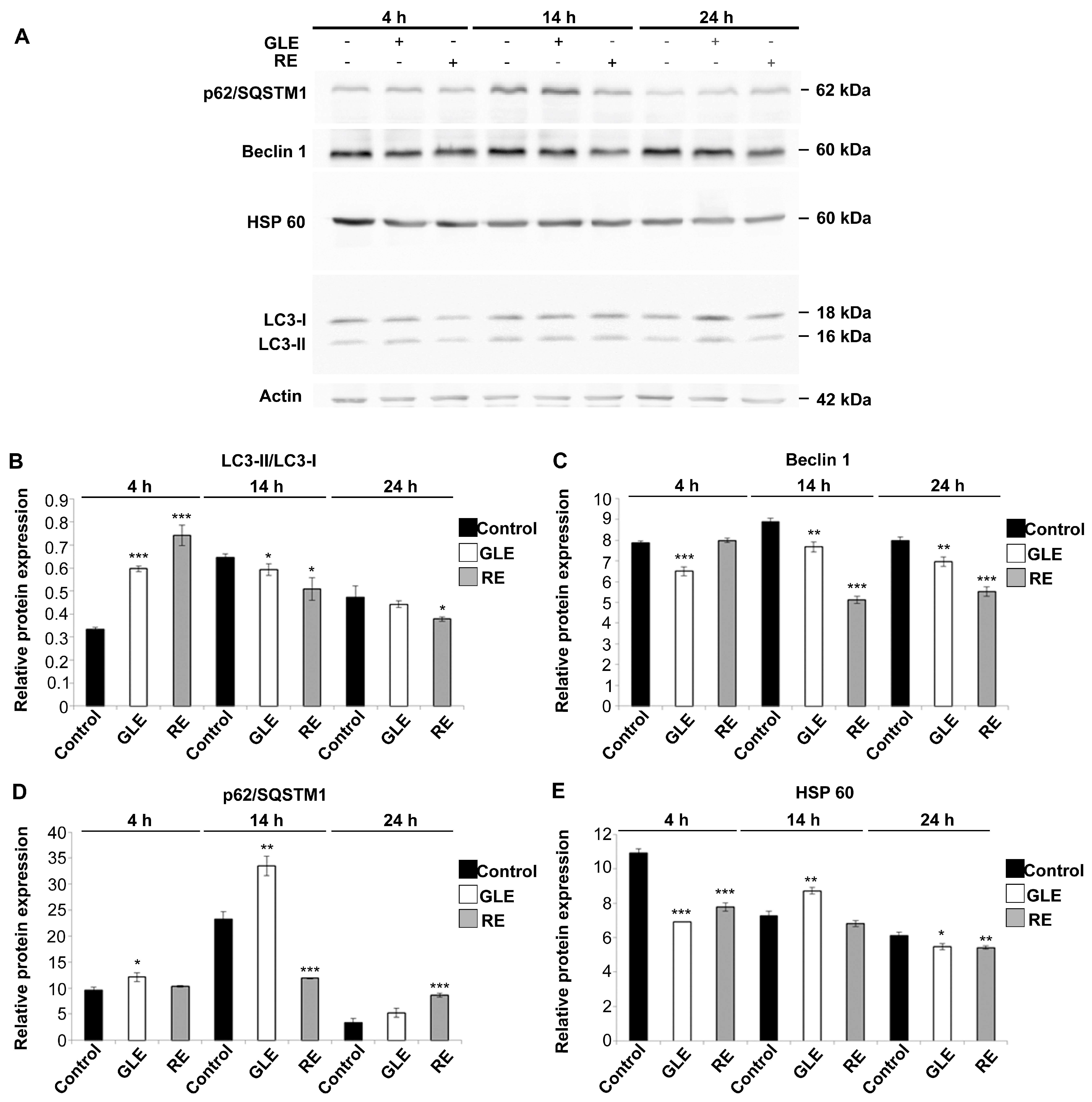
3.5. Indices of Autophagic Activity in HepG2 Cells Exposed to P. oceanica Extracts
3.6. Inhibition of the Locomotory Ability of HepG2 Cells Exposed to P. oceanica Extracts
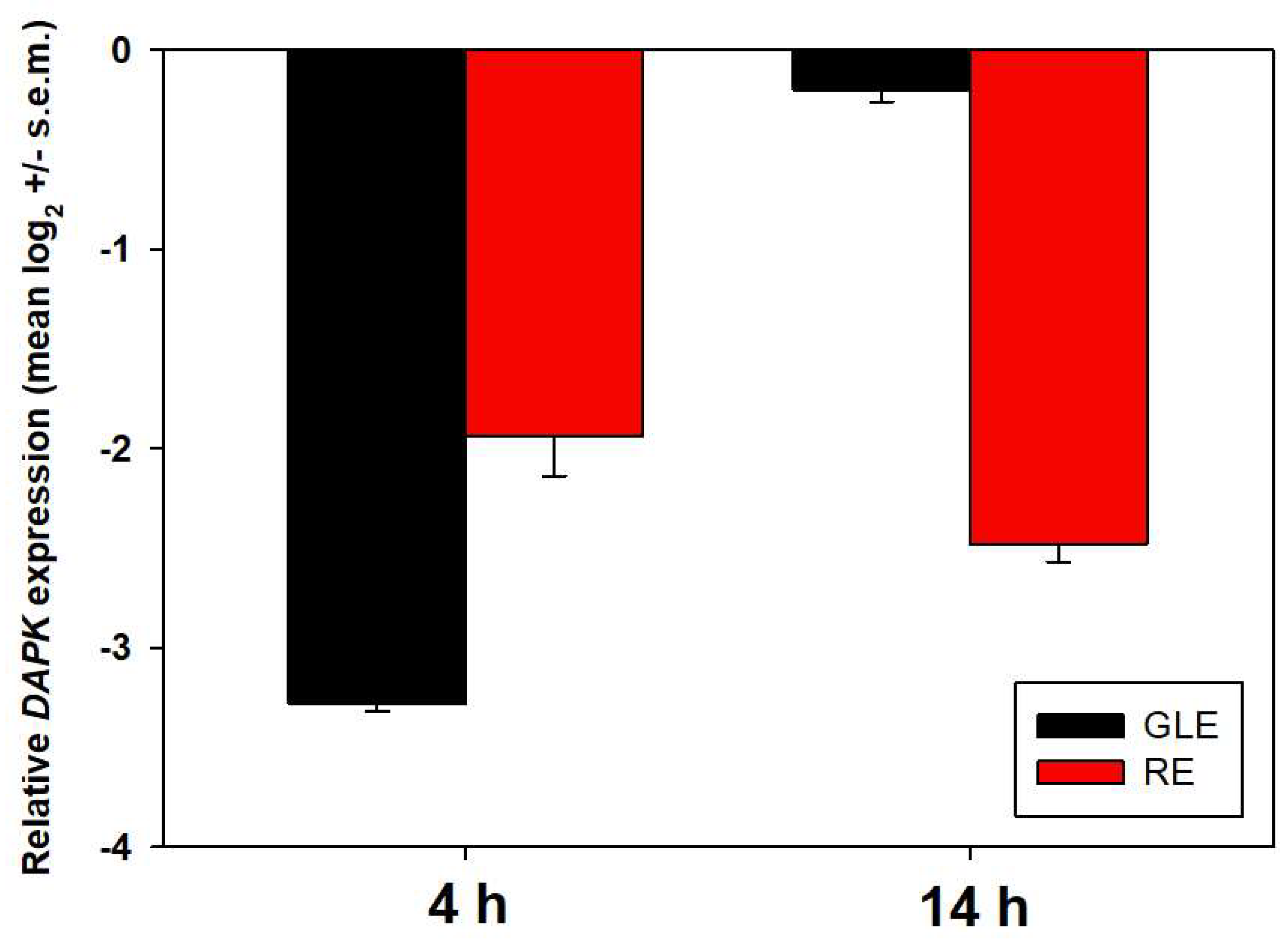
3.7. Potential Protein Contributors to the Observed GLE- and RE-Cytotoxic Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bellissimo, G.; Sirchia, B.; Ruvolo, V. Monitoring of Posidonia oceanica meadows in the Sicilian coasts under the Water Framework Directive (WFD). In Eighth International Symposium “Monitoring of Mediterranean Coastal Areas. Problems and Measurement Techniques”; Carboni, D., De Vincenzi, M., Bonora, L., Eds.; Firenze University Press: Firenze, Italy, 2020; pp. 510–518. [Google Scholar]
- Bernard, P.; Pesando, D. Antibacterial and Antifungal Activity of Extracts from the Rhizomes of the Mediterranean Seagrass Posidonia oceanica (L.) Delile. Bot. Mar. 1989, 32, 85–88. [Google Scholar] [CrossRef]
- Berfad, M.A.; Alnour, T.M.S. Phytochemical analysis and Antibacterial activity of the 5 different extract from the seagrasses Posidonia oceanica. J. Med. Plants Stud. 2014, 2, 15–18. [Google Scholar]
- Gokce, G.; Haznedaroglu, M.Z. Evaluation of antidiabetic, antioxidant and vasoprotective effects of Posidonia oceanica extract. J. Ethnopharmacol. 2008, 115, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Cornara, L.; Pastorino, G.; Borghesi, B.; Salis, A.; Clericuzio, M.; Marchetti, C.; Damonte, G.; Burlando, B. Posidonia oceanica (L.) Delile Ethanolic Extract Modulates Cell Activities with Skin Health Applications. Mar. Drugs 2018, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Vasarri, M.; Leri, M.; Barletta, E.; Ramazzotti, M.; Marzocchini, R.; Degl’Innocenti, D. Anti-inflammatory properties of the marine plant Posidonia oceanica (L.) Delile. J. Ethnopharmacol. 2020, 247, 112252. [Google Scholar] [CrossRef]
- Vasarri, M.; Barletta, E.; Ramazzotti, M.; Degl’Innocenti, D. In vitro anti-glycation activity of the marine plant Posidonia oceanica (L.) Delile. J. Ethnopharmacol. 2020, 259, 112960. [Google Scholar] [CrossRef]
- Messina, C.M.; Arena, R.; Manuguerra, S.; Pericot, Y.; Curcuraci, E.; Kerninon, F.; Renda, G.; Hellio, C.; Santulli, A. Antioxidant Bioactivity of Extracts from Beach Cast Leaves of Posidonia oceanica (L.) Delile. Mar. Drugs 2021, 19, 560. [Google Scholar] [CrossRef]
- Barletta, E.; Ramazzotti, M.; Fratianni, F.; Pessani, D.; Degl’Innocenti, D. Hydrophilic extract from Posidonia oceanica inhibits activity and expression of gelatinases and prevents HT1080 human fibrosarcoma cell line invasion. Cell Adhes. Migr. 2015, 9, 422–431. [Google Scholar] [CrossRef]
- Leri, M.; Ramazzotti, M.; Vasarri, M.; Peri, S.; Barletta, E.; Pretti, C.; Degl’Innocenti, D. Bioactive Compounds from Posidonia oceanica (L.) Delile Impair Malignant Cell Migration through Autophagy Modulation. Mar. Drugs 2018, 16, 137. [Google Scholar] [CrossRef]
- Vasarri, M.; Leri, M.; Barletta, E.; Pretti, C.; Degl’Innocenti, D. Posidonia oceanica (L.) Delile Dampens Cell Migration of Human Neuroblastoma Cells. Mar. Drugs 2021, 19, 579. [Google Scholar] [CrossRef]
- Farid, M.M.; Marzouk, M.M.; Hussein, S.R.; Elkhateeb, A.; Abdel-Hameed, E.S. Comparative study of Posidonia oceanica L.: LC/ESI/MS analysis, cytotoxic activity and chemosystematic significance. J. Mater. Environ. Sci. 2018, 9, 1676–1682. [Google Scholar]
- Asafo-Agyei, K.O.; Samant, H. Hepatocellular Carcinoma. In StatPearls [Internet]; [Updated 8 May 2022]; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Philips, C.A.; Rajesh, S.; Nair, D.C.; Ahamed, R.; Abduljaleel, J.K.; Augustine, P. Hepatocellular Carcinoma in 2021: An Exhaustive Update. Cureus 2021, 13, e19274. [Google Scholar] [CrossRef] [PubMed]
- Brar, T.S.; Hilgenfeldt, E.; Soldevila-Pico, C. Etiology and Pathogenesis of Hepatocellular Carcinoma. In Precision Molecular Pathology of Liver Cancer. Molecular Pathology Library; Liu, C., Ed.; Springer: Cham, Switzerland, 2018; pp. 1–15. [Google Scholar]
- Farzaneh, Z.; Vosough, M.; Agarwal, T.; Farzaneh, M. Critical signaling pathways governing hepatocellular carcinoma behavior; small molecule-based approaches. Cancer Cell Int. 2021, 21, 208. [Google Scholar] [CrossRef]
- Kaal, J.; Serrano, O.; Nieropd, K.G.J.; Schellekense, J.; Martínez Cortizas, A.; Mateo, M.A. Molecular composition of plant parts and sediment organic matter in a Mediterranean seagrass (Posidonia oceanica) mat. Aquat. Bot. 2016, 113, 50–61. [Google Scholar] [CrossRef]
- Astudillo-Pascual, M.; Domínguez, I.; Aguilera, P.A.; Garrido Frenich, A. New Phenolic Compounds in Posidonia oceanica Seagrass: A Comprehensive Array Using High Resolution Mass Spectrometry. Plants 2021, 10, 864. [Google Scholar] [CrossRef]
- Donato, M.T.; Tolosa, L.; Gómez-Lechón, M.J. Culture and Functional Characterization of Human Hepatoma HepG2 Cells. Meth. Mol. Biol. 2015, 1250, 77–93. [Google Scholar]
- Luparello, C.; Branni, R.; Abruscato, G.; Lazzara, V.; Drahos, L.; Arizza, V.; Mauro, M.; Di Stefano, V.; Vazzana, M. Cytotoxic capability and the associated proteomic profile of cell-free coelomic fluid extracts from the edible sea cucumber Holothuria tubulosa on HepG2 liver cancer cells. EXCLI J. 2022, 21, 722–743. [Google Scholar]
- Luparello, C.; Branni, R.; Abruscato, G.; Lazzara, V.; Sugár, S.; Arizza, V.; Mauro, M.; Di Stefano, V.; Vazzana, M. Biological and Proteomic Characterization of the Anti-Cancer Potency of Aqueous Extracts from Cell-Free Coelomic Fluid of Arbacia lixula Sea Urchin in an In Vitro Model of Human Hepatocellular Carcinoma. J. Mar. Sci. Eng. 2022, 10, 1292. [Google Scholar] [CrossRef]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef]
- Chou, T.C.; Martin, N. CompuSyn for Drug Combination User’s Guide: A Computer Program for Quantitation of Synergism and Antagonism in Drug Combinations, and the Determination of IC50 and ED50 Values. ComboSyn Inc.: Paramus, NJ, USA, 2004. Available online: https://combosyn.com/uat/pdf/CompuSyn_users_guide.pdf (accessed on 16 January 2023).
- Librizzi, M.; Longo, A.; Chiarelli, R.; Amin, J.; Spencer, J.; Luparello, C. Cytotoxic effects of Jay Amin hydroxamic acid (JAHA), a ferrocene-based class I histone deacetylase inhibitor, on triple-negative MDA-MB231 breast cancer cells. Chem. Res. Toxicol. 2012, 25, 2608–2616. [Google Scholar] [CrossRef]
- Ramos, A.A.; Prata-Sena, M.; Castro-Carvalho, B.; Dethoup, T.; Buttachon, S.; Kijjoa, A.; Rocha, E. Potential of four marine-derived fungi extracts as anti-proliferative and cell death-inducing agents in seven human cancer cell lines. Asian Pac. J. Trop. Med. 2015, 8, 798–806. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Asaro, D.M.L.; Cruciata, I.; Hassell-Hart, S.; Sansook, S.; Spencer, J.; Caradonna, F. Cytotoxic activity of the histone deacetylase 3-selective inhibitor Pojamide on MDA-MB-231 triple-negative breast cancer cells. Int. J. Mol. Sci. 2019, 20, 804. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Ragona, D.; Asaro, D.M.L.; Lazzara, V.; Affranchi, F.; Celi, M.; Arizza, V.; Vazzana, M. Cytotoxic Potential of the Coelomic Fluid Extracted from the Sea Cucumber Holothuria tubulosa against Triple-Negative MDA-MB231 Breast Cancer Cells. Biology 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Ragona, D.; Asaro, D.M.L.; Lazzara, V.; Affranchi, F.; Arizza, V.; Vazzana, M. Cell-Free Coelomic Fluid Extracts of the Sea Urchin Arbacia lixula Impair Mitochondrial Potential and Cell Cycle Distribution and Stimulate Reactive Oxygen Species Production and Autophagic Activity in Triple-Negative MDA-MB231 Breast Cancer Cells. J. Mar.Sci. Eng. 2020, 8, 261. [Google Scholar] [CrossRef]
- Chiarelli, R.; Martino, C.; Roccheri, M.C.; Cancemi, P. Toxic effects induced by vanadium on sea urchin embryos. Chemosphere 2021, 274, 129843. [Google Scholar] [CrossRef] [PubMed]
- Aranda, P.S.; LaJoie, D.M.; Jorcyk, C.L. Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis 2012, 33, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Sirchia, R.; Lo Sasso, B. Midregion PTHrP regulates Rip1 and caspase expression in MDA-MB231 breast cancer cells. Breast Cancer Res. Treat. 2008, 111, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Longo, A.; Vetrano, M. Exposure to cadmium chloride influences astrocyte-elevated gene-1 (AEG-1) expression in MDA-MB231 human breast cancer cells. Biochimie 2012, 94, 207–213. [Google Scholar] [CrossRef]
- Ronchetti, S.A.; Miler, E.A.; Duvilanski, B.H.; Cabilla, J.P. Cadmium mimics estrogen-driven cell proliferation and prolactin secretion from anterior pituitary cells. PLoS ONE 2013, 8, e81101. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Zhou, Y.; Liu, J.; Ou, X.; Li, M.; Long, X.; Ye, J.; Yu, G. AngII induces HepG2 cells to activate epithelial-mesenchymal transition. Exp. Ther. Med. 2018, 16, 3471–3477. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Chao, X.; Zhou, X.; Zheng, G.; Dong, C.; Zhang, W.; Song, X.; Jin, T. Osthole induces G2/M cell cycle arrest and apoptosis in human hepatocellular carcinoma HepG2 cells. Pharm. Biol. 2014, 52, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; He, Z.; Liu, J.; Wu, J.; Tan, G.; Jiang, J.; Su, Z.; Cao, M. Paris polyphylla 26 triggers G2/M phase arrest and induces apoptosis in HepG2 cells via inhibition of the Akt signaling pathway. J. Int. Med. Res. 2019, 47, 1685–1695. [Google Scholar] [CrossRef] [PubMed]
- Golestani Eimani, B.; Sanati, M.H.; Houshmand, M.; Ataei, M.; Akbarian, F.; Shakhssalim, N. Expression and prognostic significance of bcl-2 and bax in the progression and clinical outcome of transitional bladder cell carcinoma. Cell J. 2014, 15, 356–363. [Google Scholar]
- Kale, J.; Osterlund, E.; Andrews, D. BCL-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 2018, 25, 65–80. [Google Scholar] [CrossRef]
- Singh, R.; Czaja, M.J. Regulation of hepatocyte apoptosis by oxidative stress. J. Gastroenterol. Hepatol. 2007, 22 (Suppl. S1), S45–S48. [Google Scholar] [CrossRef]
- Podar, K.; Raab, M.S.; Tonon, G.; Sattler, M.; Barilà, D.; Zhang, J.; Tai, Y.T.; Yasui, H.; Raje, N.; DePinho, R.A.; et al. Up-regulation of c-Jun inhibits proliferation and induces apoptosis via caspase-triggered c-Abl cleavage in human multiple myeloma. Cancer Res. 2007, 67, 1680–1688. [Google Scholar] [CrossRef]
- Yuan, Z.; Gong, S.; Luo, J.; Zheng, Z.; Song, B.; Ma, S.; Guo, J.; Hu, C.; Thiel, G.; Vinson, C.; et al. Opposing roles for ATF2 and c-Fos in c-Jun-mediated neuronal apoptosis. Mol. Cell. Biol. 2009, 29, 2431–2442. [Google Scholar] [CrossRef]
- Gao, F.; Zhou, L.; Li, M.; Liu, W.; Yang, S.; Li, W. Inhibition of ERKs/Akt-Mediated c-Fos Expression Is Required for Piperlongumine-Induced Cyclin D1 Downregulation and Tumor Suppression in Colorectal Cancer Cells. Onco Targets Ther. 2020, 13, 5591–5603. [Google Scholar] [CrossRef]
- Yang, C.W.; Lee, Y.Z.; Hsu, H.Y.; Wu, C.M.; Chang, H.Y.; Chao, Y.S.; Lee, S.J. c-Jun-mediated anticancer mechanisms of tylophorine. Carcinogenesis 2013, 34, 1304–1314. [Google Scholar] [CrossRef]
- Schlossberg, H.; Zhang, Y.; Dudus, L.; Engelhardt, J.F. Expression of c-fos and c-jun during hepatocellular remodeling following ischemia/reperfusion in mouse liver. Hepatology 1996, 23, 1546–1555. [Google Scholar] [CrossRef] [PubMed]
- Auten, R.; Davis, J. Oxygen Toxicity and Reactive Oxygen Species: The Devil is in the Details. Pediatr. Res. 2009, 66, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Zaidieh, T.; Smith, J.R.; Ball, K.E.; An, Q. ROS as a novel indicator to predict anticancer drug efficacy. BMC Cancer 2019, 19, 1224. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Briedé, J.J.; Jennen, D.G.; Van Summeren, A.; Saritas-Brauers, K.; Schaart, G.; Kleinjans, J.C.; de Kok, T.M. Increased mitochondrial ROS formation by acetaminophen in human hepatic cells is associated with gene expression changes suggesting disruption of the mitochondrial electron transport chain. Toxicol. Lett. 2015, 234, 139–150. [Google Scholar] [CrossRef]
- Belyaeva, E.A.; Dymkowska, D.; Wieckowski, M.R.; Wojtczak, L. Mitochondria as an important target in heavy metal toxicity in rat hepatoma AS-30D cells. Toxicol. Appl. Pharmacol. 2008, 231, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Kenny, T.C.; Craig, A.J.; Villanueva, A.; Germain, D. Mitohormesis Primes Tumor Invasion and Metastasis. Cell Rep 2019, 27, 2292–2303.e6. [Google Scholar] [CrossRef]
- Bjørkøy, G.; Lamark, T.; Johansen, T. p62/SQSTM1: A missing link between protein aggregates and the autophagy machinery. Autophagy 2006, 2, 138–139. [Google Scholar] [CrossRef]
- Decuypere, J.P.; Parys, J.B.; Bultynck, G. Regulation of the autophagic bcl-2/beclin 1 interaction. Cells 2012, 3, 284–312. [Google Scholar] [CrossRef]
- Pace, A.; Barone, G.; Lauria, A.; Martorana, A.; Piccionello, A.P.; Pierro, P.; Terenzi, A.; Almerico, A.M.; Buscemi, S.; Campanella, C.; et al. Hsp60, a novel target for antitumor therapy: Structure-function features and prospective drugs design. Curr. Pharm. Des. 2013, 19, 2757–2764. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar]
- Singh, P.; Ravanan, P.; Talwar, P. Death Associated Protein Kinase 1 (DAPK1): A Regulator of Apoptosis and Autophagy. Front. Mol. Neurosci. 2016, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Farag, A.K.; Roh, E.J. Death-associated protein kinase (DAPK) family modulators: Current and future therapeutic outcomes. Med. Res. Rev. 2019, 39, 349–385. [Google Scholar] [CrossRef] [PubMed]
- Yogev, O.; Goldberg, R.; Anzi, S.; Yogev, O.; Shaulian, E. Jun proteins are starvation-regulated inhibitors of autophagy. Cancer Res. 2010, 70, 2318–2327. [Google Scholar] [CrossRef]
- Xie, X.; Zhu, H.; Zhang, J.; Wang, M.; Zhu, L.; Guo, Z.; Shen, W.; Wang, D. Solamargine inhibits the migration and invasion of HepG2 cells by blocking epithelial-to-mesenchymal transition. Oncol. Lett. 2017, 14, 447–452. [Google Scholar] [CrossRef]
- Zheng, J.; Shao, Y.; Jiang, Y.; Chen, F.; Liu, S.; Yu, N.; Zhang, D.; Liu, X.; Zou, L. Tangeretin inhibits hepatocellular carcinoma proliferation and migration by promoting autophagy-related BECLIN1. Cancer Manag. Res. 2019, 11, 5231–5242. [Google Scholar] [CrossRef] [PubMed]
- Furth, E.E.; Sprecher, H.; Fisher, E.A.; Fleishman, H.D.; Laposata, M. An in vitro model for essential fatty acid deficiency: HepG2 cells permanently maintained in lipid-free medium. J. Lipid Res. 1992, 33, 1719–1726. [Google Scholar] [CrossRef] [PubMed]
- Desquiret, V.; Loiseau, D.; Jacques, C.; Douay, O.; Malthièry, Y.; Ritz, P.; Roussel, D. Dinitrophenol-induced mitochondrial uncoupling in vivo triggers respiratory adaptation in HepG2 cells. Biochim. Biophys. Acta 2006, 1757, 21–30. [Google Scholar] [CrossRef]
- Lazzara, V.; Arizza, V.; Luparello, C.; Mauro, M.; Vazzana, M. Bright Spots in The Darkness of Cancer: A Review of Starfishes-Derived Compounds and Their Anti-Tumor Action. Mar. Drugs 2019, 17, 617. [Google Scholar] [CrossRef]
- Luparello, C.; Mauro, M.; Lazzara, V.; Vazzana, M. Collective Locomotion of Human Cells, Wound Healing and Their Control by Extracts and Isolated Compounds from Marine Invertebrates. Molecules 2020, 25, 2471. [Google Scholar] [CrossRef]
- Luparello, C.; Mauro, M.; Arizza, V.; Vazzana, M. Histone Deacetylase Inhibitors from Marine Invertebrates. Biology 2020, 9, 429. [Google Scholar] [CrossRef]
- Mauro, M.; Lazzara, V.; Punginelli, D.; Arizza, V.; Vazzana, M. Antitumoral compounds from vertebrate sister group: A review of Mediterranean ascidians. Dev. Comp. Immunol. 2020, 108, 103669. [Google Scholar] [CrossRef] [PubMed]
- Punginelli, D.; Schillaci, D.; Mauro, M.; Deidun, A.; Barone, G.; Arizza, V.; Vazzana, M. The potential of antimicrobial peptides isolated from freshwater crayfish species in new drug development: A review. Dev. Comp. Immunol. 2022, 126, 104258. [Google Scholar] [CrossRef] [PubMed]
- Ammar, N.M.; Hassan, H.A.; Mohammed, M.A.; Serag, A.; Abd El-Alim, S.H.; Elmotasem, H.; El Raey, M.; El Gendy, A.N.; Sobeh, M.; Abdel-Hamid, A.Z. Metabolomic profiling to reveal the therapeutic potency of Posidonia oceanica nanoparticles in diabetic rats. RSC Adv. 2021, 11, 8398–8410. [Google Scholar] [CrossRef] [PubMed]
- Maciel, L.G.; do Carmo, M.A.V.; Azevedo, L.; Daguer, H.; Molognoni, L.; de Almeida, M.M.; Granato, D.; Rosso, N.D. Hibiscus sabdariffa anthocyanins-rich extract: Chemical stability, in vitro antioxidant and antiproliferative activities. Food Chem. Toxicol. 2018, 113, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Alonso, C.; Taroncher, M.; Castaldo, L.; Izzo, L.; Rodríguez-Carrasco, Y.; Ritieni, A.; Ruiz, M.J. Effect of Phenolic Extract from Red Beans (Phaseolus vulgaris L.) on T-2 Toxin-Induced Cytotoxicity in HepG2 Cells. Foods 2022, 11, 1033. [Google Scholar] [CrossRef]
- Wei, S.; Sun, Y.; Wang, L.; Zhang, T.; Hu, W.; Bao, W.; Mao, L.; Chen, J.; Li, H.; Wen, Y.; et al. Hyperoside suppresses BMP-7-dependent PI3K/AKT pathway in human hepatocellular carcinoma cells. Ann. Transl. Med. 2021, 9, 1233. [Google Scholar] [CrossRef]
- Akbar, S.; Ishtiaq, S.; Jahangir, M.; Elhady, S.S.; Bogari, H.A.; Alahdal, A.M.; Ashour, M.L.; Youssef, F.S. Evaluation of The Antioxidant, Antimicrobial, and Anticancer Activities of Dicliptera bupleuroides Isolated Compounds Using In Vitro and In Silico Studies. Molecules 2021, 26, 7196. [Google Scholar] [CrossRef]
- Ibrahim, T.A.; El Dib, R.A.; Al-Youssef, H.M.; Amina, M. Chemical composition and antimicrobial and cytotoxic activities of Antidesm abunius L. Pak. J. Pharm. Sci. 2019, 32, 153–163. [Google Scholar]
- Huang, H.M.; Ho, C.Y.; Chang, G.R.; Shia, W.Y.; Lai, C.H.; Chao, C.H.; Wang, C.M. HPLC/ESI-MS and NMR Analysis of Chemical Constitutes in Bioactive Extract from the Root Nodule of Vaccinium emarginatum. Pharmaceuticals 2021, 14, 1098. [Google Scholar] [CrossRef]
- Zhou, M.N.; Liu, P.; Jing, S.J.; Sun, M.; Li, X.; Zhang, W.; Liu, B. Chemical constituents of Scrophulariae Radix and their antitumor activities in vitro. Zhongguo Zhong Yao Za Zhi 2022, 47, 111–121. [Google Scholar]
- Razali, N.; Mat Junit, S.; Ariffin, A.; Ramli, N.S.; Abdul Aziz, A. Polyphenols from the extract and fraction of T. indica seeds protected HepG2 cells against oxidative stress. BMC Complement. Altern. Med. 2015, 15, 438. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.H.; Yu, C.H.; Chang, Y.P.; Lin, Y.T.; Huang, C.J.; Kuo, Y.H.; Tsai, P.J. Protective Effect of Caffeic Acid Derivatives on tert-Butyl Hydroperoxide-Induced Oxidative Hepato-Toxicity and Mitochondrial Dysfunction in HepG2 Cells. Molecules 2017, 22, 702. [Google Scholar] [CrossRef] [PubMed]
- Samari, H.R.; Seglen, P.O. Inhibition of hepatocytic autophagy by adenosine, aminoimidazole-4-carboxamide riboside, and N6-mercaptopurine riboside. Evidence for involvement of amp-activated protein kinase. J. Biol. Chem. 1998, 273, 23758–23763. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Chen, Y.; Xu, K.; Li, Y.; Riaz, F.; Lu, K.; Chen, Q.; Du, X.; Wu, L.; Cao, D.; et al. Cholesterol-induced leucine aminopeptidase 3 (LAP3) upregulation inhibits cell autophagy in pathogenesis of NAFLD. Aging 2022, 14, 3259–3275. [Google Scholar] [CrossRef]
- Udupa, P.; Kumar, A.; Parit, R.; Ghosh, D.K. Acyl-CoA binding protein regulates nutrient-dependent autophagy. Metabolism 2022, 155338. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhang, X.; Bai, J.; Long, L.; Liu, D.; Zhou, Y. PGM1 suppresses colorectal cancer cell migration and invasion by regulating the PI3K/AKT pathway. Cancer Cell Int. 2022, 22, 201. [Google Scholar] [CrossRef]
- Yang, J.; Li, Q.; Yang, H.; Yan, L.; Yang, L.; Yu, L. Overexpression of human CUTA isoform2 enhances the cytotoxicity of copper to HeLa cells. Acta Biochim. Pol. 2008, 55, 411–415. [Google Scholar] [CrossRef]
- Shi, S.; Kang, X.J.; Zhou, Z.; He, Z.M.; Zheng, S.; He, S.S. Excessive mechanical stress-induced intervertebral disc degeneration is related to Piezo1 overexpression triggering the imbalance of autophagy/apoptosis in human nucleus pulpous. Arthritis Res. Ther. 2022, 24, 119. [Google Scholar] [CrossRef]
- Hermes, M.; Osswald, H.; Kloor, D. Role of S-adenosylhomocysteine hydrolase in adenosine-induced apoptosis in HepG2 cells. Exp. Cell Res. 2007, 313, 264–283. [Google Scholar] [CrossRef]
- Li, Q.; Mao, L.; Wang, R.; Zhu, L.; Xue, L. Overexpression of S-adenosylhomocysteine hydrolase (SAHH) in esophageal squamous cell carcinoma (ESCC) cell lines: Effects on apoptosis, migration and adhesion of cells. Mol. Biol. Rep. 2014, 41, 2409–2417. [Google Scholar] [CrossRef]
- Huang, W.; Li, N.; Zhang, Y.; Wang, X.; Yin, M.; Lei, Q.Y. AHCYL1 senses SAH to inhibit autophagy through interaction with PIK3C3 in an MTORC1-independent manner. Autophagy 2022, 18, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Schlattner, U.; Tokarska-Schlattner, M.; Epand, R.M.; Boissan, M.; Lacombe, M.L.; Kagan, V.E. NME4/nucleoside diphosphate kinase D in cardiolipin signaling and mitophagy. Lab. Investig. 2018, 98, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Mediavilla, M.G.; Di Venanzio, G.A.; Guibert, E.E.; Tiribelli, C. Heterologous ferredoxin reductase and flavodoxin protect Cos-7 cells from oxidative stress. PLoS ONE 2010, 5, e13501. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Choi, Y.H.; Choi, C.G.; Nam, T.J. Effects of the cyclophilin-type peptidylprolyl cis-trans isomerase from Pyropia yezoensis against hydrogen peroxide-induced oxidative stress in HepG2 cells. Mol. Med. Rep. 2017, 15, 4132–4138. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.N.; Rani, R.; Kokane, A.D.; Ghosh, D.K.; Tomar, S.; Sharma, A.K. Characterization of a cytoplasmic 2-Cys peroxiredoxin from Citrus sinensis and its potential role in protection from oxidative damage and wound healing. Int. J. Biol. Macromol. 2022, 209 Pt A, 1088–1099. [Google Scholar] [CrossRef]
- Couto, N.; Wood, J.; Barber, J. The role of glutathione reductase and related enzymes on cellular redox homoeostasis network. Free Radic Biol Med 2016, 95, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Elhusseiny, S.M.; El-Mahdy, T.S.; Awad, M.F.; Elleboudy, N.S.; Farag, M.M.S.; Yassein, M.A.; Aboshanab, K.M. Proteome Analysis and In Vitro Antiviral, Anticancer and Antioxidant Capacities of the Aqueous Extracts of Lentinula edodes and Pleurotus ostreatus Edible Mushrooms. Molecules 2021, 26, 4623. [Google Scholar] [CrossRef]
- Aniya, Y.; Imaizumi, N. Mitochondrial glutathione transferases involving a new function for membrane permeability transition pore regulation. Drug Metab. Rev. 2011, 43, 292–299. [Google Scholar] [CrossRef]
- Younus, H. Therapeutic potentials of superoxide dismutase. Int. J. Health Sci. 2018, 12, 88–93. [Google Scholar]
- Punginelli, D.; Catania, V.; Abruscato, G.; Luparello, C.; Vazzana, M.; Mauro, M.; Cunsolo, V.; Saletti, R.; Di Francesco, A.; Arizza, V.; et al. New Bioactive Peptides from the Mediterranean Seagrass Posidonia oceanica (L.) Delile and Their Impact on Antimicrobial Activity and Apoptosis of Human Cancer Cells. Int. J. Mol. Sci. 2023, 24, 5650. [Google Scholar] [CrossRef]
- Jeon, J.S.; Kwon, S.; Ban, K.; Kwon Hong, Y.; Ahn, C.; Sung, J.S.; Choi, I. Regulation of the Intracellular ROS Level Is Critical for the Antiproliferative Effect of Quercetin in the Hepatocellular Carcinoma Cell Line HepG2. Nutr. Cancer 2018, 71, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Tan, X.; Liang, J.; Wu, S.; Liu, J.; Zhang, Q.; Zhu, R. A reduction in reactive oxygen species contributes to dihydromyricetin-induced apoptosis in human hepatocellular carcinoma cells. Sci. Rep. 2014, 4, 7041. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef] [PubMed]
- Morel, E.; Mehrpour, M.; Botti, J.; Dupont, N.; Hamai, A.; Nascimbeni, A.C.; Codogno, P. Autophagy: A Druggable Process. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 375–398. [Google Scholar] [CrossRef]
- Yun, C.W.; Lee, S.H. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef]
- Luparello, C. Marine Animal-Derived Compounds and Autophagy Modulation in Breast Cancer Cells. Foundations 2021, 1, 3–20. [Google Scholar] [CrossRef]
- Tsapras, P.; Nezis, I.P. Caspase involvement in autophagy. Cell Death Differ. 2017, 24, 1369–1379. [Google Scholar] [CrossRef]
- Pang, X.; Gao, X.; Liu, F.; Jiang, Y.; Wang, M.; Li, Q.; Li, Z. Xanthoangelol modulates Caspase-1-dependent pyroptotic death among hepatocellular carcinoma cells with high expression of GSDMD. J. Funct. Foods 2021, 84, 104577. [Google Scholar] [CrossRef]
- Sun, Q.; Fan, J.; Billiar, T.R.; Scott, M.J. Inflammasome and autophagy regulation-a two-way street. Mol Med 2017, 23, 188–195. [Google Scholar] [CrossRef]
- Vigneswara, V.; Ahmed, Z. The Role of Caspase-2 in Regulating Cell Fate. Cells 2020, 9, 1259. [Google Scholar] [CrossRef]
- Tsagarakis, N.J.; Drygiannakis, I.; Batistakis, A.G.; Kolios, G.; Kouroumalis, E.A. Octreotide induces caspase activation and apoptosis in human hepatoma HepG2 cells. World J. Gastroenterol. 2011, 17, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Shalini, S.; Dorstyn, L.; Wilson, C.; Puccini, J.; Ho, L.; Kumar, S. Impaired antioxidant defence and accumulation of oxidative stress in caspase-2-deficient mice. Cell Death Differ. 2012, 19, 1370–1380. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Sharma, L.K.; Vanegas, D.; Callaway, D.A.; Bai, Y.; Lechleiter, J.D.; Herman, B. A nonapoptotic role for CASP2/caspase 2: Modulation of autophagy. Autophagy 2014, 10, 1054–1070, Erratum in Autophagy 2017, 13, 637. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, G.Y.; Bae, S.J.; Yoo, Y.H.; Choi, Y.H. Induction of apoptosis by isothiocyanate sulforaphane in human cervical carcinoma HeLa and hepatocarcinoma HepG2 cells through activation of caspase-3. Oncol. Rep. 2007, 18, 181–187. [Google Scholar] [CrossRef]
- Rust, C.; Wild, N.; Bernt, C.; Vennegeerts, T.; Wimmer, R.; Beuers, U. Bile acid-induced apoptosis in hepatocytes is caspase-6-dependent. J. Biol. Chem. 2009, 284, 2908–2916. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Lu, Y.H.; Xie, J.H.; Wang, F.; Zou, J.N.; Yang, J.S.; Xing, Y.Y.; Xi, T. Downregulation of survivin and activation of caspase-3 through the PI3K/Akt pathway in ursolic acid-induced HepG2 cell apoptosis. Anticancer Drugs 2009, 20, 249–258. [Google Scholar] [CrossRef]
- Valionyte, E.; Yang, Y.; Griffiths, S.A.; Bone, A.T.; Barrow, E.R.; Sharma, V.; Lu, B.; Luo, S. The caspase-6-p62 axis modulates p62 droplets based autophagy in a dominant-negative manner. Cell Death Differ. 2022, 29, 1211–1227. [Google Scholar] [CrossRef]






| Gene (Primer) | Sequence (5′→ 3′) | Reference |
|---|---|---|
| BAD (sense) | GTTCCAGATCCCAGAGTTTGAGC | [31] |
| BAD (antisense) | TTAAAGGAGTCCACAAACTCGTCACT | |
| BAX (sense) | ATGGACGGTCCGGGGAGCAGC | [31] |
| BAX (antisense) | CCCCAGTTGAAGTTGCCGTCAG | |
| BCL2 (sense) | GCCTTTGTGGAACTGTACGGC | [31] |
| BCL2 (antisense) | GGCAGTAAATAGCTGATTCGACGTT | |
| DAPK (sense) | GATGGCAACATGCCTATCGTG | [31] |
| DAPK (antisense) | GATGAAGAGTCCTCGGTGCGTAT | |
| JUN (sense) | CCCCAAGATCCTGAAACAGA | [32] |
| JUN (antisense) | CCGTTGCTGGACTGGATTAT | |
| FOS (sense) | CCAACTTTATCCCCACGGTGAC | [33] |
| FOS (antisense) | TGGCAATCTCGGTCTGCAAC | |
| ACTB (sense) | GGAAGGTGGACAGCGAGGCC | [34] |
| ACTB (antisense) | GTGACGTGGACATCCGCAAAG |
| Polyphenol | GLE (μg/g) | BLE (μg/g) | RE (μg/g) |
|---|---|---|---|
| Delphinidin-3-glucoside | n.q | - | 11.52 |
| Quercetin 3-O-galactoside | n.q | - | 10.81 |
| Procyanidin dimer B type isomer 2 | n.q | - | 0.20 |
| Procyanidin dimer B type isomer 3 | n.q | n.q | 0.30 |
| Pro-Cyanidin-Dimer-B | n.q | n.q | - |
| Cyanidin 3-O-glucoside | - | n.q | - |
| Vanillic acid | - | - | 0.6 |
| Gallic acid | n.q | - | n.q. |
| Kaempferol 3-O-glucoside | - | - | n.q. |
| Kaempferol | n.q | - | - |
| Kaempferol 7-O-hexuronide | - | - | n.q. |
| Procyanidin trimer B type | - | - | n.q. |
| Gallic acid ethyl ester | - | - | n.q. |
| Catechin | n.q | n.q | n.q. |
| Epicatechin | - | - | n.q. |
| Myricetin | - | - | n.q. |
| Peonidin 3-O-hexoside isomer | - | - | n.q. |
| Malvidin 3-O-pentoside | - | - | n.q. |
| Quercetin 3-O-hexuronide | - | - | n.q. |
| Quercetin 3-O-(6”-malonyl) hexoside | n.q | n.q | - |
| Resveratrol tetramer | - | - | n.q. |
| Caffeic acid methyl ester | 0.37 | - | - |
| Caffeic acid | n.q | n.q | - |
| p-Coumaric acid | n.q. | - | - |
| Ellagic acid | n.q | - | - |
| p-Hydroxybenzoic acid | - | 0.29 | - |
| Ferulic acid | - | n.q | - |
| Myricetin | - | n.q | - |
| Myricetin 3-O-hexoside | - | n.q | - |
| Petunidin 3-O-(6”-acetyl) hexoside | - | n.q | - |
| Accession Number/ Protein Description | GLE Amount | RE Amount |
|---|---|---|
| A0A843WDB0 ACB domain-containing protein | 5.34 × 104 | 3.72 × 105 |
| A0A1D1ZBB9 Adenosine kinase (Fragment) | 4.68 × 104 | 3.92 × 104 |
A0A1D1YQU6 Adenosylhomocysteinase (Fragment) A0A1D1YZX9 2-Cys peroxiredoxin BAS1-like, chloroplastic (Fragment) A0A0K9PQ40 Ferredoxin-NADP reductase, chloroplastic A0A1D1ZGY1 Glutathione reductase A0A0K9P699 Glutathione transferase A0A0K9P881 Leucyl aminopeptidase A0A843XPL2 Mechanosensitive ion channel protein A0A0K9Q3S1 Nucleoside-diphosphate kinase A0A0K9P7Q7 Peptidyl-prolyl cis-trans isomerase F8U875 Phosphoglucomutase (alpha-D-glucose- 1,6-bisphosphate-dependent) A0A0K9PHA0 Protein CutA 1, chloroplastic A0A1D1XUN0 Superoxide dismutase | 1.06 × 105 5850 1.37 × 105 0 0 9.56 × 104 7.32 × 106 3.87 × 104 0 0 0 3.92 × 105 | 6.39 × 105 6.58 × 104 0 1.31 × 105 2.01 × 105 4.23 × 104 5.62 × 106 2.31 × 105 1.96 × 105 1.22× 105 4.93 × 105 1.95× 106 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abruscato, G.; Chiarelli, R.; Lazzara, V.; Punginelli, D.; Sugár, S.; Mauro, M.; Librizzi, M.; Di Stefano, V.; Arizza, V.; Vizzini, A.; et al. In Vitro Cytotoxic Effect of Aqueous Extracts from Leaves and Rhizomes of the Seagrass Posidonia oceanica (L.) Delile on HepG2 Liver Cancer Cells: Focus on Autophagy and Apoptosis. Biology 2023, 12, 616. https://doi.org/10.3390/biology12040616
Abruscato G, Chiarelli R, Lazzara V, Punginelli D, Sugár S, Mauro M, Librizzi M, Di Stefano V, Arizza V, Vizzini A, et al. In Vitro Cytotoxic Effect of Aqueous Extracts from Leaves and Rhizomes of the Seagrass Posidonia oceanica (L.) Delile on HepG2 Liver Cancer Cells: Focus on Autophagy and Apoptosis. Biology. 2023; 12(4):616. https://doi.org/10.3390/biology12040616
Chicago/Turabian StyleAbruscato, Giulia, Roberto Chiarelli, Valentina Lazzara, Diletta Punginelli, Simon Sugár, Manuela Mauro, Mariangela Librizzi, Vita Di Stefano, Vincenzo Arizza, Aiti Vizzini, and et al. 2023. "In Vitro Cytotoxic Effect of Aqueous Extracts from Leaves and Rhizomes of the Seagrass Posidonia oceanica (L.) Delile on HepG2 Liver Cancer Cells: Focus on Autophagy and Apoptosis" Biology 12, no. 4: 616. https://doi.org/10.3390/biology12040616
APA StyleAbruscato, G., Chiarelli, R., Lazzara, V., Punginelli, D., Sugár, S., Mauro, M., Librizzi, M., Di Stefano, V., Arizza, V., Vizzini, A., Vazzana, M., & Luparello, C. (2023). In Vitro Cytotoxic Effect of Aqueous Extracts from Leaves and Rhizomes of the Seagrass Posidonia oceanica (L.) Delile on HepG2 Liver Cancer Cells: Focus on Autophagy and Apoptosis. Biology, 12(4), 616. https://doi.org/10.3390/biology12040616


_Kwok.png)







