Can Probiotics, Particularly Limosilactobacillus fermentum UCO-979C and Lacticaseibacillus rhamnosus UCO-25A, Be Preventive Alternatives against SARS-CoV-2?
Simple Summary
Abstract
1. Introduction
1.1. SARS-CoV-2 and Probiotics
1.2. SARS-CoV-2: A High-Prevalence Threat to Global Public Health
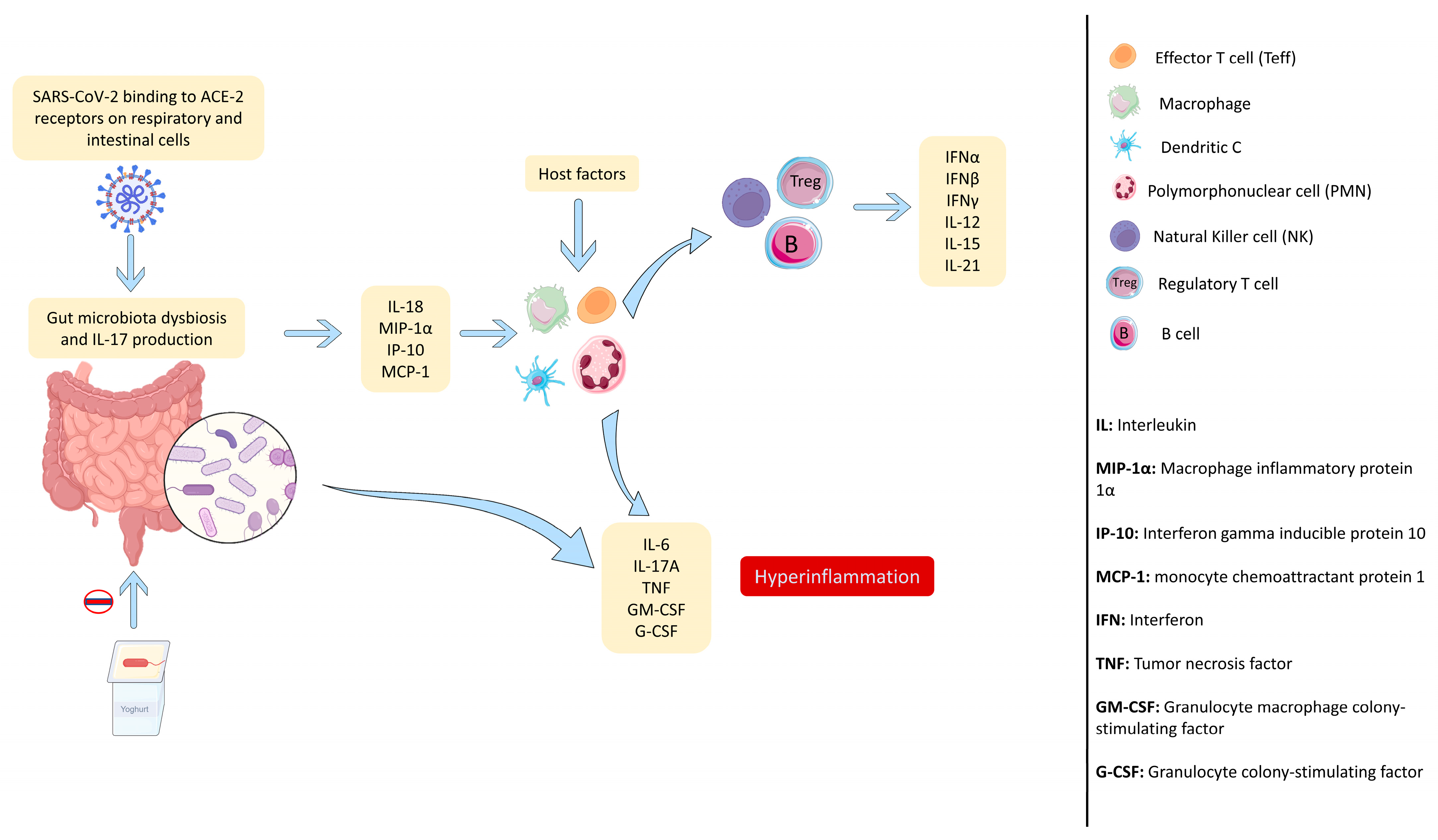
1.3. SARS-CoV-2 Infection Generates an Uncontrolled Immune Response
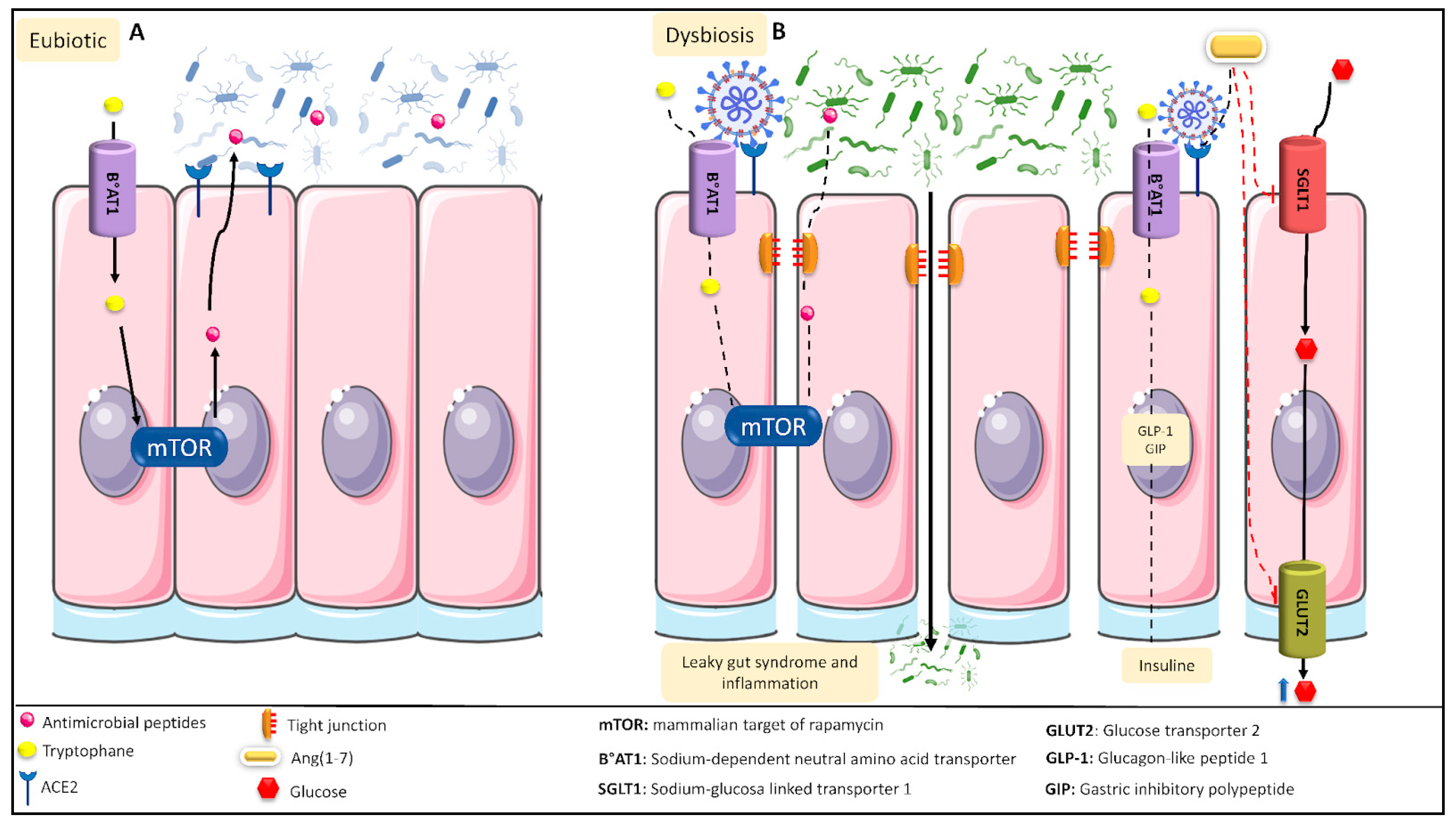
1.4. SARS-CoV-2 Infection Causes Intestinal Dysbiosis Associated with a Loss in Intestinal Mucosa
1.5. Current Strategies to Deal with COVID-19
1.5.1. Vaccines, the Only Preventive Therapy to Deal with COVID-19
1.5.2. Current Treatments to Control SARS-CoV-2 Infection
1.5.3. Co-Adjuvant Treatments against COVID-19
2. Materials and Methods
3. Results
3.1. Probiotics, an Alternative
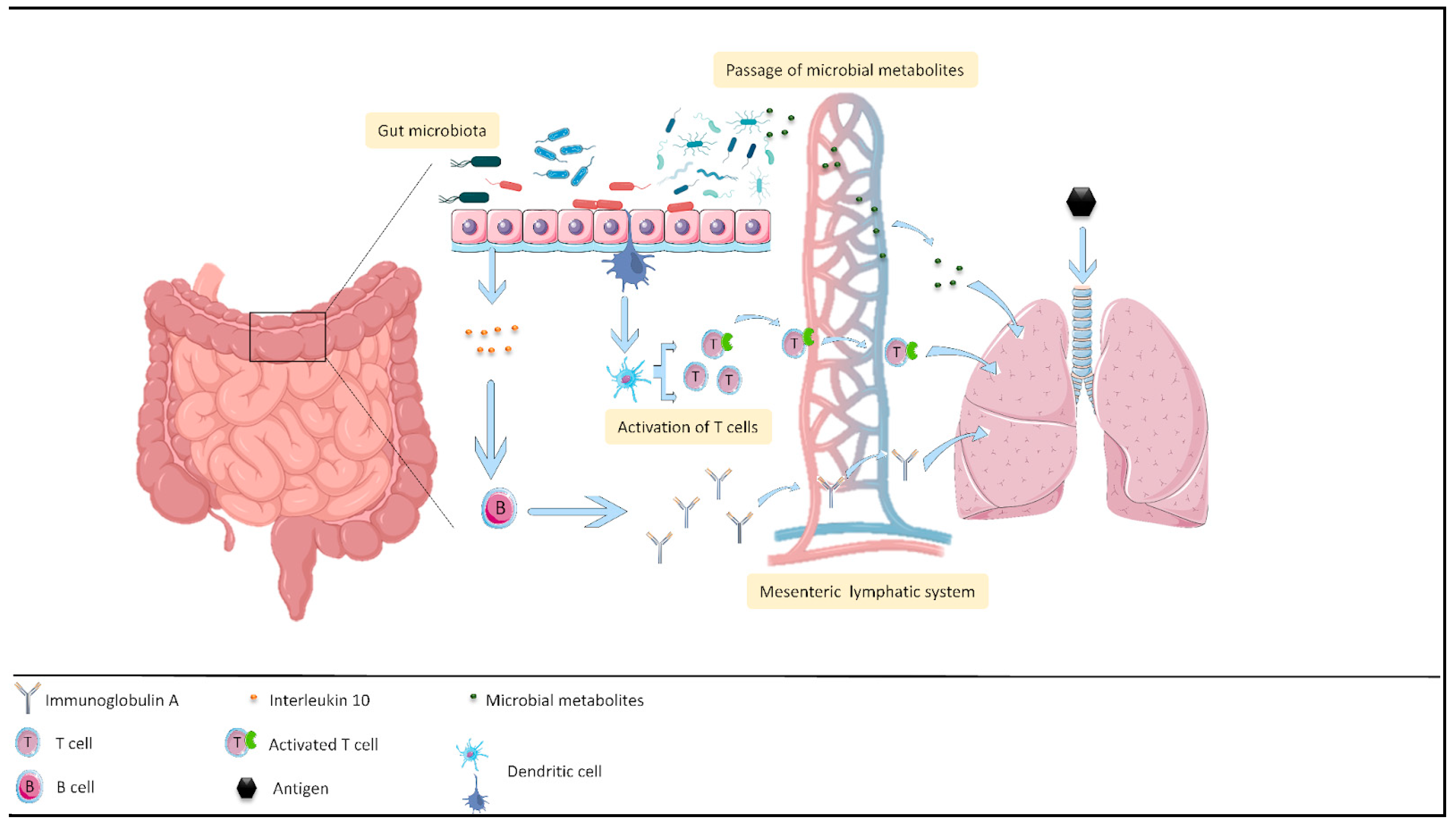
3.2. Effect of Probiotics on the Intestine–Lung Axis
3.3. Some Probiotics Might Be a Complement to Current Therapies
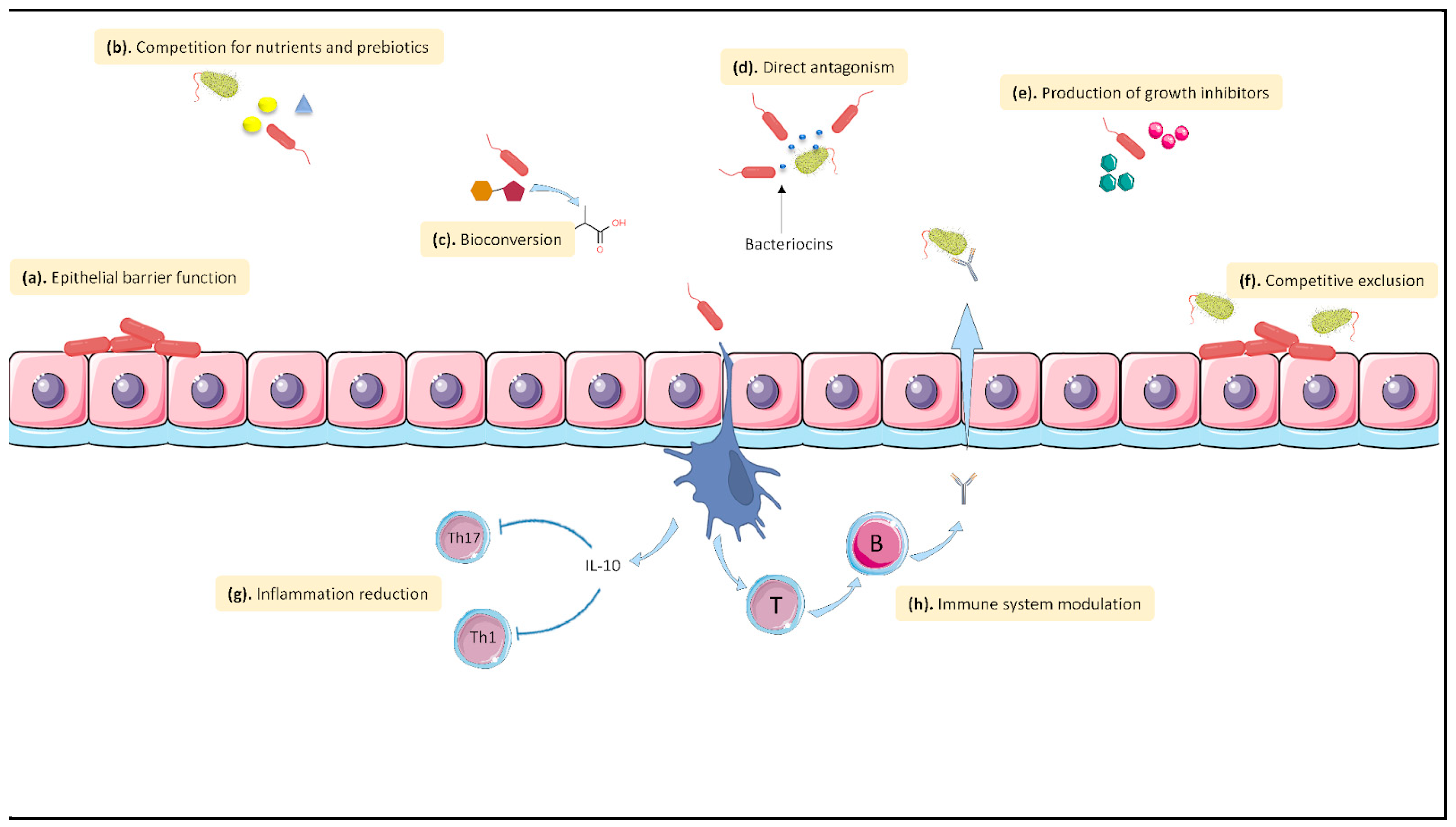
3.4. Probiotics May Antagonize SARS-CoV-2 Infection
3.5. Possible Targets and Metabolites of Probiotics Related to SARS-CoV-2 Infection
3.5.1. Possible Targets
RBD
3CLpro
Non-Structural Proteins (NSPs)
3.5.2. Possible Metabolites of Probiotics with SARS-CoV-2 Inhibitory Activity
Bacteriocins from Lactic Acid Bacteria
Microbial Peptides
Short-Chain Fatty Acids (SCFA)
3.6. Intellectual Property Associated with Probiotics and COVID-19
3.7. The Use of Probiotics Could Reduce the Economic Costs Associated with the Control of Respiratory Diseases
3.8. Limosilactobacillus Fermentum UCO-979C and Lacticaseibacillus Rhamnosus UCO-25A Strains Exhibit Immunological Characteristics That Are of Interest Due to Their Possible Use against SARS-CoV-2
| Factor | SARS-CoV-2 | L. fermentum UCO-979C | L. rhamnosus UCO-25A | References | |
|---|---|---|---|---|---|
| Immune response | Cytokine storm | Increased serum levels of IL-2, IL-7, IL-8, IL-9, IL-10, IL-17, G-CSF, GM-CSF, IFN-γ, TNFα, IP10, MCP1, MIP1A, and MIP1B, but particularly increases IL-6 and IL-1 levels | It modulates intestinal and systemic cytokine levels | It modulates intestinal and systemic cytokine levels | [115,116,117,119] |
| IL-6 | Deregulates IL-6 levels | Reduces the proinflammatory response, decreasing IL6 It possesses mixed stimulating/anti-inflammatory effects at the intestinal level, increasing IL-6 | Reduces the proinflammatory response, increasing gastrointestinal IL-6 | [115,116,117,119] | |
| IL-10 | Deregulates IL-10 levels | Reduces inflammation in the affected gastric tissue, increasing IL-10 | Shows anti-inflammatory activity, increasing IL-10 | [115,116,117,119] | |
| Macrophages | Macrophage activation syndrome (MAS) | Macrophages (THP-1 cell line) show anti-inflammatory activity, reducing TNF-α and increasing IL-10 as well as IFN-γ Stimulates the activation of peritoneal macrophages | Macrophages (THP-1 cell line) show anti-inflammatory activity, reducing TNF-α as well as IL-8 and increasing IL-10 as well as IFN-γ Increases the activity of peritoneal macrophages | [115,116,117,118,119] | |
| T cells | Causes inadequate behavior of IFN and MHC expression pathways in macrophages, NK cells, and dendritic cells, delaying the activation as well as response of T lymphocytes and allowing increased viral replication | Stimulates the activation of T CD4+ cells | Increases the activity of T cells | [115,116,117,118,119] | |
| B cells | The alterations to the T response and its delayed activation, caused by viral interference on the innate response, causes an inadequate follicular B response | Reduces the number of immature B cells in Peyer’s patches | Increases the activity of B cells | [114,115,116,117,118,119] | |
| Intestinal microbiota | Intestinal dysbiosis | Dysbiosis of the microbiota | In an animal model (Mongolian gerbils), these probiotic strains were able to colonize as well as survive in the gastrointestinal tract and maintain their viability for a long period (10 to 15 days), favoring their probiotic capacity Capable of reducing and protecting from intestinal inflammation, as well as inhibiting the adhesion and invasion of pathogens (H. pylori) Help to restore the intestinal microbiota and reduce severe gastrointestinal symptoms | [114,115,116,117,118,119] | |
| It can infect the gastrointestinal tract and actively replicate in it | |||||
| In patients infected by SARS-CoV-2, 5 to 10% develop intestinal symptoms, such as diarrhea, vomiting, and abdominal pain | |||||
| Intestinal immune response | Large release of proinflammatory cytokines and recruitment of neutrophils, macrophages, and other cell types which contribute to uncontrolled systemic inflammation and a cytokine storm Increased IL-33 and IL-8 levels in fecal samples of COVID-19 patients due to intestinal involvement in addition to a decrease in IL-1β, TNF-α, and IL-6 cytokines | At the gastrointestinal level (AGS cell line), it reduces the proinflammatory response (reduces TNF-α, IL1-β, IL-6, and IL-8) | At the gastrointestinal level (AGS cell line), it reduces the proinflammatory response (reduces TNF-α, IL1-β, and IL-8) and increases IL-6 | [114,115,116,117,119] | |
| At the intestinal level (swine intestinal cells and murine model), mixed stimulating/ anti-inflammatory activity Reduces the expression of inflammatory factors, such as CXCL8, CXCL9, CXCL10, CXCL11, C1S, and C3, and it increases the expression of IL-6, CCL8, C1R, and CFB | In the murine model, it increases the activity of peritoneal macrophages and intestinal IgA levels | [114,115,116,117,118,119] | |||
| At the gastric level in a murine model, it decreases the inflammation of the affected tissue, in addition to an increase in IFN-γ and IL-10 as well as a decrease in serum TNF-α and IL-8 | |||||
3.9. In Silico Metagenomic Analysis of Metabolites Present in Immunobiotic Strains Limosilactobacillus Fermentum UCO-979C and Lacticaseibacillus Rhamnosus UCO-25A with a Possible Effect on COVID-19 Treatment
| Probiotic Strain | Metabolite | Gene | Related Gene UniProt Code (Organism) | Function Related to Pathogenicity | References |
|---|---|---|---|---|---|
| L. fermentum UCO-979C | Short-chain fatty acid | 3-hydroxy-isobutyrate dehydrogenase | P31937 (human) | Participates in the 3- hidroxybutyrate synthesis pathway in bacteria | [126] |
| Thiolases | D7GV33 (butyrate-producing bacteria) | Participates in the butyrate synthesis pathway via acetyl-CoA | [121] | ||
| Bacteriocins | Genes related to the linocin M18 family | V0A9Z1 (E. coli) | Bacteriocin that is capable of stimulating the humoral immune response in the lungs | [122] | |
| Secondary bile acids | Bile acid dehydratase | P19412 (Clostridium scindens) | Participates in the biosynthesis of bile acids, which stimulate the immune response associated with T cells | [125] | |
| L. rhamnosus UCO-25A | Short-chain fatty acid | Genes related to the synthesis of hydroxyglutaryl | A0A2P6WFN2 (acidobacteria bacterium) | Participates in the butyrate synthesis pathway via glutarate | [121] |
| Thiolases | D7GV33 (butyrate-producing bacteria) | Participates in the butyrate synthesis pathway via acetyl-CoA | [121] | ||
| Genes related to their biosynthesis | P54616 (Bacillus subtilis) | Participates in the synthesis of short-chain fatty acids, which have immunomodulatory functions | [100,101,102] | ||
| Bacteriocins | Bacteriocin | P83002 (Lactococcus lactis subsp. lactis) | May be able to prevent the entry of SARS-CoV-2 into host cells and exhibit antiviral activity | [91,122] | |
| Genes related to secretion | P22519 (E. coli) | Participates in the secretion of bacteriocins from the probiotic strains to exert their antiviral function | [91,122] | ||
| Secondary bile acids | Genes related to transport | P32369 (Clostridium scindens) | Participates in the transport of secondary bile acids, which stimulate the immune system | [123,124] |
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Forchette, L.; Sebastian, W.; Liu, T. A Comprehensive Review of COVID-19 Virology, Vaccines, Variants, and Therapeutics. Curr. Med. Sci. 2021, 41, 1037–1051. [Google Scholar] [CrossRef] [PubMed]
- Dhar, D.; Mohanty, A. Gut microbiota and Covid-19-possible link and implications. Virus Res. 2020, 285, 198018. [Google Scholar] [CrossRef]
- Fiolet, T.; Kherabi, Y.; MacDonald, C.J.; Ghosn, J.; Peiffer-Smadja, N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: A narrative review. Clin. Microbiol. Infect. 2022, 28, 202–221. [Google Scholar] [CrossRef]
- Todorov, S.D.; Tagg, J.R.; Ivanova, I.V. Could Probiotics and Postbiotics Function as “Silver Bullet” in the Post-COVID-19 Era? Probiotics Antimicrob. Proteins 2021, 13, 1499–1507. [Google Scholar] [CrossRef]
- Sundararaman, A.; Ray, M.; Ravindra, P.V.; Halami, P.M. Role of probiotics to combat viral infections with emphasis on COVID-19. Appl. Microbiol. Biotechnol. 2020, 104, 8089–8104. [Google Scholar] [CrossRef]
- Fanos, V.; Pintus, M.C.; Pintus, R.; Marcialis, M.A. Lung microbiota in the acute respiratory disease: From coronavirus to metabolomics. J. Pediatr. Neonatal Individ. Med. (JPNIM) 2020, 9, e090139. [Google Scholar] [CrossRef]
- Bingula, R.; Filaire, M.; Radosevic-Robin, N.; Bey, M.; Berthon, J.Y.; Bernalier-Donadille, A.; Vasson, M.P.; Filaire, E. Desired Turbulence? Gut-Lung Axis, Immunity, and Lung Cancer. J. Oncol. 2017, 2017, 5035371. [Google Scholar] [CrossRef] [PubMed]
- Altadill, T.; Espadaler-Mazo, J.; Liong, M.T. Effects of a Lactobacilli Probiotic on Reducing Duration of URTI and Fever, and Use of URTI-Associated Medicine: A Re-Analysis of a Randomized, Placebo-Controlled Study. Microorganisms 2021, 9, 528. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, V.; Cerda, J.; Le Corre, N.; Medina, R.; Ferrés, M. Caracterización clínica y epidemiológica de infección asociada a atención en salud por virus influenza en pacientes críticos. Rev. Chil. Infectol. 2019, 36, 274–282. [Google Scholar] [CrossRef]
- Pastrián-Soto, G. Bases Genéticas y Moleculares del COVID-19 (SARS-CoV-2). Mecanismos de Patogénesis y de Respuesta Inmune. Int. J. Odontostomat. 2020, 14, 331–337. [Google Scholar] [CrossRef]
- Mahooti, M.; Miri, S.M.; Abdolalipour, E.; Ghaemi, A. The immunomodulatory effects of probiotics on respiratory viral infections: A hint for COVID-19 treatment? Microb. Pathog. 2020, 148, 104452. [Google Scholar] [CrossRef]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Inf. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Cifras: Situación Nacional de COVID-19 en Chile. Available online: https://www.gob.cl/coronavirus/cifrasoficiales (accessed on 30 October 2022).
- Leclerc, Q.J.; Fuller, N.M.; Knight, L.E.; CMMID COVID-19 Working Group; Funk, S.; Knight, G.M. What settings have been linked to SARS-CoV-2 transmission clusters? Wellcome Open Res. 2020, 5, 83. [Google Scholar] [CrossRef]
- Liu, M.; Caputi, T.L.; Dredze, M.; Kesselheim, A.S.; Ayers, J.W. Internet searches for unproven COVID-19 therapies in the United States. JAMA Intern. Med. 2020, 180, 1116–1118. [Google Scholar] [CrossRef] [PubMed]
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Zhang, R.; Li, Y.; Zhang, A.L.; Wang, Y.; Molina, M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. USA 2020, 117, 14857–14863. [Google Scholar] [CrossRef]
- Ozma, M.A.; Maroufi, P.; Khodadadi, E.; Köse, Ş.; Esposito, I.; Ganbarov, K.; Dao, S.; Esposito, S.; Dal, T.; Zeinalzadeh, E.; et al. Clinical manifestation, diagnosis, prevention and control of SARS-CoV-2 (COVID-19) during the outbreak period. Infez. Med. 2020, 28, 153–165. [Google Scholar] [PubMed]
- Maguiña Vargas, C.; Gastelo Acosta, R.; Tequen Bernilla, A. El nuevo Coronavirus y la pandemia del Covid-19. Rev. Med. Herediana 2020, 31, 125–131. [Google Scholar] [CrossRef]
- Pan, L.; Mu, M.; Yang, P.; Sun, Y.; Wang, R.; Yan, J.; Li, P.; Hu, B.; Wang, J.; Hu, C.; et al. Clinical Characteristics of COVID-19 Patients With Digestive Symptoms in Hubei, China: A Descriptive, Cross-Sectional, Multicenter Study. Am. J. Gastroenterol. 2020, 115, 766–773. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.; Shao, C.; Huang, J.; Gan, J.; Huang, X.; Bucci, E.; Piacentini, M.; Ippolito, G.; Melino, G. COVID-19 infection: The perspectives on immune responses. Cell Death Differ. 2020, 27, 1451–1454. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; HLH Across Speciality Collaboration, UK. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Sanz, J.M.; Gómez Lahoz, A.M.; . Martín, R.O. Papel del sistema inmune en la infección por el SARS-CoV-2: Inmunopatología de la COVID-19 [Role of the immune system in SARS-CoV-2 infection: Immunopathology of COVID-19]. Medicine 2021, 13, 1917–1931. [Google Scholar] [CrossRef]
- McElvaney, O.J.; Hobbs, B.D.; Qiao, D.; McElvaney, O.F.; Moll, M.; McEvoy, N.L.; Clarke, J.; O’Connor, E.; Walsh, S.; Cho, M.H.; et al. A linear prognostic score based on the ratio of interleukin-6 to interleukin-10 predicts outcomes in COVID-19. eBioMedicine 2020, 61, 103026. [Google Scholar] [CrossRef]
- Kindler, E.; Thiel, V. SARS-CoV and IFN: Too little, too late. Cell Host Microbe 2016, 19, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.; Kitazawa, H. The Modulation of Mucosal Antiviral Immunity by Immunobiotics: Could They Offer Any Benefit in the SARS-CoV-2 Pandemic? Front. Physiol. 2020, 11, 699. [Google Scholar] [CrossRef]
- Kaneko, N.; Kuo, H.H.; Boucau, J.; Farmer, J.R.; Allard-Chamard, H.; Mahajan, V.S.; Piechocka-Trocha, A.; Lefteri, K.; Osborn, M.; Bals, J.; et al. Massachusetts Consortium on Pathogen Readiness Specimen Working Group. Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell 2020, 183, 143–157.e13. [Google Scholar] [CrossRef]
- Ibarrondo, F.J.; Fulcher, J.A.; Goodman-Meza, D.; Elliott, J.; Hofmann, C.; Hausner, M.A.; Ferbas, K.G.; Tobin, N.H.; Aldrovandi, G.M.; Yang, O.O. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild Covid-19. N. Engl. J. Med. 2020, 383, 1085–1087. [Google Scholar] [CrossRef] [PubMed]
- Härdtner, C.; Mörke, C.; Walther, R.; Wolke, C.; Lendeckel, U. High glucose activates the alternative ACE2/Ang-(1-7)/Mas and APN/Ang IV/IRAP RAS axes in pancreatic β-cells. Int. J. Mol. Med. 2013, 32, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D.; et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 422. [Google Scholar] [CrossRef] [PubMed]
- Instituto de Salud Pública (ISP). Fases de Desarrollo de las Vacunas. Available online: https://www.ispch.cl/anamed/farmacovigilancia/vacunas/fases-de-desarrollo-de-las-vacunas/ (accessed on 10 November 2022).
- Monasterio, F. Cómo Avanzan las Vacunas Contra el Covid-19 en Chile y el Mundo. Pauta. Available online: https://www.pauta.cl/ciencia-y-tecnologia/vacunacion-covid-chile-mundo-desarrollo-fechas (accessed on 10 November 2022).
- Center for Disease Control and Prevention (CDC). Pfizer—BioNTech COVID-19 Vaccine. Available online: https://www.cdc.gov/vaccines/covid-19/info-by-product/pfizer/index.html (accessed on 10 November 2022).
- Food and Drug Administration (FDA). Pfizer-BioNTech COVID-19 Vaccine Frequently Asked Questions. Available online: https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/pfizer-biontech-covid-19-vaccine-frequently-asked-questions (accessed on 10 November 2022).
- Pfizer-BioNTech. Hoja Informativa para Proveedores de la Salud Que Administren la Vacuna (Proveedores de Vacunación). Available online: https://www.minsal.cl/wp-content/uploads/2020/12/Informaci%C3%B3n-para-prescribir-FDA-para-profesionales_vacuna-Pfizer-BioNTECH-COVID-19.pdf (accessed on 10 November 2022).
- Singh, P.K.; Kulsum, U.; Rufai, S.B.; Mudliar, S.R.; Singh, S. Mutations in SARS-CoV-2 Leading to Antigenic Variations in Spike Protein: A Challenge in Vaccine Development. J. Lab. Physicians 2020, 12, 154–160. [Google Scholar] [CrossRef] [PubMed]
- The Lancet Microbe. COVID-19 vaccines: The pandemic will not end overnight. Lancet Microbe 2021, 2, E1. [Google Scholar] [CrossRef] [PubMed]
- Serafin, M.B.; Bottega, A.; Foletto, V.S.; da Rosa, T.F.; Hörner, A.; Hörner, R. Drug repositioning is an alternative for the treatment of coronavirus COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105969. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.S.; Lee, P.I.; Hsueh, P.R. Treatment options for COVID-19: The reality and challenges. J. Microbiol. Immunol. Infect. 2020, 53, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Chugh, H.; Awasthi, A.; Agarwal, Y.; Gaur, R.K.; Dhawan, G.; Chandra, R. A comprehensive review on potential therapeutics interventions for COVID-19. Eur. J. Pharmacol. 2021, 890, 173741. [Google Scholar] [CrossRef]
- Mansur, J.L.; Tajer, C.; Mariani, J.; Inserra, F.; Ferder, L.; Manucha, W. Vitamin D high doses supplementation could represent a promising alternative to prevent or treat COVID-19 infection. Clin. Investig. Arterioscler. 2020, 32, 267–277. [Google Scholar] [CrossRef]
- Boretti, A.; Banik, B.K. Intravenous vitamin C for reduction of cytokines storm in acute respiratory distress syndrome. PharmaNutrition 2020, 12, 100190. [Google Scholar] [CrossRef]
- Oyagbemi, A.A.; Ajibade, T.O.; Aboua, Y.G.; Gbadamosi, I.T.; Adedapo, A.D.A.; Aro, A.O.; Adejumobi, O.A.; Thamahane-Katengua, E.; Omobowale, T.O.; Falayi, O.O.; et al. Potential health benefits of zinc supplementation for the management of COVID-19 pandemic. J. Food Biochem. 2021, 45, e13604. [Google Scholar] [CrossRef]
- Manzano-Santana, P.; Peñarreta, J.; Chóez-Guaranda, I.; Barragán, A.; Orellana-Manzano, A.; Rastrelli, L. Potential bioactive compounds of medicinal plants against new Coronavirus (SARS-CoV-2): A review. Bionatura 2021, 6, 1653–1658. [Google Scholar] [CrossRef]
- Mirzaei, R.; Attar, A.; Papizadeh, S.; Jeda, A.S.; Hosseini-Fard, S.R.; Jamasbi, E.; Kazemi, S.; Amerkani, S.; Talei, G.R.; Moradi, P.; et al. The emerging role of probiotics as a mitigation strategy against coronavirus disease 2019 (COVID-19). Arch. Virol. 2021, 166, 1819–1840. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernandez-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. bioRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Garg, R. Probiotics. Indian J. Med. Microbiol. 2006, 27, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Clancy, R. Immunobiotics and the probiotic evolution. FEMS Immunol. Med. Microbiol. 2003, 38, 9–12. [Google Scholar] [CrossRef]
- Szajewska, H.; Kołodziej, M.; Gieruszczak-Białek, D.; Skórka, A.; Ruszczyński, M.; Shamir, R. Systematic review with meta-analysis: Lactobacillus rhamnosus GG for treating acute gastroenteritis in children—A 2019 update. Aliment. Pharmacol. Ther. 2019, 49, 1376–1384. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Goldenberg, J.Z.; Humphrey, C.; El Dib, R.; Johnston, B.C. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst. Rev. 2019, 4, CD004827. [Google Scholar] [CrossRef] [PubMed]
- Chiu, L.; Bazin, T.; Truchetet, M.E.; Schaeverbeke, T.; Delhaes, L.; Pradeu, T. Protective Microbiota: From Localized to Long-Reaching Co-Immunity. Front. Immunol. 2017, 8, 1678. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Mande, S.S. Diet, Microbiota and Gut-Lung Connection. Front. Microbiol. 2018, 9, 2147. [Google Scholar] [CrossRef]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef]
- Li, Y.C.; Chen, Y.; Du, J. Critical roles of intestinal epithelial vitamin D receptor signaling in controlling gut mucosal inflammation. J. Steroid Biochem. Mol. Biol. 2015, 148, 179–183. [Google Scholar] [CrossRef]
- Del Pinto, R.; Ferri, C.; Cominelli, F. Vitamin d axis in inflammatory bowel diseases: Role, current uses and future perspectives. Int. J. Mol. Sci. 2017, 18, 2360. [Google Scholar] [CrossRef]
- El Hage, R.; Hernandez-Sanabria, E.; Van de Wiele, T. Emerging trends in “smart probiotics”: Functional consideration for the development of novel health and industrial applications. Front. Microbiol. 2017, 8, 1889. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulou, E.; Bezirtzoglou, E. Probiotics in Medicine: A Long Debate. Front. Immunol. 2020, 11, 2192. [Google Scholar] [CrossRef] [PubMed]
- Gallacher, D.J.; Kotecha, S. Respiratoy microbiome of new-born infants. Front. Pediatr. 2016, 4, 10. [Google Scholar] [CrossRef]
- Zafar, N.; Aslam, M.; Ali, A.; Khatoon, A.; Nazir, A.; Tanveer, Q.; Bilal, M.; Kanwar, R.; Qadeer, A.; Sikandar, M.; et al. Probiotics: Helpful for the prevention of COVID-19? BMRAT 2020, 7, 4086–4099. [Google Scholar] [CrossRef]
- D’Ettorre, G.; Ceccarelli, G.; Marazzato, M.; Campagna, G.; Pinacchio, C.; Alessandri, F.; Mastroianni, C. Challenges in the Management of SARS-CoV2 Infection: The Role of Oral Bacteriotherapy as Complementary Therapeutic Strategy to Avoid the Progression of COVID-19. Front. Med. 2020, 7, 389. [Google Scholar] [CrossRef] [PubMed]
- Olveira, G.; González-Molero, I. Actualización de probióticos, prebióticos y simbióticos en nutrición clínica. Endocrinol. Nutr. 2016, 63, 482–494. [Google Scholar] [CrossRef]
- Zelaya, H.; Alvarez, S.; Kitazawa, H.; Villena, J. Respiratory antiviral immunity and immunobiotics: Beneficial effects on inflammation-coagulation interaction during influenza virus infection. Front. Immunol. 2016, 7, 633. [Google Scholar] [CrossRef]
- Wang, K.; Ran, L.; Yan, T.; Niu, Z.; Kan, Z.; Zhang, Y.; Yang, Y.; Xie, L.; Huang, S.; Yu, Q.; et al. Anti-TGEV miller strain infection effect of Lactobacillus plantarum supernatant based on the JAK-STAT1 signaling pathway. Front. Microbiol. 2019, 10, 2540. [Google Scholar] [CrossRef]
- Waki, N.; Matsumoto, M.; Fukui, Y.; Suganuma, H. Effects of probiotic Lactobacillus brevis KB 290 on incidence of influenza infection among schoolchildren: An open-label pilot study. Lett. Appl. Microbiol. 2014, 59, 565–571. [Google Scholar] [CrossRef]
- Yasui, H.; Kiyoshima, J.; Hori, T. Reduction of influenza virus titer and protection against influenza virus infection in infant mice fed Lactobacillus casei Shirota. Clin. Diagn. Lab. Immunol. 2004, 11, 675–679. [Google Scholar] [CrossRef]
- Shimizu, K.; Yamada, T.; Ogura, H.; Mohri, T.; Kiguchi, T.; Fujimi, S.; Asahara, T.; Yamada, T.; Ojima, M.; Ikeda, M.; et al. Synbiotics modulate gut microbiota and reduce enteritis and ventilator-associated pneumonia in patients with sepsis: A randomized controlled trial. Crit. Care 2018, 22, 239. [Google Scholar] [CrossRef]
- Leyer, G.J.; LI, S.; Mubasher, M.E.; Reifer, C.; Ouwehand, A.C. Probiotic effects on cold and influenza-like symptom incidence and duration in children. Pediatrics 2009, 124, e172–e179. [Google Scholar] [CrossRef]
- Kawashima, T.; Hayashi, K.; Kosaka, A.; Kawashima, M.; Igarashi, T.; Tsutsui, H.; Tsuji, N.M.; Nishimura, I.; Hayashi, T.; Obata, A. Lactobacillus plantarum strain YU from fermented foods activates Th1 and protective immune responses. Int. Immunopharmacol. 2011, 11, 2017–2024. [Google Scholar] [CrossRef] [PubMed]
- Maeda, N.; Nakamura, R.; Hirose, Y.; Murosaki, S.; Yamamoto, Y.; Kase, T.; Yoshikai, Y. Oral administration of heat-killed Lactobacillus plantarum L-137 enhances protection against influenza virus infection by stimulation of type I interferon production in mice. Int. Immunopharmacol. 2009, 9, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Olivares, M.; Díaz-Ropero, M.P.; Sierra, S.; Lara-Villoslada, F.; Fonollá, J.; Navas, M.; Rodríguez, J.M.; Xaus, J. Oral intake of Lactobacillus fermentum CECT5716 enhances the effects of influenza vaccination. Nutrition 2007, 23, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Boge, T.; Rémigy, M.; Vaudaine, S.; Tanguy, J.; Bourdet-Sicard, R.; van der Werf, S. A probiotic fermented dairy drink improves antibody response to influenza vaccination in the elderly in two randomised controlled trials. Vaccine 2009, 27, 5677–5684. [Google Scholar] [CrossRef]
- Singh, K.; Rao, A. Probiotics: A potential immunomodulator in COVID-19 infection management. Nutr. Res. 2021, 87, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.J.; Ku, K.B.; Kim, H.S.; Moon, H.W.; Jeong, G.U.; Hwang, I.; Yoon, G.Y.; Lee, S.; Lee, S.; Ahn, D.G.; et al. Receptor-binding domain of SARS-CoV-2 spike protein efficiently inhibits SARS-CoV-2 infection and attachment to mouse lung. Int. J. Biol. Sci. 2021, 17, 3786–3794. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Anwar, F.; Altayb, H.N.; Al-Abbasi, F.A.; Al-Malki, A.L.; Kamal, M.A.; Kumar, V. Antiviral effects of probiotic metabolites on COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 4175–4184. [Google Scholar] [CrossRef]
- Bacha, U.; Barrila, J.; Velazquez-Campoy, A.; Leavitt, S.A.; Freire, E. Identification of Novel Inhibitors of the SARS Coronavirus Main Protease 3CLpro. Biochemistry 2004, 43, 4906–4912. [Google Scholar] [CrossRef] [PubMed]
- Balmeh, N.; Mahmoudi, S.; Allahyari Fard, N. Manipulated bio antimicrobial peptides from probiotic bacteria as proposed drugs for COVID-19 disease. Inform. Med. Unlocked 2021, 23, 100515. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Macchiagodena, M.; Pagliai, M.; Procacci, P. Identification of potential binders of the main protease 3CLpro of the COVID-19 via structure-based ligand design and molecular modeling. Chem Phys Lett 2020, 750, 137489. [Google Scholar] [CrossRef]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef]
- Newman, J.A.; Douangamath, A.; Yadzani, S.; Yosaatmadja, Y.; Aimon, A.; Brandão-Neto, J.; Dunnett, L.; Gorrie-Stone, T.; Skyner, R.; Fearon, D.; et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021, 12, 4848. [Google Scholar] [CrossRef]
- Yazdi, A.K.; Pakarian, P.; Perveen, S.; Hajian, T.; Santhakumar, V.; Bolotokova, A.; Li, F.; Vedadi, M. Kinetic Characterization of SARS-CoV-2 nsp13 ATPase Activity and Discovery of Small-Molecule Inhibitors. ACS Infect. Dis. 2022, 8, 1533–1542. [Google Scholar] [CrossRef]
- Sui, C.; Xiao, T.; Zhang, S.; Zeng, H.; Zheng, Y.; Liu, B.; Xu, G.; Gao, C.; Zhang, Z. SARS-CoV-2 NSP13 Inhibits Type I IFN Production by Degradation of TBK1 via p62-Dependent Selective Autophagy. J. Immunol. 2022, 208, 753–761. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019, 10, 2342. [Google Scholar] [CrossRef]
- Ahn, D.G.; Choi, J.K.; Taylor, D.R.; Oh, J.W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012, 157, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- Naydenova, K.; Muir, K.W.; Wu, L.F.; Zhang, Z.; Coscia, F.; Peet, M.J.; Castro-Hartmann, P.; Qian, P.; Sader, K.; Dent, K.; et al. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. USA 2021, 118, e2021946118. [Google Scholar] [CrossRef] [PubMed]
- Gautier, T.; David-Le Gall, S.; Sweidan, A.; Tamanai-Shacoori, Z.; Jolivet-Gougeon, A.; Loréal, O.; Bousarghin, L. Next-Generation Probiotics and Their Metabolites in COVID-19. Microorganisms 2021, 9, 941. [Google Scholar] [CrossRef]
- Nguyen, Q.V.; Chong, L.C.; Hor, Y.-Y.; Lew, L.-C.; Rather, I.A.; Choi, S.-B. Role of Probiotics in the Management of COVID-19: A Computational Perspective. Nutrients 2022, 14, 274. [Google Scholar] [CrossRef]
- Rather, I.A.; Choi, S.-B.; Kamli, M.R.; Hakeem, K.R.; Sabir, J.S.M.; Park, Y.-H.; Hor, Y.-Y. Potential Adjuvant Therapeutic Effect of Lactobacillus plantarum Probio-88 Postbiotics against SARS-COV-2. Vaccines 2021, 9, 1067. [Google Scholar] [CrossRef] [PubMed]
- Erol, I.; Kotil, S.E.; Fidan, O.; Yetiman, A.E.; Durdagi, S.; Ortakci, F. In Silico Analysis of Bacteriocins from Lactic Acid Bacteria against SARS-CoV-2. Probiotics Antimicrob. Proteins 2023, 15, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef]
- Souza, P.F.N.; vanTilburg, M.F.; Mesquita, F.P.; Amaral, J.L.; Lima, L.B.; Montenegro, R.C.; Lopes, F.E.S.; Martins, R.X.; Vieira, L.; Farias, D.F.; et al. Neutralizing Effect of Synthetic Peptides toward SARS-CoV-2. ACS Omega 2022, 7, 16222–16234. [Google Scholar] [CrossRef]
- Chen, J.; Li, S.; Lei, Z.; Tang, Q.; Mo, L.; Zhao, X.; Xie, F.; Zi, D.; Tan, J. Inhibition of SARS-CoV-2 pseudovirus invasion by ACE2 protecting and Spike neutralizing peptides: An alternative approach to COVID19 prevention and therapy. Int. J. Biol. Sci. 2021, 17, 2957–2969. [Google Scholar] [CrossRef]
- Mahmud, S.; Biswas, S.; Kumar Paul, G.; Mita, M.A.; Afrose, S.; Robiul Hasan, M.; Sharmin Sultana Shimu, M.; Uddin, M.A.R.; Salah Uddin, M.; Zaman, S.; et al. Antiviral peptides against the main protease of SARS-CoV-2: A molecular docking and dynamics study. Arab. J. Chem. 2021, 14, 103315. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, X.; Li, B.; Lu, C.; Yang, S.; Ling, J.; Chen, H.; Huang, J.; He, B. A database of anti-coronavirus peptides. Sci. Data 2022, 9, 294. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.; Chowdhury, T.; Chakraborty, R.; Mandal, S.M. Probiotics-Derived Peptides and Their Immunomodulatory Molecules Can Play a Preventive Role against Viral Diseases Including COVID-19. Probiotics Antimicrob. Proteins 2021, 13, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Moens, F.; Van den Abbeele, P.; Basit, A.W.; Dodoo, C.; Chatterjee, R.; Smith, B.; Gaisford, S. A four-strain probiotic exerts positive immunomodulatory effects by enhancing colonic butyrate production in vitro. Int. J. Pharm. 2019, 555, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.T.; Cresci, G.A.M. The Immunomodulatory Functions of Butyrate. J. Inflamm. Res. 2021, 14, 6025–6041. [Google Scholar] [CrossRef]
- Chakraborty, K.; Raundhal, M.; Chen, B.B.; Morse, C.; Tyurina, Y.Y.; Khare, A.; Oriss, T.B.; Huff, R.; Lee, J.S.; St Croix, C.M.; et al. The mito-DAMP cardiolipin blocks IL-10 production causing persistent inflammation during bacterial pneumonia. Nat. Commun. 2017, 8, 13944. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Pattaroni, C.; Lopez-Mejia, I.C.; Riva, E.; Pernot, J.; Ubags, N.; Fajas, L.; Nicod, L.P.; Marsland, B.J. Dietary Fiber Confers Protection against Flu by Shaping Ly6c− Patrolling Monocyte Hematopoiesis and CD8+ T Cell Metabolism. Immunity 2018, 48, 992–1005. [Google Scholar] [CrossRef]
- Li, J.; Richards, E.M.; Handberg, E.M.; Pepine, C.J.; Raizada, M.K. Butyrate Regulates COVID-19–Relevant Genes in Gut Epithelial Organoids From Normotensive Rats. Hypertension 2021, 77, e13–e16. [Google Scholar] [CrossRef]
- de Vrese, M.; Winkler, P.; Rautenberg, P.; Harder, T.; Noah, C.; Laue, C.; Ott, S.; Hampe, J.; Schreiber, S.; Heller, K.; et al. Effect of Lactobacillus gasseri PA 16/8, Bifidobacterium longum SP 07/3, B. bifidum MF 20/5 on common cold episodes: A double blind, randomized, controlled trial. Clin. Nutr. 2005, 24, 481–491. [Google Scholar] [CrossRef]
- Brown, R.L.; Sequeira, R.P.; Clarke, T.B. The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun 2017, 8, 1512. [Google Scholar] [CrossRef]
- Berggren, A.; Lazou Ahrén, I.; Larsson, N.; Önning, G. Randomised, double-blind and placebo-controlled study using new probiotic lactobacilli for strengthening the body immune defence against viral infections. Eur. J. Nutr. 2011, 50, 203–210. [Google Scholar] [CrossRef]
- Lenoir-Wijnkoop, I.; Merenstein, D.; Korchagina, D.; Broholm, C.; Sanders, M.; Tancredi, D. Probiotics reduce health care cost and societal impact of flu-like respiratory tract infections in the USA: An economic modeling study. Front. Pharmacol. 2019, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- The Clinic ¿Cuánto Cuesta Enfermarse de COVID? Available online: https://www.litoralpress.cl/sitio/MediosOnline_Detalles.cshtml?lpkey=.B.O/.N.%C3%9Cm.N.V5.%C3%9C.B/h.C.J1e0.Rn.Y/.Bb.Rp.C.%C3%9Czogn5/s.Redh.Xp.O.Q.%C3%96 (accessed on 30 October 2022).
- Presupuesto del Trabajo y la Recuperación. Available online: https://www.gob.cl/presupuesto2021/ (accessed on 30 October 2022).
- Ceccarelli, G.; Borrazzo, C.; Pinacchio, C.; Santinelli, L.; Innocenti, G.P.; Cavallari, E.N.; Celani, L.; Marazzato, M.; Alessandri, F.; Ruberto, F.; et al. Oral bacteriotherapy in patients with COVID-19: A Retrospective Cohort Study. Front. Nutr. 2021, 7, 613928. [Google Scholar] [CrossRef] [PubMed]
- García, A.; Henríquez, P.; Retamal, C.; Pineda, S.; Delgado Sen, C.; González, C. Propiedades probióticas de Lactobacillus spp aislados de biopsias gástricas de pacientes con y sin infección por Helicobacter pylori. Rev. Med. Chile 2009, 137, 369–376. [Google Scholar] [CrossRef]
- Sanhueza, E.; Paredes, E.; González, C.; García, A. Effect of pH in the survival of Lactobacillus salivarius strain UCO-979C wild type and the pH acid acclimated variant. Electron. J. Biotechnol. 2015, 18, 343–346. [Google Scholar] [CrossRef]
- Merino, J.; García, A.; Pastene, E.; Salas, A.; Saez, K.; González, C. Lactobacillus fermentum UCO-979C strongly inhibited Helicobacter pylori SS1 in Meriones unguiculatus. Benef. Microbes 2018, 9, 625–627. [Google Scholar] [CrossRef]
- García, A.; Navarro, K.; Sanhueza, E.; Pineda, S.; Pastene, E.; Quezada, M.; Henríquez, K.; Karlyshev, A.; Villena, J.; González, C. Characterization of Lactobacillus fermentum UCO-979C, a probiotic strain with a potent anti-Helicobacter pylori activity. Electron. J. Biotechnol. 2017, 25, 75–83. [Google Scholar] [CrossRef]
- Garcia-Castillo, V.; Zelaya, H.; Ilabaca, A.; Espinoza-Monje, M.; Komatsu, R.; Albarracin, L.; Kitazawa, H.; García, A.; Villena, J. Lactobacillus fermentum UCO-979C beneficially modulates the innate immune response triggered by Helicobacter pylori infection. Benef. Microbes 2018, 9, 829–841. [Google Scholar] [CrossRef] [PubMed]
- García-Castillo, V.; Komatsu, R.; Clua, P.; Indo, Y.; Takagi, M.; Salva, S.; Islam, M.A.; Alvarez, S.; Takahashi, H.; Garcia-Cancino, A.; et al. Evaluation of the immunomodulatory activities of the probiotic strain Lactobacillus fermentum UCO-979C. Front. Immunol. 2019, 10, 1376. [Google Scholar] [CrossRef]
- Garcia-Castillo, V.; Marín-Vega, A.M.; Ilabaca, A.; Albarracín, L.; Marcial, G.; Kitazawa, H.; Garcia-Cancino, A.; Villena, J. Characterization of the immunomodulatory and anti-Helicobacter pylori properties of the human gastric isolate Lactobacillus rhamnosus UCO-25A. Biofouling 2019, 35, 922–937. [Google Scholar] [CrossRef]
- Salas-Jara, M.J.; Sanhueza, E.; Retamal-Díaz, A.; González, C.; Urrutia, H.; García, A. Probiotic Lactobacillus fermentum UCO-979C biofilm formation on AGS and Caco-2 cells and Helicobacter pylori inhibition. Biofouling 2016, 32, 1245–1257. [Google Scholar] [CrossRef]
- García-Castillo, V.; Marcial, G.; Albarracín, L.; Tomokiyo, M.; Clua, P.; Takahashi, H.; Kitazawa, H.; Garcia-Cancino, A.; Villena, J. The exopolysaccharide of Lactobacillus fermentum UCO-979C is partially involved in its immunomodulatory effect and its ability to improve the resistance against Helicobacter pylori infection. Microorganisms 2020, 8, 479. [Google Scholar] [CrossRef]
- Carrión, P.; Corral, M.C.; Victoriano, M.; Gutiérrez-Zamorano, C.; Sánchez-Alonso, K.; Parra-Sepúlveda, C.; Saez, K.; García-Cancino, A. Efecto del consumo del probiótico Lactobacillus fermentum UCO-979C en peso, masa grasa, masa magra, circunferencia de la cintura y hábitos alimenticios. Rev. Esp. Nutr. Comunitaria 2021, 27, 49–54. [Google Scholar] [CrossRef]
- Vital, M.; Howe, A.C.; Tiedje, J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 2014, 5, e00889. [Google Scholar] [CrossRef] [PubMed]
- McClean, S.; Healy, M.E.; Collins, C.; Carberry, S.; O’Shaughnessy, L.; Dennehy, R.; Adams, Á.; Kennelly, H.; Corbett, J.M.; Carty, F.; et al. Linocin and OmpW Are Involved in Attachment of the Cystic Fibrosis-Associated Pathogen Burkholderia cepacia Complex to Lung Epithelial Cells and Protect Mice against Infection. Infect. Immun. 2016, 84, 1424–1437. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.B.; Guo, C.J.; Violante, S.; et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2020, 581, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Hagan, T.; Cortese, M.; Rouphael, N.; Boudreau, C.; Linde, C.; Maddur, M.S.; Das, J.; Wang, H.; Guthmiller, J.; Zheng, N.Y.; et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell 2019, 178, 1313–1328. [Google Scholar] [CrossRef]
- Bhowmik, S.; Chiu, H.P.; Jones, D.H.; Chiu, H.J.; Miller, M.D.; Xu, Q.; Farr, C.L.; Ridlon, J.M.; Wells, J.E.; Elsliger, M.A.; et al. Structure and functional characterization of a bile acid 7α dehydratase BaiE in secondary bile acid synthesis. Proteins 2016, 84, 316–331. [Google Scholar] [CrossRef]
- Shiraki, M.; Endo, T.; Saito, T. Fermentative production of (R)-(−)-3-hydroxybutyrate using 3-hydroxybutyrate dehydrogenase null mutant of Ralstonia eutropha and recombinant Escherichia coli. J. Biosci. Bioeng. 2006, 102, 529–534. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef]
| Probiotic | Function of the Probiotic | Reference |
|---|---|---|
| Lactobacillus casei DN-114 001; fermented beverage DanActive/Actimel, Danone. | Reduces the incidence and duration of respiratory tract infections. | [62] |
| Lactobacillus gasseri PA 16/8, Bifidobacterium longum SP 07/3, and B. bifidum MF 20/5; TriBion Harmonis, Merck. | Reduces the duration and acuteness of influenza-type infections. | [63] |
| Lactobacillus rhamnosus GG; Culturelle or other brands. | For healthy digestion and a strong intestinal barrier. Anticipation of a respiratory tract viral infection. | [64] |
| Lactobacillus plantarum DR7. | Immunologic stimulation reduces infections of the superior respiratory tract. | [65] |
| Bifidobacterium breve Yakult and Lactobacillus casei Shirota; available as fermented beverages. | Reduces the prevalence of ventilator-associated pneumonia. The oral administration of L. casei Shirota to neonatal and infant mice showed that a boost of the immature immune system can play a positive role in the protection against influenza virus infection. | [4,50,66] |
| Bifidobacterium longum BB536; Morinaga, sold in many formulations. | Stimulates the innate immune response and protects against influenza. | [67] |
| Lactobacillus rhamnosus CRL1505. | Administered in yogurt, it improves mucosal immunity and reduces the incidence as well as severity of intestinal and respiratory viral infections. | [26] |
| Lactobacillus acidophilus NCFM. | Reduces the occurrence of influenza-like symptoms. | [4,68] |
| Lactobacillus plantarum YU, Lactobacillus plantarum L-137. | Stimulates the immune system of a host by lactic acid bacteria (LAB) against the influenza virus H1N1 and provides protective effects against some respiratory virus infections in mouse models and humans. | [4,69,70] |
| Lactobacillus fermentum CECT5716, Lactobacillus casei DN-114 001. | Probiotics have enhanced the effects of the influenza virus vaccine. | [4,71,72] |
| Lactobacillus rhamnosus GG, Lactobacillus casei (including the Shirota strain), Lactobacillus plantarum, Bifidobacterium lactis Bb-12, and various strains of Bifidobacterium longum. | Significant reduction in the prevalence of upper respiratory infections and flu-related symptoms. | [4,73] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdebenito-Navarrete, H.; Fuentes-Barrera, V.; Smith, C.T.; Salas-Burgos, A.; Zuniga, F.A.; Gomez, L.A.; García-Cancino, A. Can Probiotics, Particularly Limosilactobacillus fermentum UCO-979C and Lacticaseibacillus rhamnosus UCO-25A, Be Preventive Alternatives against SARS-CoV-2? Biology 2023, 12, 384. https://doi.org/10.3390/biology12030384
Valdebenito-Navarrete H, Fuentes-Barrera V, Smith CT, Salas-Burgos A, Zuniga FA, Gomez LA, García-Cancino A. Can Probiotics, Particularly Limosilactobacillus fermentum UCO-979C and Lacticaseibacillus rhamnosus UCO-25A, Be Preventive Alternatives against SARS-CoV-2? Biology. 2023; 12(3):384. https://doi.org/10.3390/biology12030384
Chicago/Turabian StyleValdebenito-Navarrete, Héctor, Victor Fuentes-Barrera, Carlos T. Smith, Alexis Salas-Burgos, Felipe A. Zuniga, Leonardo A. Gomez, and Apolinaria García-Cancino. 2023. "Can Probiotics, Particularly Limosilactobacillus fermentum UCO-979C and Lacticaseibacillus rhamnosus UCO-25A, Be Preventive Alternatives against SARS-CoV-2?" Biology 12, no. 3: 384. https://doi.org/10.3390/biology12030384
APA StyleValdebenito-Navarrete, H., Fuentes-Barrera, V., Smith, C. T., Salas-Burgos, A., Zuniga, F. A., Gomez, L. A., & García-Cancino, A. (2023). Can Probiotics, Particularly Limosilactobacillus fermentum UCO-979C and Lacticaseibacillus rhamnosus UCO-25A, Be Preventive Alternatives against SARS-CoV-2? Biology, 12(3), 384. https://doi.org/10.3390/biology12030384







