Kanamycin and Ofloxacin Activate the Intrinsic Resistance to Multiple Antibiotics in Mycobacterium smegmatis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media
2.2. MIC Determination
2.3. Search for Maximum Non-Inhibiting Antibiotics Concentrations
2.4. Drug Susceptibility Testing
2.5. Mycobacterial RNA Isolation and Real-Time qPCR
3. Results
3.1. Minimal Inhibiting Antibiotic Concentrations Determination for M. smegmatis
3.2. The Determination of Maximum Non-Inhibiting Concentrations
3.3. Evaluation of the Intrinsic Drug Cross-Resistance Induction
3.4. Selection of Candidate Genes Potentially Involved in Cross-Resistance
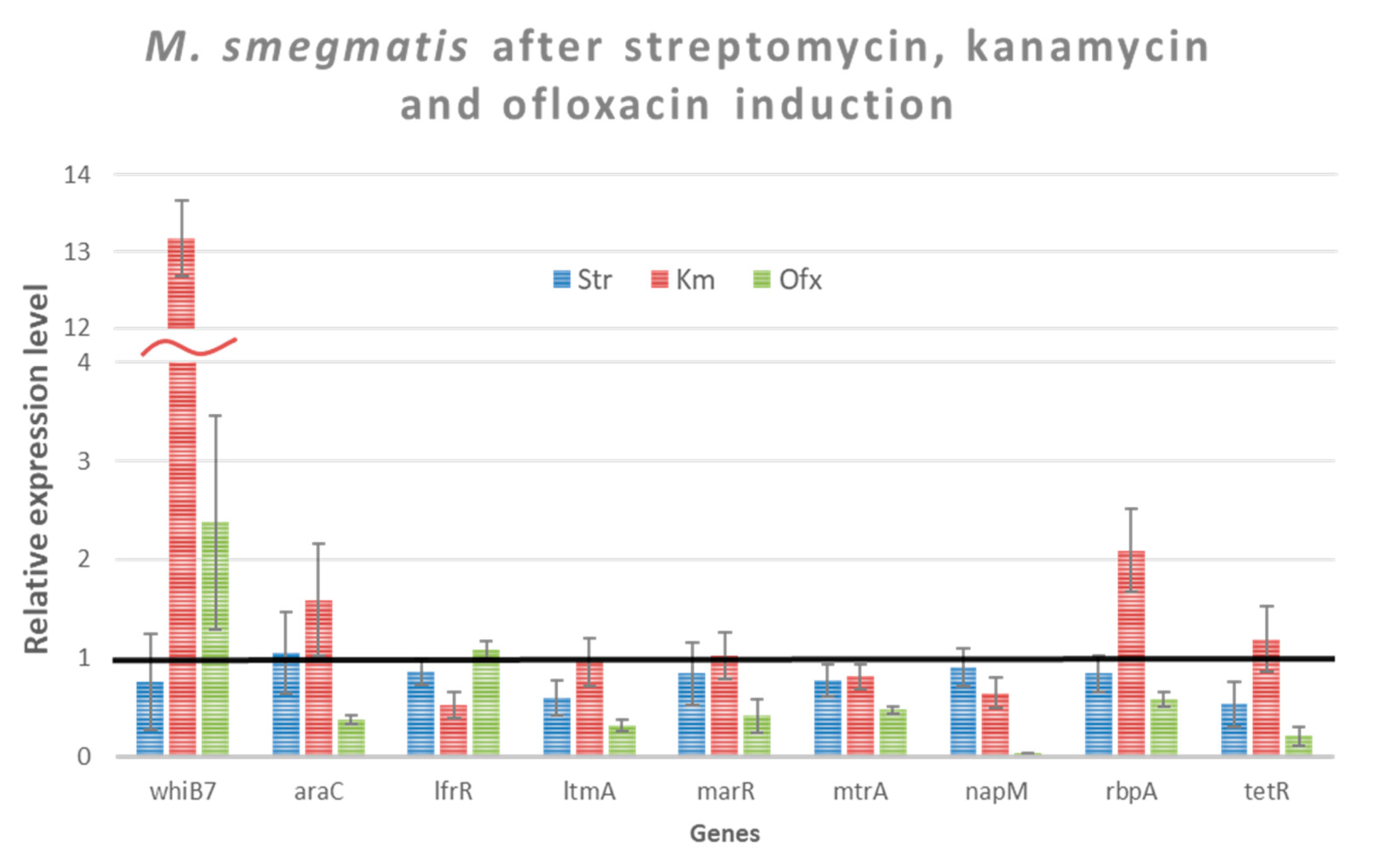
3.5. Study of Gene Expression of Intrinsic Cross-Resistance of M. smegmatis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Primer Name | 5′-3′ Sequence | Target |
|---|---|---|
| WhiB7 | CATGTCGGTGACCCGGATCTGT | MSMEG_1953 |
| WhiB7-R | ACAATGCTTCCGCGGTCGAG | |
| NapM | GCGTTCTCCTACGGTTCGCTCTATC | MSMEG_6903 |
| NapM-R | GAAGCCGTCGTCGGAGTAGTTCTG | |
| LtmA | ACGAGAAATAGGTGTAGGCGGTTGC | MSMEG_6479 |
| LtmA-R | TGTCCGATCCGGCCATGGAGT | |
| TetR | GCCGATACCGCCGATCTGTTCAT | MSMEG_4022 |
| TetR-R | CTTCGCCAGTTCTGCGTTCGAAAT | |
| RbpA | CGAGGAGTTCGACGTACCTTTCGC | MSMEG_3858 |
| RbpA-R | CTTGATCAGGTCGAGACGCTCCTTG | |
| MtrA | GCAAACCACGGCAGGTGTTTACTC | MSMEG_1874 |
| MtrA-R | TTGTATCCCACTCCTCGAACGGTCA | |
| LfrR | CCGATCGTGCTGTTCGTCTACTACG | MSMEG_6223 |
| LfrR-R | CCTCATAACCGGCCTGCATCAGT | |
| MarR | CGGCGACCTGGCAAGTGTCAT | MSMEG_6508 |
| MarR-R | AGTGTCGACGCGGTGTTGGG | |
| AraC | CTCCCAACGGTGTGCACTTCCA | MSMEG_0307 |
| AraC-R | GGTGAAACCTCTTGCCGCCACT | |
| qsigAs-sm-f | CGAGCTTGTTGATCACCTCGACCAT | sigA |
| qsigAs-sm-r | CTCGACCTCATCCAGGAAGGCAAC | |
| qPolAs-sm-f | GGTCTGGTTGAACGTCGTGTGGATG | polA |
| qPolAs-sm-r | GCTGGAGATGCCGAAGACCAAGAAG |
| Antibiotics (µg/disc) | M. smegmatis mc2 155−2 without Induction | SD | M. smegmatis mc2 155−2 with Induction | SD |
|---|---|---|---|---|
| Streptomycin induction | ||||
| Rifampicin 100 | 10.3 | 1.1 | 7.8 | 0.3 |
| Ofloxacine induction | ||||
| Netilmicin 10 | 39.3 | 1.3 | 32.7 | 1.5 |
| Meropenem 10 | 28.3 | 1.5 | 22.7 | 1.2 |
| Kanamycin induction | ||||
| Imipenem 10 | 32.0 | 1.0 | 27.7 | 0.6 |
| Spiramycin 100 | 33.3 | 1.2 | 28.0 | 1.6 |
| Norfloxacin 10 | 26.5 | 0.6 | 21.5 | 1.7 |
| Tetracycline induction | ||||
| Ofloxacin 5 | 28.3 | 0.6 | 19.7 | 1.2 |
| Linezolid 10 | 33.7 | 0.6 | 28.3 | 1.5 |
| Kanamycin 30 | 31.7 | 0.6 | 28.0 | 0.1 |
| Oxytetracycline 30 | 39.7 | 1.5 | 34.7 | 1.5 |
| Azithromycin 15 | 26.3 | 1.2 | 19.3 | 1.2 |
| Levofloxacin 5 | 35.3 | 2.1 | 26.7 | 1.2 |
| Ciprofloxacin 1 | 22.3 | 1.2 | 16.3 | 1.5 |
| Tetracycline 30 | 38.3 | 1.2 | 30.3 | 1.5 |
| Lomefloxacin 10 | 25.3 | 0.6 | 21.7 | 0.6 |
References
- Cantón, R.; Morosini, M.-I. Emergence and Spread of Antibiotic Resistance Following Exposure to Antibiotics. FEMS Microbiol. Rev. 2011, 35, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.A.; Westman, E.L.; Wright, G.D. The Antibiotic Resistome: What ‘s New? Curr. Opin. Microbiol. 2014, 21, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, Z.; Suo, Y.; Cui, Y.; Zhang, F.; Shi, C.; Shi, X. Effect of Sublethal Concentrations of Ceftriaxone on Antibiotic Susceptibility of Multiple Antibiotic-Resistant Salmonella Strains. FEMS Microbiol. Lett. 2019, 366, fny283. [Google Scholar] [CrossRef] [PubMed]
- Wistrand-Yuen, E.; Knopp, M.; Hjort, K.; Koskiniemi, S.; Berg, O.G.; Andersson, D.I. Evolution of High-Level Resistance during Low-Level Antibiotic Exposure. Nat. Commun. 2018, 9, 1599. [Google Scholar] [CrossRef]
- Uddin, M.J.; Ahn, J. Characterization of β-Lactamase- and Efflux Pump-Mediated Multiple Antibiotic Resistance in Salmonella Typhimurium. Food Sci. Biotechnol. 2018, 27, 921–928. [Google Scholar] [CrossRef]
- Verbrugghe, E.; Van Parys, A.; Haesendonck, R.; Leyman, B.; Boyen, F.; Haesebrouck, F.; Pasmans, F. Subtherapeutic Tetracycline Concentrations Aggravate Salmonella Typhimurium Infection by Increasing Bacterial Virulence. J. Antimicrob. Chemother. 2016, 71, 2158–2166. [Google Scholar] [CrossRef]
- Waack, U.; Nicholson, T.L. Subinhibitory Concentrations of Amoxicillin, Lincomycin, and Oxytetracycline Commonly Used to Treat Swine Increase Streptococcus Suis Biofilm Formation. Front. Microbiol. 2018, 9, 2707. [Google Scholar] [CrossRef]
- Prieto Martin Gil, S.; Tajuelo, A.; López-Siles, M.; McConnell, M.J. Subinhibitory Concentrations of Clinically-Relevant Antimicrobials Affect Resistance-Nodulation-Division Family Promoter Activity in Acinetobacter Baumannii. Front. Microbiol. 2021, 12, 780201. [Google Scholar] [CrossRef]
- Gullberg, E.; Cao, S.; Berg, O.G.; Ilbäck, C.; Sandegren, L.; Hughes, D.; Andersson, D.I. Selection of Resistant Bacteria at Very Low Antibiotic Concentrations. PLoS Pathog. 2011, 7, e1002158. [Google Scholar] [CrossRef]
- Stanton, I.C.; Murray, A.K.; Zhang, L.; Snape, J.; Gaze, W.H. Evolution of Antibiotic Resistance at Low Antibiotic Concentrations Including Selection below the Minimal Selective Concentration. Commun. Biol. 2020, 3, 467. [Google Scholar] [CrossRef]
- Sandegren, L. Selection of Antibiotic Resistance at Very Low Antibiotic Concentrations. Ups. J. Med. Sci. 2014, 119, 103–107. [Google Scholar] [CrossRef] [PubMed]
- D‘Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the Antibiotic Resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.; Ghosh, P. A Complex Regulatory Network Controlling Intrinsic Multidrug Resistance in Mycobacterium Smegmatis. Mol. Microbiol. 2014, 91, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Thompson, C.J. Foundations of Antibiotic Resistance in Bacterial Physiology: The Mycobacterial Paradigm. Trends Microbiol. 2006, 14, 304–312. [Google Scholar] [CrossRef]
- McArthur, A.G.; Wright, G.D. Bioinformatics of Antimicrobial Resistance in the Age of Molecular Epidemiology. Curr. Opin. Microbiol. 2015, 27, 45–50. [Google Scholar] [CrossRef]
- Reeves, A.Z.; Campbell, P.J.; Sultana, R.; Malik, S.; Murray, M.; Plikaytis, B.B.; Shinnick, T.M.; Posey, J.E. Aminoglycoside Cross-Resistance in Mycobacterium Tuberculosis Due to Mutations in the 5 ‘ Untranslated Region of WhiB7. Antimicrob. Agents Chemother. 2013, 57, 1857–1865. [Google Scholar] [CrossRef]
- Gillespie, S.H.; Basu, S.; Dickens, A.L.; O ‘Sullivan, D.M.; McHugh, T.D. Effect of Subinhibitory Concentrations of Ciprofloxacin on Mycobacterium Fortuitum Mutation Rates. J. Antimicrob. Chemother. 2005, 56, 344–348. [Google Scholar] [CrossRef]
- CFR. Code of Federal Regulations Title 21. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?CFRPart=556&showFR=1&subpartNode=21:6.0.1.1.18.2 (accessed on 6 March 2023).
- Gilbert, N. World‘s Rivers “Awash with Dangerous Levels of Antibiotics”. The Guardian 2019.World ‘s Rivers ‘Awash with Dangerous Levels of Antibiotics. Largest Global Study Finds the Drugs in Two-Thirds of Test Sites in 72 Countries. Guardian, 27 May 2019. Available online: https://www.theguardian.com/society/2019/may/27/worlds-rivers-awash-with-dangerous-levels-of-antibiotics (accessed on 6 March 2023).
- Altaf, M.; Miller, C.H.; Bellows, D.S.; O ‘Toole, R. Evaluation of the Mycobacterium Smegmatis and BCG Models for the Discovery of Mycobacterium Tuberculosis Inhibitors. Tuberc. Edinb. Scotl. 2010, 90, 333–337. [Google Scholar] [CrossRef]
- Dandapani, S.; Rosse, G.; Southall, N.; Salvino, J.M.; Thomas, C.J. Selecting, Acquiring, and Using Small Molecule Libraries for High-Throughput Screening. Curr. Protoc. Chem. Biol. 2012, 4, 177–191. [Google Scholar] [CrossRef]
- Etienne, G.; Laval, F.; Villeneuve, C.; Dinadayala, P.; Abouwarda, A.; Zerbib, D.; Galamba, A.; Daffé, M. The Cell Envelope Structure and Properties of Mycobacterium Smegmatis Mc(2)155: Is There a Clue for the Unique Transformability of the Strain? Microbiol. Read. Engl. 2005, 151, 2075–2086. [Google Scholar] [CrossRef]
- Bekker, O.B.; Sokolov, D.N.; Luzina, O.A.; Komarova, N.I.; Gatilov, Y.V.; Andreevskaya, S.N.; Smirnova, T.G.; Maslov, D.A.; Chernousova, L.N.; Salakhutdinov, N.F.; et al. Synthesis and Activity of (+)-Usnic Acid and (−)-Usnic Acid Derivatives Containing 1,3-Thiazole Cycle against Mycobacterium Tuberculosis. Med. Chem. Res. 2015, 24, 2926–2938. [Google Scholar] [CrossRef]
- Erkmen, O. Practice 18—Antibiotic Sensitivity Test Technique. In Laboratory Practices in Microbiology; Erkmen, O., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 181–186. ISBN 978-0-323-91017-0. [Google Scholar]
- Haimes, J.; Kelley, M. Demonstration of a ΔΔCq Calculation Method to Compute Relative Gene Expression from QPCR Data. Semantic Scholar. 2015. Available online: https://api.semanticscholar.org/CorpusID:28223471 (accessed on 1 January 2023).
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.A.; van der Laan, T.; Akkerman, O.W.; Bolhuis, M.S.; de Lange, W.C.M.; Kosterink, J.G.W.; van der Werf, T.S.; Alffenaar, J.W.C.; van Soolingen, D. In Vitro Susceptibility of Mycobacterium Tuberculosis to Amikacin, Kanamycin, and Capreomycin. Antimicrob. Agents Chemother. 2018, 62, e01724-17. [Google Scholar] [CrossRef] [PubMed]
- Cohen, K.A.; Stott, K.E.; Munsamy, V.; Manson, A.L.; Earl, A.M.; Pym, A.S. Evidence for Expanding the Role of Streptomycin in the Management of Drug-Resistant Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2020, 64, e00860-20. [Google Scholar] [CrossRef] [PubMed]
- Crowle, A.J.; Elkins, N.; May, M.H. Effectiveness of Ofloxacin against Mycobacterium Tuberculosis and Mycobacterium Avium, and Rifampin against M. Tuberculosis in Cultured Human Macrophages. Am. Rev. Respir. Dis. 1988, 137, 1141–1146. [Google Scholar] [CrossRef]
- Hobby, G.L.; Lenert, T.F. Antituberculous Activity of Tetracycline and Related Compounds. Am. Rev. Tuberc. 1955, 72, 367–372. [Google Scholar]
- Blokpoel, M.C.J.; Murphy, H.N.; O ‘Toole, R.; Wiles, S.; Runn, E.S.C.; Stewart, G.R.; Young, D.B.; Robertson, B.D. Tetracycline-Inducible Gene Regulation in Mycobacteria. Nucleic Acids Res. 2005, 33, e22. [Google Scholar] [CrossRef]
- Miotto, P.; Sorrentino, R.; De Giorgi, S.; Provvedi, R.; Cirillo, D.M.; Manganelli, R. Transcriptional Regulation and Drug Resistance in Mycobacterium Tuberculosis. Front. Cell. Infect. Microbiol. 2022, 12, 990312. [Google Scholar] [CrossRef]
- Evangelopoulos, D.; Gupta, A.; Lack, N.A.; Maitra, A.; ten Bokum, A.M.C.; Kendall, S.; Sim, E.; Bhakta, S. Characterisation of a Putative AraC Transcriptional Regulator from Mycobacterium Smegmatis. Tuberc. Edinb. Scotl. 2014, 94, 664–671. [Google Scholar] [CrossRef]
- Burian, J.; Thompson, C.J. Regulatory Genes Coordinating Antibiotic-Induced Changes in Promoter Activity and Early Transcriptional Termination of the Mycobacterial Intrinsic Resistance Gene WhiB7. Mol. Microbiol. 2018, 107, 402–415. [Google Scholar] [CrossRef]
- Morris, R.P.; Nguyen, L.; Gatfield, J.; Visconti, K.; Nguyen, K.; Schnappinger, D.; Ehrt, S.; Liu, Y.; Heifets, L.; Pieters, J.; et al. Ancestral Antibiotic Resistance in Mycobacterium Tuberculosis. Proc. Natl. Acad. Sci. USA 2005, 102, 12200–12205. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, M.; He, Z.-G. Novel TetR Family Transcriptional Factor Regulates Expression of Multiple Transport-Related Genes and Affects Rifampicin Resistance in Mycobacterium Smegmatis. Sci. Rep. 2016, 6, 27489. [Google Scholar] [CrossRef] [PubMed]
- Sander, P.; De Rossi, E.; Böddinghaus, B.; Cantoni, R.; Branzoni, M.; Böttger, E.C.; Takiff, H.; Rodriquez, R.; Lopez, G.; Riccardi, G. Contribution of the Multidrug Efflux Pump LfrA to Innate Mycobacterial Drug Resistance. FEMS Microbiol. Lett. 2000, 193, 19–23. [Google Scholar] [CrossRef]
- Li, W.; He, Z.-G. LtmA, a Novel Cyclic Di-GMP-Responsive Activator, Broadly Regulates the Expression of Lipid Transport and Metabolism Genes in Mycobacterium Smegmatis. Nucleic Acids Res. 2012, 40, 11292–11307. [Google Scholar] [CrossRef]
- Gong, Z.; Li, H.; Cai, Y.; Stojkoska, A.; Xie, J. Biology of MarR Family Transcription Factors and Implications for Targets of Antibiotics against Tuberculosis. J. Cell. Physiol. 2019, 234, 19237–19248. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zeng, J.; Zhang, H.; He, Z.-G. The Characterization of Conserved Binding Motifs and Potential Target Genes for M. Tuberculosis MtrAB Reveals a Link between the Two-Component System and the Drug Resistance of M. Smegmatis. BMC Microbiol. 2010, 10, 242. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, H.; Cui, T.; Zhou, X.; Jia, Y.; Zhang, H.; He, Z.-G. NapM, a New Nucleoid-Associated Protein, Broadly Regulates Gene Expression and Affects Mycobacterial Resistance to Anti-Tuberculosis Drugs. Mol. Microbiol. 2016, 101, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Morichaud, Z.; Chen, S.; Leonetti, J.-P.; Brodolin, K. Mycobacterium Tuberculosis RbpA Protein Is a New Type of Transcriptional Activator That Stabilizes the σ A-Containing RNA Polymerase Holoenzyme. Nucleic Acids Res. 2012, 40, 6547–6557. [Google Scholar] [CrossRef]
- Deng, W.; Li, C.; Xie, J. The Underling Mechanism of Bacterial TetR/AcrR Family Transcriptional Repressors. Cell. Signal. 2013, 25, 1608–1613. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Microbiological Effects of Sublethal Levels of Antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Ter Kuile, B.H.; Kraupner, N.; Brul, S. The Risk of Low Concentrations of Antibiotics in Agriculture for Resistance in Human Health Care. FEMS Microbiol. Lett. 2016, 363, fnw210. [Google Scholar] [CrossRef] [PubMed]
- Gudda, F.O.; Waigi, M.G.; Odinga, E.S.; Yang, B.; Carter, L.; Gao, Y. Antibiotic-Contaminated Wastewater Irrigated Vegetables Pose Resistance Selection Risks to the Gut Microbiome. Environ. Pollut. 2020, 264, 114752. [Google Scholar] [CrossRef] [PubMed]
- Lorian, V.; De Freitas, C.C. Minimal Antibiotic Concentrations of Aminoglycosides and Beta-Lactam Antibiotics for Some Gram-Negative Bacilli and Gram-Positive Cocci. J. Infect. Dis. 1979, 139, 599–603. [Google Scholar] [CrossRef]
- Tornqvist, I.O.; Holm, S.E.; Cars, O. Pharmacodynamic Effects of Subinhibitory Antibiotic Concentrations. Scand. J. Infect. Dis. Suppl. 1990, 74, 94–101. [Google Scholar]
- Sharma, A.K.; Chatterjee, A.; Gupta, S.; Banerjee, R.; Mandal, S.; Mukhopadhyay, J.; Basu, J.; Kundu, M. MtrA, an Essential Response Regulator of the MtrAB Two-Component System, Regulates the Transcription of Resuscitation-Promoting Factor B of Mycobacterium Tuberculosis. Microbiol. Read. Engl. 2015, 161, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.B.; Arnipalli, S.R.; Ziouzenkova, O. Antibiotics in Food Chain: The Consequences for Antibiotic Resistance. Antibiotics 2020, 9, 688. [Google Scholar] [CrossRef] [PubMed]
- Hashempour-Baltork, F.; Hosseini, H.; Shojaee-Aliabadi, S.; Torbati, M.; Alizadeh, A.M.; Alizadeh, M. Drug Resistance and the Prevention Strategies in Food Borne Bacteria: An Update Review. Adv. Pharm. Bull. 2019, 9, 335–347. [Google Scholar] [CrossRef]
- Vidovic, N.; Vidovic, S. Antimicrobial Resistance and Food Animals: Influence of Livestock Environment on the Emergence and Dissemination of Antimicrobial Resistance. Antibiotics 2020, 9, 52. [Google Scholar] [CrossRef]
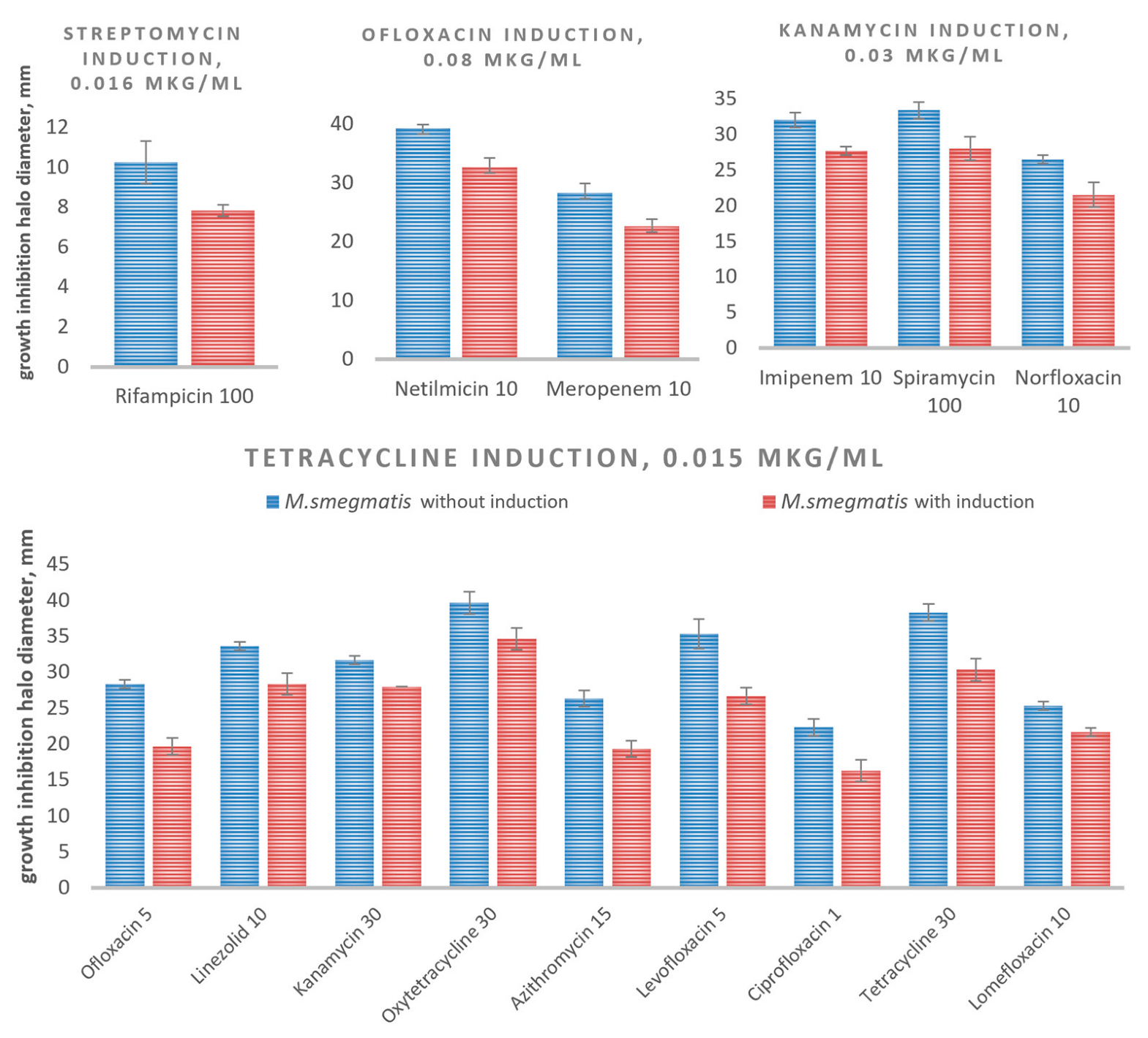
| Antibiotics | MIC, μg/mL | Ref. | |
|---|---|---|---|
| M. smegmatis mc2 155−2 | M. tuberculosis | ||
| Kanamycin | 3.2 ± 0.2 | 4 | [27] |
| Streptomycin | 0.8 ± 0.1 | 1 | [28] |
| Ofloxacin | 0.32 ± 0.03 | 1.25 | [29] |
| Tetracycline | 0.06 ± 0.01 | 0.55 | [30] |
| Chemical Class | Antibiotic | Proportion of MIC, Concentration, µg/mL | OD600 after 24 h of Incubation |
|---|---|---|---|
| Aminoglycosides | Kanamycin | Control Sample * | ~0.2 |
| MIC—3.2 | ~0.012 | ||
| ½ MIC—1.6 | ~0.06 | ||
| 1/60 MIC—0.05 | ~0.17 | ||
| 1/120 MIC—0.03 | ~0.21 | ||
| Streptomycin | Control Sample * | ~0.21 | |
| MIC—0.8 | ~0.01 | ||
| ½ MIC—0.4 | ~0.023 | ||
| 1/25 MIC—0.2 | ~0.07 | ||
| 1/50 MIC—0.016 | ~0.2 | ||
| Fluoroquinolones | Ofloxacin | Control Sample * | ~0.25 |
| MIC—0.32 | ~0.02 | ||
| 1/4 MIC—0.08 | ~0.25 | ||
| Tetracyclines | Tetracycline | Control Sample * | ~0.22 |
| MIC—0.06 | ~0.02 | ||
| ½ MIC—0.03 | ~0.12 | ||
| 1/4 MIC—0.015 | ~0.23 |
| Gene Name (Locus Tag) | Predicted Function | Drug Resistance Phenotype | Ref. |
|---|---|---|---|
| whiB7 (MSMEG_1953) | Transcriptional factor | Aminoglycosides, macrolides, tetracyclines, fluoroquinolones, phenicols, β-lactams | [34,35] |
| tetR (MSMEG_4022) | Transcriptional factor | Rifampin | [32,36] |
| araC (MSMEG_0307) | Transcriptional factor | Rifampin, kanamycin, chloramphenicol | [33] |
| lfrR (MSMEG_6223) | Transcriptional factor | Fluoroquinolones | [37] |
| ltmA (MSMEG_6479) | c-di-GMP-depended Transcriptional factor | Rifampin, isoniazid | [38] |
| marR (MSMEG_6508) | Transcriptional factor | Isoniazid, Rifampin, ethambutol, kanamycin | [39] |
| mtrA (MSMEG_1874) | Transcriptional factor | Isoniazid, streptomycin, Rifampin | [40] |
| napM (MSMEG_6903) | Transcriptional factor | Rifampin, ethambutol | [41] |
| rbpA (MSMEG_3858) | Transcriptional factor | Rifampin | [42] |
| Antibiotic | Transcription Level Increase (Fold) | Transcription Level Decrease (Fold) |
|---|---|---|
| Kanamycin | whiB7 (13.2) | |
| rbpA (2.08) | ||
| Ofloxacin | whiB7 (2.4) | napM (27.42) |
| tetR (4.88) | ||
| araC (2.67) | ||
| ltmA (3.17) | ||
| marR (2.39) | ||
| mtrA (2.12) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vatlin, A.A.; Bekker, O.B.; Shur, K.V.; Ilyasov, R.A.; Shatrov, P.A.; Maslov, D.A.; Danilenko, V.N. Kanamycin and Ofloxacin Activate the Intrinsic Resistance to Multiple Antibiotics in Mycobacterium smegmatis. Biology 2023, 12, 506. https://doi.org/10.3390/biology12040506
Vatlin AA, Bekker OB, Shur KV, Ilyasov RA, Shatrov PA, Maslov DA, Danilenko VN. Kanamycin and Ofloxacin Activate the Intrinsic Resistance to Multiple Antibiotics in Mycobacterium smegmatis. Biology. 2023; 12(4):506. https://doi.org/10.3390/biology12040506
Chicago/Turabian StyleVatlin, Aleksey A., Olga B. Bekker, Kirill V. Shur, Rustem A. Ilyasov, Petr A. Shatrov, Dmitry A. Maslov, and Valery N. Danilenko. 2023. "Kanamycin and Ofloxacin Activate the Intrinsic Resistance to Multiple Antibiotics in Mycobacterium smegmatis" Biology 12, no. 4: 506. https://doi.org/10.3390/biology12040506
APA StyleVatlin, A. A., Bekker, O. B., Shur, K. V., Ilyasov, R. A., Shatrov, P. A., Maslov, D. A., & Danilenko, V. N. (2023). Kanamycin and Ofloxacin Activate the Intrinsic Resistance to Multiple Antibiotics in Mycobacterium smegmatis. Biology, 12(4), 506. https://doi.org/10.3390/biology12040506







