Animal Disease Models and Patient-iPS-Cell-Derived In Vitro Disease Models for Cardiovascular Biology—How Close to Disease?
Abstract
Simple Summary
Abstract
1. Introduction
2. Animal Disease Models
2.1. Zebrafish
2.2. Rodents
2.3. Pigs
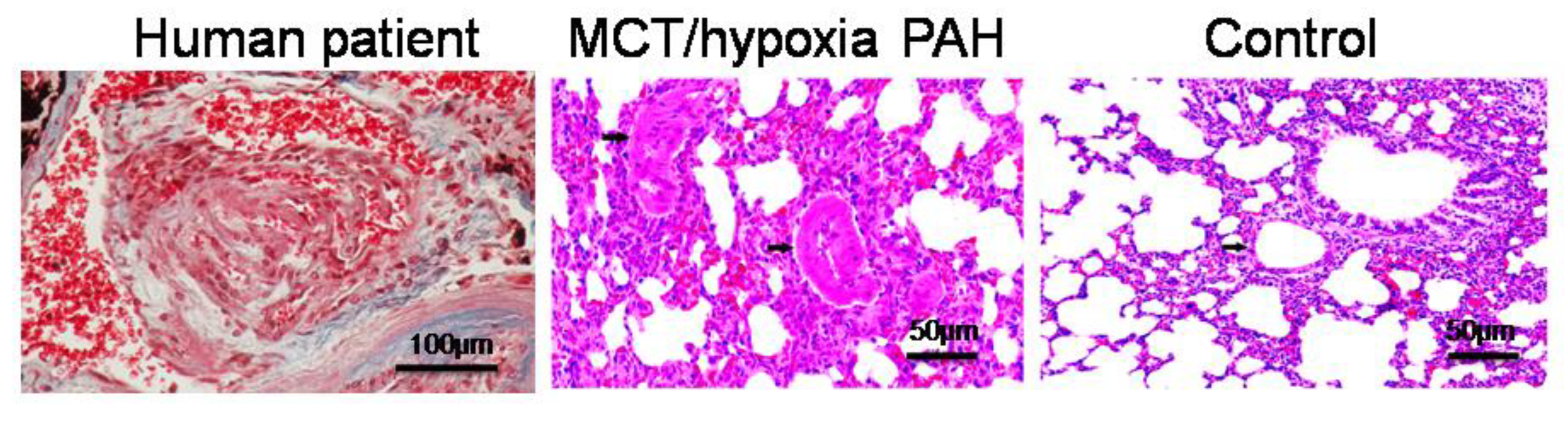
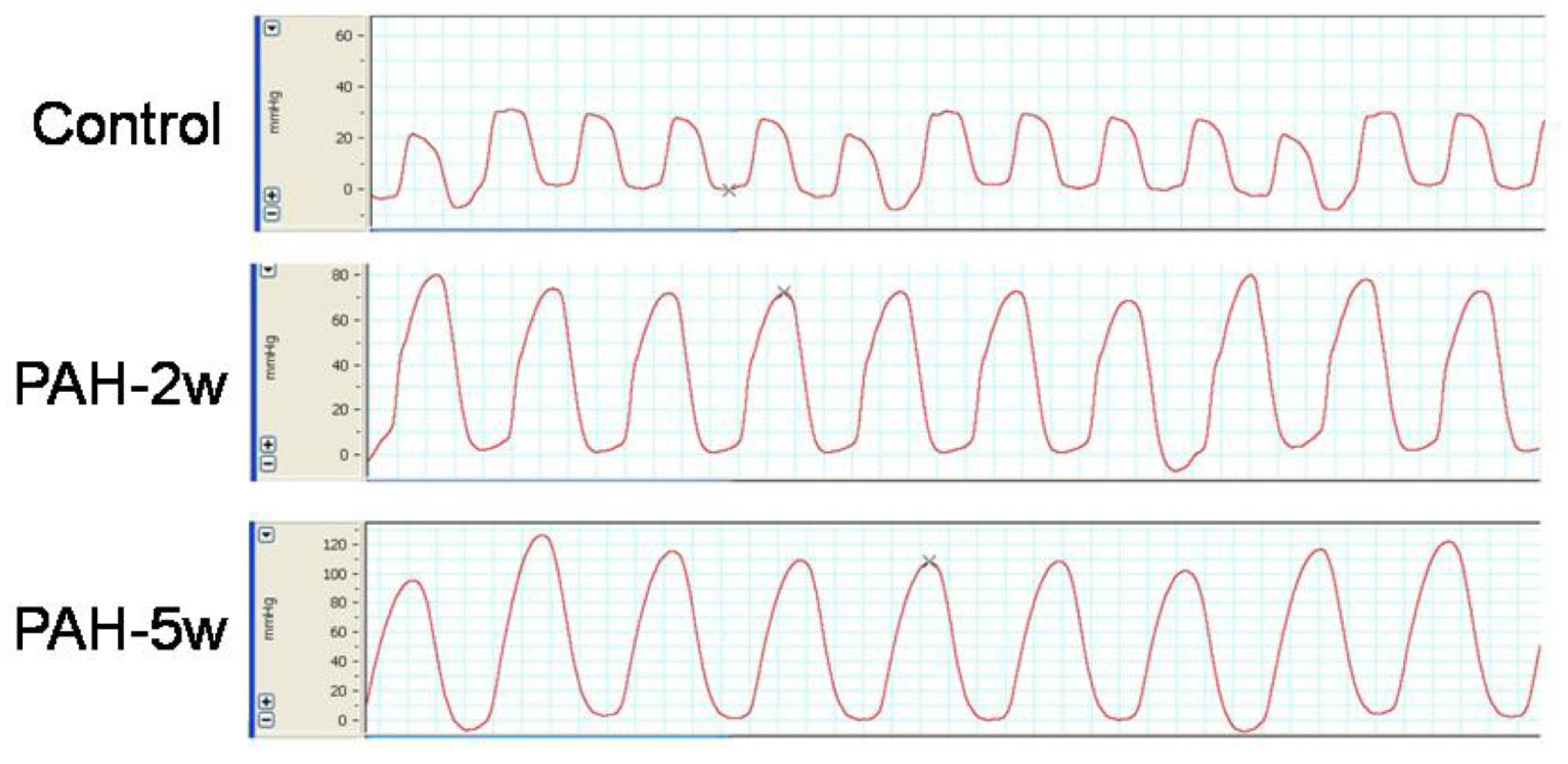
2.4. PAH Model
2.5. Making Animal Disease Models More Similar to Human Diseases
3. In Vitro Disease Models
3.1. Engineered Heart Tissues and Organoids
3.2. Multi-Lineage Differentiation of iPSCs
4. Humanized Animals
Humanized Mice
5. Computer Models
5.1. Machine Learning (ML)
5.2. Deep Learning
5.3. Experimental Data Validation for ML
6. Summary
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data availability Statement
Conflicts of Interest
References
- Petetta, F.; Ciccocioppo, R. Public perception of laboratory animal testing: Historical, philosophical, and ethical view. Addict. Biol. 2020, 26, e12991. [Google Scholar] [CrossRef]
- Darwin, C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life; John Murray: London, UK, 1859. [Google Scholar]
- Hoareau, M.; El Kholti, N.; Debret, R.; Lambert, E. Zebrafish as a Model to Study Vascular Elastic Fibers and Associated Pathologies. Int. J. Mol. Sci. 2022, 23, 2102. [Google Scholar] [CrossRef] [PubMed]
- Poon, K.L.; Brand, T. The zebrafish model system in cardiovascular research: A tiny fish with mighty prospects. Glob. Cardiol. Sci. Pract. 2013, 2013, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Gauvrit, S.; Bossaer, J.; Lee, J.; Collins, M.M. Modeling Human Cardiac Arrhythmias: Insights from Zebrafish. J. Cardiovasc. Dev. Dis. 2022, 9, 13. [Google Scholar] [CrossRef]
- Johny, E.; Dutta, P. Left Coronary Artery Ligation: A Surgical Murine Model of Myocardial Infarction. J. Vis. Exp. 2022, 186, e64387. [Google Scholar]
- Tuder, R.M.; Archer, S.L.; Dorfmüller, P.; Erzurum, S.C.; Guignabert, C.; Michelakis, E.; Rabinovitch, M.; Schermuly, R.; Stenmark, K.R.; Morrell, N.W. Relevant Issues in the Pathology and Pathobiology of Pulmonary Hypertension. J. Am. Coll. Cardiol. 2013, 62, D4–D12. [Google Scholar] [CrossRef] [PubMed]
- Ranchoux, B.; Antigny, F.; Rucker-Martin, C.; Hautefort, A.; Péchoux, C.; Bogaard, H.J.; Dorfmüller, P.; Remy, S.; Lecerf, F.; Planté, S.; et al. Endothelial-to-Mesenchymal Transition in Pulmonary Hypertension. Circulation 2015, 131, 1006–1018. [Google Scholar] [CrossRef]
- Galie, N.; Humbert, M.; Vachiery, J.L.; Gibbs, S.; Lang, I.; Torbicki, A.; Simonneau, G.; Peacock, A.; VonkNoordegraaf, A.; Beghetti, M.; et al. 2015 ESC/ERS guidelinesfor the diagnosis and treatment of pulmonary hypertension: The joint taskforce for the diagnosis and treatment of pulmonary hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society(ERS): Endorsed by: Association for European Pediatric and Congenital Cardiology (AEPC), International Society for Heart and Lung Transplantation(ISHLT). Eur. Heart J. 2016, 37, 67–119. [Google Scholar]
- Bueno-Beti, C.; Hadri, L.; Hajjar, R.J.; Sassi, Y. The Sugen 5416/Hypoxia Mouse Model of Pulmonary Arterial Hypertension. Methods Mol. Biol. 2018, 1816, 243–252. [Google Scholar]
- Yuan, K.; Shamskhou, E.A.; Orcholski, M.E.; Nathan, A.; Reddy, S.; Honda, H.; Mani, V.; Zeng, Y.; Ozen, M.O.; Wang, L.; et al. Loss of Endothelium-Derived Wnt5a Is Associated With Reduced Pericyte Recruitment and Small Vessel Loss in Pulmonary Arterial Hypertension. Circulation 2019, 139, 1710–1724. [Google Scholar] [CrossRef]
- Zhang, J.; He, Y.; Yan, X.; Chen, S.; He, M.; Lei, Y.; Zhang, J.; Gongol, B.; Gu, M.; Miao, Y.; et al. MicroRNA-483 amelioration of experimental pulmonary hypertension. EMBO Mol. Med. 2020, 12, e11303. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, N.; Satoh, K.; Kurosawa, R.; Yaoita, N.; Elias-Al-Mamun, M.; Siddique, M.A.H.; Omura, J.; Satoh, T.; Nogi, M.; Sunamura, S.; et al. Selenoprotein P Promotes the Development of Pulmonary Arterial Hypertension: Possible Novel Therapeutic Target. Circulation 2018, 138, 600–623. [Google Scholar] [CrossRef] [PubMed]
- Westöö, C.; Norvik, C.; Peruzzi, N.; van der Have, O.; Lovric, G.; Jeremiasen, I.; Tran, P.-K.; Mokso, R.; de Jesus Perez, V.; Brunnström, H.; et al. Distinct types of plexiform lesions identified by synchrotron-based phase-contrast micro-CT. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2021, 321, L17–L28. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, S.; Yoshida, Y. Recent progress of iPSC technology in cardiac diseases. Arch. Toxicol. 2021, 95, 3633–3650. [Google Scholar] [CrossRef]
- Tanaka, A.; Yuasa, S.; Node, K.; Fukuda, K. Cardiovascular Disease Modeling Using Patient-Specific Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2015, 16, 18894–18922. [Google Scholar] [CrossRef]
- Kawaguchi, N.; Hayama, E.; Furutani, Y.; Nakanishi, T. ProspectiveIn VitroModels of Channelopathies and Cardiomyopathies. Stem Cells Int. 2012, 2012, 439219. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Tani, H.; Tohyama, S. Human Engineered Heart Tissue Models for Disease Modeling and Drug Discovery. Front. Cell Dev. Biol. 2022, 10, 855763. [Google Scholar] [CrossRef]
- Kawaguchi, N.; Nakanishi, T. Stem Cell Studies in Cardiovascular Biology and Medicine: A Possible Key Role of Macrophages. Biology 2022, 11, 122. [Google Scholar] [CrossRef]
- Ivashchenko, C.Y.; Pipes, G.C.; Lozinskaya, I.M.; Lin, Z.; Xiaoping, X.; Needle, S.; Grygielko, E.T.; Hu, E.; Toomey, J.R.; Lepore, J.J.; et al. Human-induced pluripotent stem cell-derived cardiomyocytes exhibit temporal changes in phenotype. Am. J. Physiol.Heart Circ. Physiol. 2013, 305, H913–H922. [Google Scholar] [CrossRef]
- Dubois, N.C.; Craft, A.M.; Sharma, P.; Elliott, D.A.; Stanley, E.G.; Elefanty, A.G.; Gramolini, A.; Keller, G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat. Biotechnol. 2011, 29, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Tohyama, S.; Hattori, F.; Sano, M.; Hishiki, T.; Nagahata, Y.; Matsuura, T.; Hashimoto, H.; Suzuki, T.; Yamashita, H.; Satoh, Y.; et al. Distinct Metabolic Flow Enables Large-Scale Purification of Mouse and Human Pluripotent Stem Cell-Derived Cardiomyocytes. Cell Stem Cell 2012, 12, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Hayama, E.; Furutani, Y.; Kawaguchi, N.; Seki, A.; Nagashima, Y.; Okita, K.; Takeuchi, D.; Matsuoka, R.; Inai, K.; Hagiwara, N.; et al. Induced Pluripotent Stem Cell-Derived Cardiomyocytes with SCN5A R1623Q Mutation Associated with Severe Long QT Syndrome in Fetuses and Neonates Recapitulates Pathophysiological Phenotypes. Biology 2021, 10, 1062. [Google Scholar] [CrossRef]
- Milan, D.J.; Kim, A.M.; Winterfield, J.R.; Jones, I.L.; Pfeufer, A.; Sanna, S.; Arking, D.E.; Amsterdam, A.H.; Sabeh, K.M.; Mably, J.D.; et al. Drug-Sensitized Zebrafish Screen Identifies Multiple Genes, Including GINS3, as Regulators of Myocardial Repolarization. Circulation 2009, 120, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Milan, D.J.; Peterson, T.A.; Ruskin, J.N.; Peterson, R.T.; MacRae, C.A. Drugs That Induce Repolarization Abnormalities Cause Bradycardia in Zebrafish. Circulation 2003, 107, 1355–1358. [Google Scholar] [CrossRef]
- Langheinrich, U.; Vacun, G.; Wagner, T. Zebrafish embryos express an orthologue of HERG and are sensitive toward a range of QT-prolonging drugs inducing severe arrhythmia☆. Toxicol. Appl. Pharmacol. 2003, 193, 370–382. [Google Scholar] [CrossRef]
- Peal, D.S.; Mills, R.W.; Lynch, S.N.; Mosley, J.M.; Lim, E.; Ellinor, P.T.; January, C.T.; Peterson, R.T.; Milan, D.J. Novel Chemical Suppressors of Long QT Syndrome Identified by an In Vivo Functional Screen. Circulation 2011, 123, 23–30. [Google Scholar] [CrossRef]
- Summerton. Uncharged nucleic acid analogs for therapeutic and diagnostic applications: Oligomers assembled from ribose-derived subunits. In Discoveries in Antisense Nucleic Acids; Brakel, C., Ed.; The Portfolio Publishing Co.: Woodlands, TX, USA, 1989; pp. 71–80. [Google Scholar]
- Lawson, N.D.; Weinstein, B.M. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. Dev. Biol. 2002, 248, 307–318. [Google Scholar] [CrossRef]
- Jin, S.-W.; Beis, D.; Mitchell, T.; Chen, J.-N.; Stainier, D.Y.R. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 2005, 132, 5199–5209. [Google Scholar] [CrossRef]
- Liu, N.; Olson, E.N. CRISPR Modeling and Correction of Cardiovascular Disease. Circ. Res. 2022, 130, 1827–1850. [Google Scholar] [CrossRef] [PubMed]
- Saulnier-Blache, J.S.; Wilson, R.; Klavins, K.; Graham, D.; Alesutan, I.; Kastenmüller, G.; Wang-Sattler, R.; Adamski, J.; Roden, M.; Rathmann, W.; et al. Ldlr−/− and ApoE−/− mice better mimic the human metabolite signature of increased carotid intima media thickness compared to other animal models of cardiovascular disease. Atherosclerosis 2018, 276, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Kuro-o, M.; Matsumura, Y.; Aizawa, H.; Kawaguchi, H.; Suga, T.; Utsugi, T.; Ohyama, Y.; Kurabayashi, M.; Kaname, T.; Kume, E.; et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 1997, 390, 45–51. [Google Scholar] [CrossRef]
- Wang, W.; He, W.; Ruan, Y.; Geng, Q. First pig-to-human heart transplantation. Innovation 2022, 3, 100223. [Google Scholar] [CrossRef] [PubMed]
- Kavarana, S.; Kwon, J.H.; Zilinskas, K.; Kang, L.; Turek, J.W.; Mohiuddin, M.M.; Rajab, T.K. Recent advances in porcine cardiac xenotransplantation: From aortic valve replacement to heart transplantation. Expert Rev. Cardiovasc. Ther. 2022, 20, 597–608. [Google Scholar] [CrossRef]
- Gabriel, G.C.; Devine, W.; Redel, B.K.; Whitworth, K.M.; Samuel, M.; Spate, L.D.; Cecil, R.F.; Prather, R.S.; Wu, Y.; Wells, K.D.; et al. Cardiovascular Development and Congenital Heart Disease Modeling in the Pig. J. Am. Heart Assoc. 2021, 10, e021631. [Google Scholar] [CrossRef]
- Abe, K.; Toba, M.; Alzoubi, A.; Ito, M.; Fagan, K.A.; Cool, C.D.; Voelkel, N.F.; McMurtry, I.F.; Oka, M. Formation of plexiform lesions in experimental severe pulmonary arterial hypertension. Circulation 2010, 121, 2747–2754. [Google Scholar] [CrossRef]
- Zhang, T.; Kawaguchi, N.; Hayama, E.; Furutani, Y.; Nakanishi, T. High expression of CXCR4 and stem cell markers in a monocrotaline and chronic hypoxia-induced rat model of pulmonary arterial hypertension. Exp. Ther. Med. 2018, 15, 4615–4622. [Google Scholar] [CrossRef]
- Zhang, T.; Kawaguchi, N.; Yoshihara, K.; Hayama, E.; Furutani, Y.; Kawaguchi, K.; Tanaka, T.; Nakanishi, T. Silibinin efficacy in a rat model of pulmonary arterial hypertension using monocrotaline and chronic hypoxia. Respir. Res. 2019, 20, 79. [Google Scholar] [CrossRef]
- Joukar, S. A comparative review on heart ion channels, action potentials and electrocardiogram in rodents and human: Extrapolation of experimental insights to clinic. Lab. Anim. Res. 2021, 37, 25. [Google Scholar] [CrossRef]
- Yu, G.; Chakrabarti, S.; Tischenko, M.; Chen, A.-L.; Wang, Z.; Cho, H.; French, B.A.; Prasad, S.V.N.; Chen, Q.; Wang, Q.K. Gene therapy targeting protein trafficking regulator MOG1 in mouse models of Brugada syndrome, arrhythmias, and mild cardiomyopathy. Sci. Transl. Med. 2022, 14, eabf3136. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Yong, S.L.; Fan, C.; Ni, Y.; Yoo, S.; Zhang, T.; Zhang, X.; Obejero-Paz, C.A.; Rho, H.J.; Ke, T.; et al. Identification of a new co-factor, MOG1, required for the full function of cardiac sodium channel Nav 1.5. J. Biol. Chem 2008, 283, 6968–6978. [Google Scholar] [CrossRef] [PubMed]
- Morotti, S.; Liu, C.; Hegyi, B.; Ni, H.; Iseppe, A.F.; Wang, L.; Pritoni, M.; Ripplinger, C.M.; Bers, D.M.; Edwards, A.G.; et al. Quantitative cross-species translators of cardiac myocyte electrophysiology: Model training, experimental validation, and applications. Sci. Adv. 2021, 7, eabg0927. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.Q.X.; Sobie, E.A. Population-based mechanistic modeling allows for quantitative predictions of drug responses across cell types. NPJ Syst. Biol. Appl. 2018, 4, 11. [Google Scholar] [CrossRef]
- Parikh, S.S.; Blackwell, D.J.; Gomez-Hurtado, N.; Frisk, M.; Wang, L.; Kim, K.; Dahl, C.P.; Fiane, A.; Tønnessen, T.; Kryshtal, D.O.; et al. Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-InducedPluripotent Stem Cell-Derived Cardiomyocytes. Circ. Res. 2017, 121, 1323–1330. [Google Scholar] [CrossRef]
- Yoshida, S.; Miyagawa, S.; Fukushima, S.; Kawamura, T.; Kashiyama, N.; Ohashi, F.; Toyofuku, T.; Toda, K.; Sawa, Y. Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Soluble Factors from Human Mesenchymal Stem Cells. Mol. Ther. 2018, 26, 2681–2695. [Google Scholar] [CrossRef]
- Chan, Y.-C.; Ting, S.; Lee, Y.-K.; Ng, K.-M.; Zhang, J.; Chen, Z.; Siu, C.-W.; Oh, S.K.W.; Tse, H.-F. Electrical Stimulation Promotes Maturation of Cardiomyocytes Derived from Human Embryonic Stem Cells. J. Cardiovasc. Transl. Res. 2013, 6, 989–999. [Google Scholar] [CrossRef]
- Giacomelli, E.; Meraviglia, V.; Campostrini, G.; Cochrane, A.; Cao, X.; van Helden, R.W.J.; Krotenberg Garcia, A.; Mircea, M.; Kostidis, S.; Davis, R.P.; et al. Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 2020, 26, 862–879. [Google Scholar] [CrossRef]
- Zhang, J.; Klos, M.; Wilson, G.F.; Herman, A.M.; Lian, X.; Raval, K.K.; Barron, M.R.; Hou, L.; Soerens, A.G.; Yu, J.; et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: The matrix sandwich method. Circ. Res. 2012, 111, 1125–1136. [Google Scholar] [CrossRef]
- Tang, X.-Y.; Wu, S.; Da Wang, D.; Chu, C.; Hong, Y.; Tao, M.; Hu, H.; Xu, M.; Guo, X.; Liu, Y. Human organoids in basic research and clinical applications. Signal Transduct. Target. Ther. 2022, 7, 168. [Google Scholar] [CrossRef]
- Heydari, Z.; Moeinvaziri, F.; Agarwal, T.; Pooyan, P.; Shpichka, A.; Maiti, T.K.; Timashev, P.; Baharvand, H.; Vosough, M. Organoids: A novel modality in disease modeling. Bio-Design Manuf. 2021, 4, 689–716. [Google Scholar] [CrossRef] [PubMed]
- Ott, H.C.; Matthiesen, T.S.; Goh, S.-K.; Black, L.D.; Kren, S.M.; Netoff, T.I.; Taylor, D.A. Perfusion-decellularized matrix: Using nature’s platform to engineer a bioartificial heart. Nat. Med. 2008, 14, 213–221. [Google Scholar] [CrossRef]
- Kitahara, H.; Yagi, H.; Tajima, K.; Okamoto, K.; Yoshitake, A.; Aeba, R.; Kudo, M.; Kashima, I.; Kawaguchi, S.; Hirano, A.; et al. Heterotopic transplantation of a decellularized and recellularized whole porcine heart. Interact. Cardiovasc. Thorac. Surg. 2016, 22, 571–579. [Google Scholar] [CrossRef]
- Tsukamoto, Y.; Akagi, T.; Akashi, M. Vascularized cardiac tissue construction with orientation by layer-by-layer method and 3D printer. Sci. Rep. 2020, 10, 5484. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.J.; Guyette, J.P.; Miki, K.; Xiao, L.; Kaur, G.; Wu, T.; Zhu, L.; Hansen, K.J.; Ling, K.H.; Milan, D.J.; et al. Human iPS-derived pre-epicardial cells direct cardiomyocyte aggregation expansion and organization in vitro. Nat. Commun. 2021, 12, 4997. [Google Scholar] [CrossRef]
- Narazaki, G.; Uosaki, H.; Teranishi, M.; Okita, K.; Kim, B.; Matsuoka, S.; Yamanaka, S.; Yamashita, J.K. Directed and Systematic Differentiation of Cardiovascular Cells From Mouse Induced Pluripotent Stem Cells. Circulation 2008, 118, 498–506. [Google Scholar] [CrossRef] [PubMed]
- So, K.H.; Han, Y.J.; Park, H.Y.; Kim, J.G.; Sung, D.J.; Bae, Y.M.; Yang, B.C.; Park, S.B.; Chang, S.K.; Kim, E.Y.; et al. Generation of functional cardiomyocytes from mouse induced pluripotent stem cells. Int. J. Cardiol. 2011, 153, 277–285. [Google Scholar] [CrossRef]
- Schenke-Layland, K.; Rhodes, K.E.; Angelis, E.; Butylkova, Y.; Heydarkhan-Hagvall, S.; Gekas, C.; Zhang, R.; Goldhaber, J.I.; Mikkola, H.K.; Plath, K.; et al. Reprogrammed Mouse Fibroblasts Differentiate into Cells of the Cardiovascular and Hematopoietic Lineages. Stem Cells 2008, 26, 1537–1546. [Google Scholar] [CrossRef]
- Ren, Y.; Lee, M.Y.; Schliffke, S.; Paavola, J.; Amos, P.J.; Ge, X.; Ye, M.; Zhu, S.; Senyei, G.; Lum, L.; et al. Small molecule Wnt inhibitors enhance the efficiency of BMP-4-directed cardiac differentiation of human pluripotent stem cells. J. Mol. Cell. Cardiol. 2011, 51, 280–287. [Google Scholar] [CrossRef]
- Ng, W.H.; Johnston, E.K.; Tan, J.J.; Bliley, J.M.; Feinberg, A.W.; Stolz, D.B.; Sun, M.; Wijesekara, P.; Hawkins, F.; Kotton, D.N.; et al. Recapitulating human cardio-pulmonary co-development using simultaneous multilineage differentiation of pluripotent stem cells. eLife 2022, 11, e67872. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Sato, H.; Kato-Itoh, M.; Goto, T.; Hara, H.; Sanbo, M.; Mizuno, N.; Kobayashi, T.; Yanagida, A.; Umino, A.; et al. Interspecies organogenesis generates autologous functional islets. Nature 2017, 542, 191–196. [Google Scholar] [CrossRef]
- Matsunari, H.; Nagashima, H.; Watanabe, M.; Umeyama, K.; Nakano, K.; Nagaya, M.; Kobayashi, T.; Yamaguchi, T.; Sumazaki, R.; Herzenberg, L.A.; et al. Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. Proc. Natl. Acad. Sci. USA 2013, 110, 4557–4562. [Google Scholar] [CrossRef] [PubMed]
- Kano, M.; Mizutani, E.; Homma, S.; Masaki, H.; Nakauchi, H. Xenotransplantation and interspecies organogenesis: Current status and issues. Front. Endocrinol. 2022, 13, 963282. [Google Scholar] [CrossRef]
- Stevens, T.L.; Manring, H.R.; Wallace, M.J.; Argall, A.; Dew, T.; Papaioannou, P.; Antwi-Boasiako, S.; Xu, X.; Campbell, S.G.; Akar, F.G.; et al. Humanized Dsp ACM Mouse Model Displays Stress-Induced Cardiac Electrical and Structural Phenotypes. Cells 2022, 11, 3049. [Google Scholar] [CrossRef] [PubMed]
- Ng, R.; Manring, H.; Papoutsidakis, N.; Albertelli, T.; Tsai, N.; See, C.J.; Li, X.; Park, J.; Stevens, T.L.; Bobbili, P.J.; et al. Patient mutations linked to arrhythmogenic cardiomyopathy enhance calpain-mediated desmoplakin degradation. JCI Insight 2019, 5, e128643. [Google Scholar] [CrossRef] [PubMed]
- Uehara, S.; Higuchi, Y.; Yoneda, N.; Kawai, K.; Yamamoto, M.; Kamimura, H.; Iida, Y.; Oshimura, M.; Kazuki, Y.; Yamazaki, H.; et al. An improved TK-NOG mouse as a novel platform for humanized liver that overcomes limitations in both male and female animals. Drug Metab. Pharmacokinet. 2021, 42, 100410. [Google Scholar] [CrossRef]
- Trayanova, N.A.; Popescu, D.M.; Shade, J.K. Machine Learning in Arrhythmia and Electrophysiology. Circ. Res. 2021, 128, 544–566. [Google Scholar] [CrossRef]
- Feeny, A.K.; Chung, M.K.; Madabhushi, A.; Attia, Z.I.; Cikes, M.; Firouznia, M.; Friedman, P.A.; Kalscheur, M.M.; Kapa, S.; Narayan, S.M.; et al. Artificial Intelligence and Machine Learning in Arrhythmias and Cardiac Electrophysiology. Circ. Arrhythmia Electrophysiol. 2020, 13, e007952. [Google Scholar] [CrossRef]
- Nagarajan, V.D.; Lee, S.L.; Robertus, J.L.; Nienaber, C.A.; Trayanova, N.A.; Ernst, S. Artificial intelligence in the diagnosis and management of arrhythmias. Eur. Heart J. 2021, 42, 3904–3916. [Google Scholar] [CrossRef]
- Ng, B.; Nayyar, S.; Chauhan, V.S. The Role of Artificial Intelligence and Machine Learning in Clinical Cardiac Electrophysiology. Can. J. Cardiol. 2022, 38, 246–258. [Google Scholar] [CrossRef]
- Kabra, R.; Israni, S.; Vijay, B.; Baru, C.; Mendu, R.; Fellman, M.; Sridhar, A.; Mason, P.; Cheung, J.W.; DiBiase, L.; et al. Emerging role of artificial intelligence in cardiac electrophysiology. Cardiovasc. Digit. Health J. 2022, 3, 263–275. [Google Scholar] [CrossRef]
- Chung, C.T.; Lee, S.; King, E.; Liu, T.; Armoundas, A.A.; Bazoukis, G.; Tse, G. Clinical significance, challenges and limitations in using artificial intelligence for electrocardiography-based diagnosis. Int. J. Arrhythmia 2022, 23, 24. [Google Scholar] [CrossRef] [PubMed]
- Cantwell, C.D.; Mohamied, Y.; Tzortzis, K.N.; Garasto, S.; Houston, C.; Chowdhury, R.A.; Ng, F.S.; Bharath, A.A.; Peters, N.S. Rethinking multiscale cardiac electrophysiology with machine learning and predictive modelling. Comput. Biol. Med. 2019, 104, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Varshneya, M.; Mei, X.; Sobie, E.A. Prediction of arrhythmia susceptibility through mathematical modeling and machine learning. Proc. Natl. Acad. Sci. USA 2021, 118, e2104019118. [Google Scholar] [CrossRef] [PubMed]
- Zolotarev, A.M.; Hansen, B.J.; Ivanova, E.A.; Helfrich, K.M.; Li, N.; Janssen, P.M.; Mohler, P.J.; Mokadam, N.A.; Whitson, B.A.; Fedorov, M.V.; et al. Optical Mapping-Validated Machine Learning Improves Atrial Fibrillation Driver Detection by Multi-Electrode Mapping. Circ. Arrhythmia Electrophysiol. 2020, 13, e008249. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Colborn, K.L.; Smith, D.E.; Xing, F.; Ghosh, D.; Rosenberg, M.A. Assessment of a Machine Learning Model Applied to Harmonized Electronic Health Record Data for the Prediction of Incident Atrial Fibrillation. JAMA Netw. Open 2020, 3, e1919396. [Google Scholar] [CrossRef] [PubMed]
- Piccini, J.P.; Russo, A.M.; Sharma, P.S.; Kron, J.; Tzou, W.; Sauer, W.; Park, D.S.; Birgersdotter-Green, U.; Frankel, D.S.; Healey, J.S.; et al. Advances in Cardiac Electrophysiology. Circ. Arrhythmia Electrophysiol. 2022, 15, e009911. [Google Scholar] [CrossRef]
- Glass, C.; Lafata, K.J.; Jeck, W.; Horstmeyer, R.; Cooke, C.; Everitt, J.; Glass, M.; Dov, D.; Seidman, M.A. The Role of Machine Learning in Cardiovascular Pathology. Can. J. Cardiol. 2021, 38, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Doi, K.; Giger, M.; Wu, Y.; Nishikawa, R.; Schmidt, R.A. Computerized detection of clustered microcalcifications in digital mammograms using a shift-invariant artificial neural network. Med. Phys. 1994, 21, 517–524. [Google Scholar] [CrossRef]
- Lima, E.M.; Ribeiro, A.H.; Paixão, G.M.M.; Ribeiro, M.H.; Pinto-Filho, M.M.; Gomes, P.R.; Oliveira, D.M.; Sabino, E.C.; Duncan, B.B.; Giatti, L.; et al. Deep neural network-estimated electrocardiographic age as a mortality predictor. Nat. Commun. 2021, 12, 5117. [Google Scholar] [CrossRef]
- Young, M.T.; Blanchard, S.M.; White, M.W.; Johnson, E.E.; Smith, W.M.; Ideker, R.E. Using an Artificial Neural Network to Detect Activations during Ventricular Fibrillation. Comput. Biomed. Res. 2000, 33, 43–58. [Google Scholar] [CrossRef]
- Martin, C.H.; Oved, A.; Chowdhury, R.A.; Ullmann, E.; Peters, N.S.; Bharath, A.A.; Varela, M. EP-PINNs: Cardiac Electrophysiology Characterisation Using Physics-Informed Neural Networks. Front. Cardiovasc. Med. 2022, 8, 768419. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-M.; Liu, C.-L.; Hu, K.-W.; Tseng, V.S.; Chang, S.-L.; Lin, Y.-J.; Lo, L.-W.; Chung, F.-P.; Chao, T.-F.; Tuan, T.-C.; et al. A Deep Learning–Enabled Electrocardiogram Model for the Identification of a Rare Inherited Arrhythmia: Brugada Syndrome. Can. J. Cardiol. 2022, 38, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Aghasafari, P.; Yang, P.C.; Kernik, D.C.; Sakamoto, K.; Kanda, Y.; Kurokawa, J.; Vorobyov, I.; Clancy, C.E. A deep learning algorithm to translate and classify cardiac electrophysiology. eLife 2021, 10, e68335. [Google Scholar] [CrossRef] [PubMed]
- Orita, K.; Sawada, K.; Koyama, R.; Ikegaya, Y. Deep learning-based quality control of cultured human-induced pluripotent stem cell-derived cardiomyocytes. J. Pharmacol. Sci. 2019, 140, 313–316. [Google Scholar] [CrossRef]
- Pang, J.K.; Chia, S.; Zhang, J.; Szyniarowski, P.; Stewart, C.; Yang, H.; Chan, W.-K.; Ng, S.Y.; Soh, B.-S. Characterizing arrhythmia using machine learning analysis of Ca2+ cycling in human cardiomyocytes. Stem Cell Rep. 2022, 17, 1810–1823. [Google Scholar] [CrossRef]
- Song, E.; Lee, Y.-S. Interpretable machine learning of action potential duration restitution kinetics in single-cell models of atrial cardiomyocytes. J. Electrocardiol. 2022, 74, 137–145. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawaguchi, N.; Nakanishi, T. Animal Disease Models and Patient-iPS-Cell-Derived In Vitro Disease Models for Cardiovascular Biology—How Close to Disease? Biology 2023, 12, 468. https://doi.org/10.3390/biology12030468
Kawaguchi N, Nakanishi T. Animal Disease Models and Patient-iPS-Cell-Derived In Vitro Disease Models for Cardiovascular Biology—How Close to Disease? Biology. 2023; 12(3):468. https://doi.org/10.3390/biology12030468
Chicago/Turabian StyleKawaguchi, Nanako, and Toshio Nakanishi. 2023. "Animal Disease Models and Patient-iPS-Cell-Derived In Vitro Disease Models for Cardiovascular Biology—How Close to Disease?" Biology 12, no. 3: 468. https://doi.org/10.3390/biology12030468
APA StyleKawaguchi, N., & Nakanishi, T. (2023). Animal Disease Models and Patient-iPS-Cell-Derived In Vitro Disease Models for Cardiovascular Biology—How Close to Disease? Biology, 12(3), 468. https://doi.org/10.3390/biology12030468






