Distinct Molecular Patterns of Two-Component Signal Transduction Systems in Thermophilic Cyanobacteria as Revealed by Genomic Identification
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Collection of Thermophilic Cyanobacteria
2.2. Identification of TCS Genes
2.3. Protein Sequences Analysis
2.4. Identification of Orthologous Proteins
2.5. Phylogeny
3. Results and Discussion
3.1. Composition of TCS Genes in Thermophilic Cyanobacteria
3.2. Genetic Organization of TCS Genes in Thermophilic Cyanobacteria
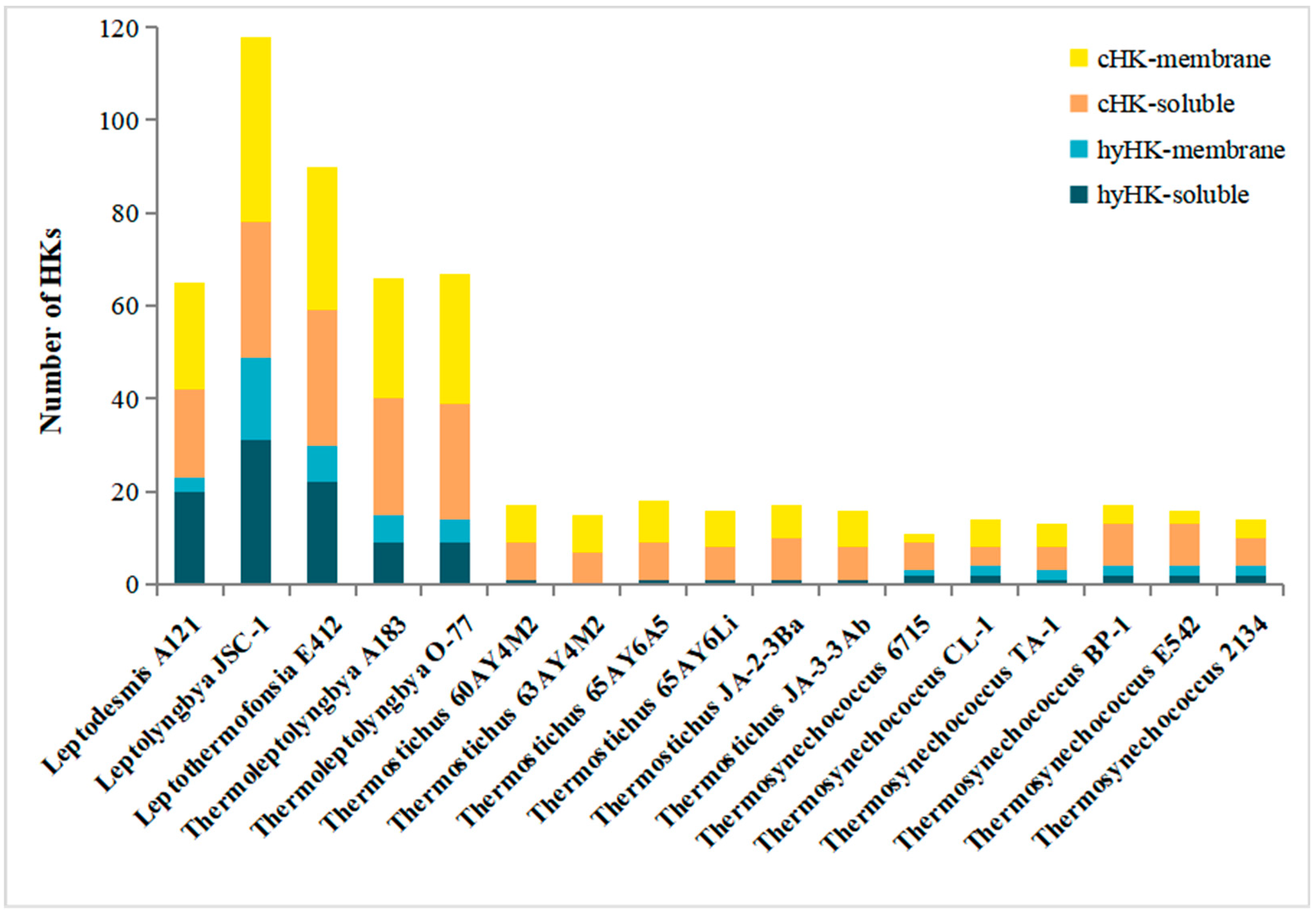
3.3. Cellular Localization of HK in Thermophilic Cyanobacteria
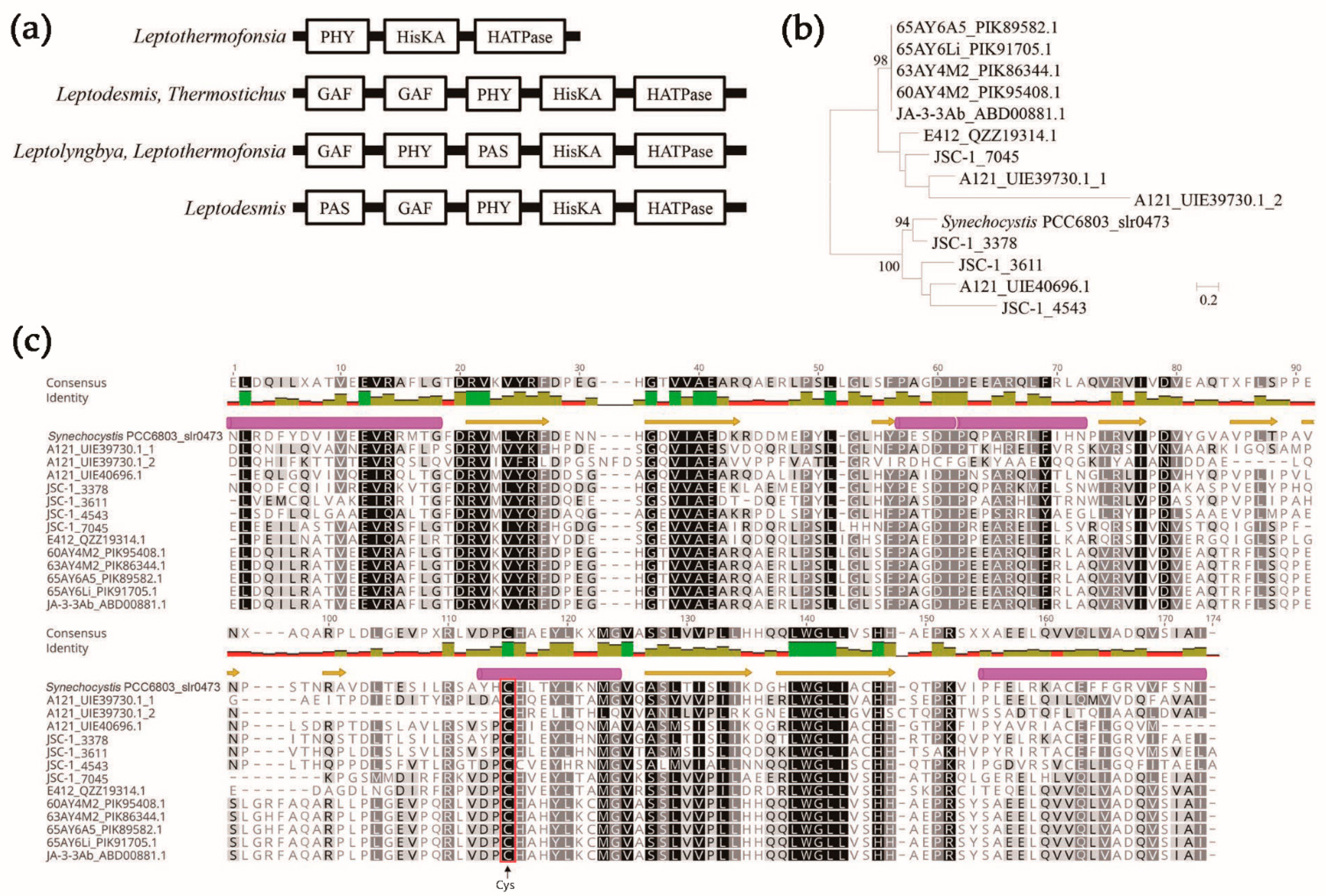
3.4. Sensing-Domain Architecture of HKs in Thermophilic Cyanobacteria
3.5. Domain Architecture of HKs in Thermophilic Cyanobacteria
3.6. Domain Architecture of RRs in Thermophilic Cyanobacteria
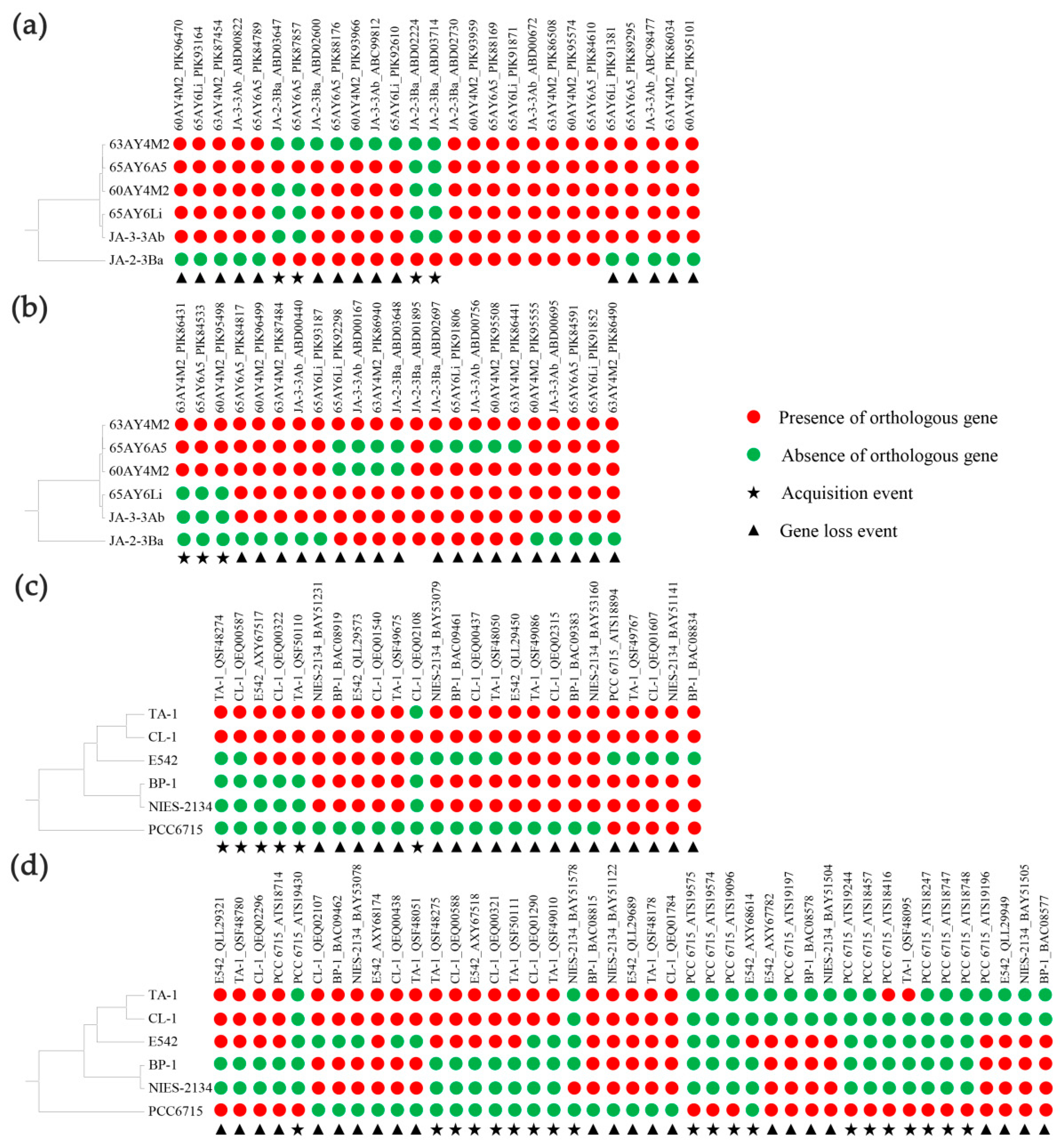
3.7. Evolutionary Conserved and Accessory TCS in Thermostichus and Thermosynechococcus
3.8. Evolutionary Origin of Accessory TCS in Thermostichus and Thermosynechococcus
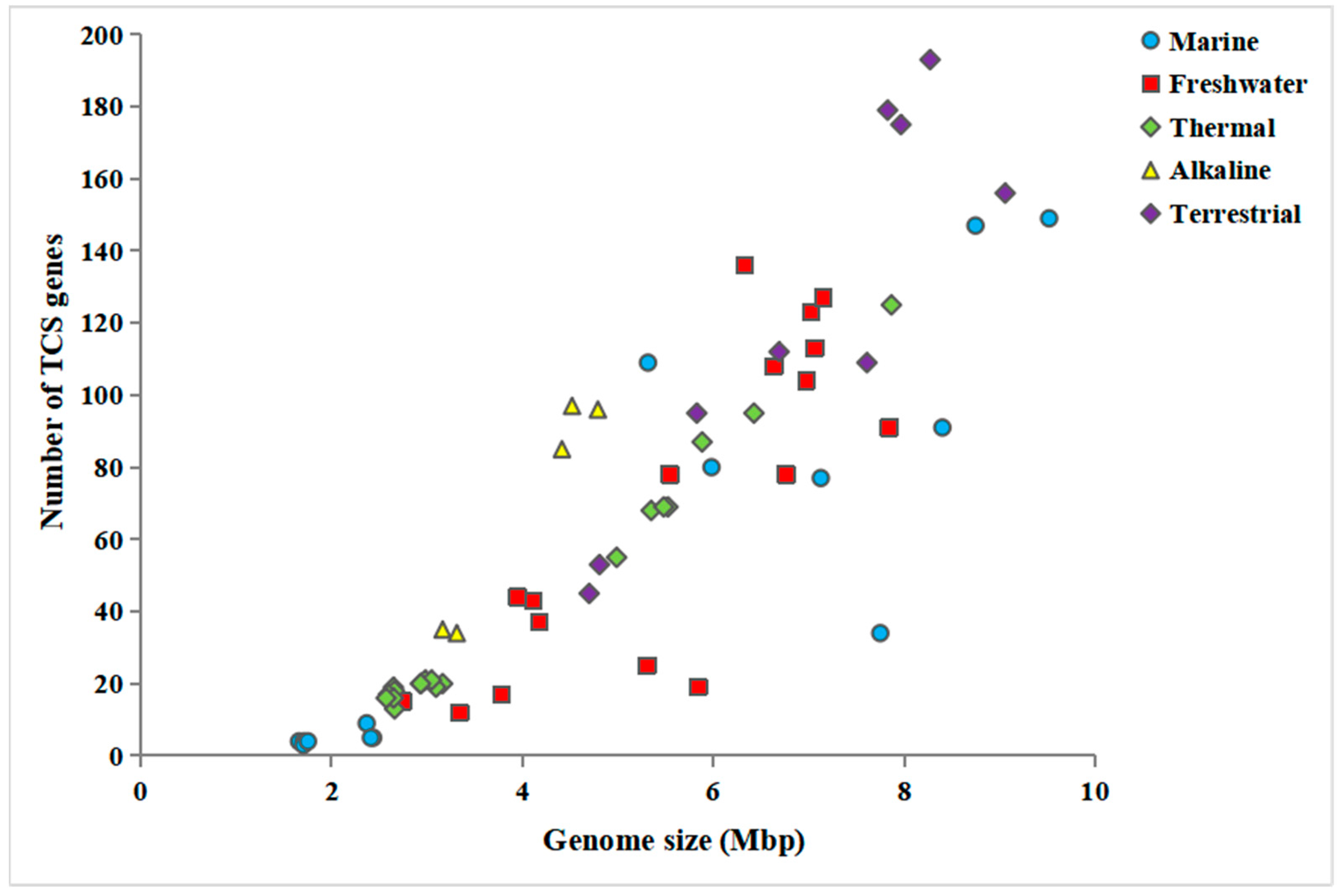
3.9. Functional and Comparative Analysis of TCS between Thermophilic and Mesophilic Cyanobacteria
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tang, J.; Liang, Y.; Jiang, D.; Li, L.; Luo, Y.; Shah, M.R.; Daroch, M. Temperature-controlled thermophilic bacterial communities in hot springs of western Sichuan, China. BMC Microbiol. 2018, 18, 134. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Du, L.-M.; Liang, Y.-M.; Daroch, M. Complete Genome Sequence and Comparative Analysis of Synechococcus sp. CS-601 (SynAce01), a Cold-Adapted Cyanobacterium from an Oligotrophic Antarctic Habitat. Int. J. Mol. Sci. 2019, 20, 152. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-Y.; Teng, W.-K.; Zhao, L.; Hu, C.-X.; Zhou, Y.-K.; Han, B.-P.; Song, L.-R.; Shu, W.-S. Comparative genomics reveals insights into cyanobacterial evolution and habitat adaptation. ISME J. 2020, 15, 211–227. [Google Scholar] [CrossRef] [PubMed]
- Kolber, Z.S.; Van Dover, C.L.; Niederman, R.A.; Falkowski, P.G. Bacterial photosynthesis in surface waters of the open ocean. Nature 2000, 407, 177–179. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Matsakas, L.; Rova, U.; Christakopoulos, P. A perspective on biotechnological applications of thermophilic microalgae and cyanobacteria. Bioresour. Technol. 2019, 278, 424–434. [Google Scholar] [CrossRef]
- Ashby, M.K.; Houmard, J. Cyanobacterial Two-Component Proteins: Structure, Diversity, Distribution, andEvolution. Microbiol. Mol. Biol. Rev. 2006, 70, 472–509. [Google Scholar] [CrossRef]
- Rai, R.; Rai, K.K.; Singh, S.; Raj, A.; Rai, L.C. Stress proteins and signal transduction in cyanobacteria. In Ecophysiology and Biochemistry of Cyanobacteria; Rastogi, R.P., Ed.; Springer Nature: Singapore, 2021; pp. 155–180. [Google Scholar]
- Rachedi, R.; Foglino, M.; Latifi, A. Stress Signaling in Cyanobacteria: A Mechanistic Overview. Life 2020, 10, 312. [Google Scholar] [CrossRef]
- Xu, W.; Wang, Y. Sequences, Domain Architectures, and Biological Functions of the Serine/Threonine and Histidine Kinases in Synechocystis sp. PCC 6803. Appl. Biochem. Biotechnol. 2019, 188, 1022–1065. [Google Scholar] [CrossRef]
- Liu, Z.; Li, H.; Wei, Y.; Chu, W.; Chong, Y.; Long, X.; Qin, S.; Shao, H. Signal transduction pathways in Synechocystis sp. PCC 6803 and biotechnological implications under abiotic stress. Crit. Rev. Biotechnol. 2013, 35, 269–280. [Google Scholar] [CrossRef]
- Ishii, E.; Eguchi, Y. Diversity in Sensing and Signaling of Bacterial Sensor Histidine Kinases. Biomolecules 2021, 11, 1524. [Google Scholar] [CrossRef]
- Pei, G.; Niu, X.; Zhou, Y.; Chen, L.; Zhang, W. Crosstalk of two-component signal transduction systems in regulating central carbohydrate and energy metabolism during autotrophic and photomixotrophic growth of Synechocystis sp. PCC 6803. Integr. Biol. 2017, 9, 485–496. [Google Scholar] [CrossRef]
- Ortet, P.; Whitworth, D.E.; Santaella, C.; Achouak, W.; Barakat, M. P2CS: Updates of the prokaryotic two-component systems database. Nucleic Acids Res. 2014, 43, D536–D541. [Google Scholar] [CrossRef]
- Gushchin, I.; Aleksenko, V.; Orekhov, P.; Goncharov, I.; Nazarenko, V.; Semenov, O.; Remeeva, A.; Gordeliy, V. Nitrate- and Nitrite-Sensing Histidine Kinases: Function, Structure, and Natural Diversity. Int. J. Mol. Sci. 2021, 22, 5933. [Google Scholar] [CrossRef]
- Bellieny-Rabelo, D.; Pretorius, W.J.S.; Moleleki, L.N. Novel Two-Component System-Like Elements Reveal Functional Domains Associated with Restriction–Modification Systems and paraMORC ATPases in Bacteria. Genome Biol. Evol. 2021, 13, evab024. [Google Scholar] [CrossRef]
- Alcorta, J.; Alarcón-Schumacher, T.; Salgado, O.; Díez, B. Taxonomic Novelty and Distinctive Genomic Features of Hot Spring Cyanobacteria. Front. Genet. 2020, 11, 568223. [Google Scholar] [CrossRef]
- Walter, J.M.; Coutinho, F.H.; Dutilh, B.E.; Swings, J.; Thompson, F.L.; Thompson, C.C. Ecogenomics and Taxonomy of Cyanobacteria Phylum. Front. Microbiol. 2017, 8, 2132. [Google Scholar] [CrossRef]
- Hossain, G.S.; Saini, M.; Miyake, R.; Ling, H.; Chang, M.W. Genetic Biosensor Design for Natural Product Biosynthesis in Microorganisms. Trends Biotechnol. 2020, 38, 797–810. [Google Scholar] [CrossRef]
- Shi, M.; Chen, L.; Zhang, W. Regulatory Diversity and Functional Analysis of Two-Component Systems in Cyanobacterium Synechocystis sp. PCC 6803 by GC-MS Based Metabolomics. Front. Microbiol. 2020, 11, 403. [Google Scholar] [CrossRef]
- Zhou, P.; Wang, L.; Liu, H.; Li, C.; Li, Z.; Wang, J.; Tan, X. CyanoOmicsDB: An integrated omics database for functional genomic analysis of cyanobacteria. Nucleic Acids Res. 2021, 50, D758–D764. [Google Scholar] [CrossRef]
- Zschiedrich, C.P.; Keidel, V.; Szurmant, H. Molecular Mechanisms of Two-Component Signal Transduction. J. Mol. Biol. 2016, 428, 3752–3775. [Google Scholar] [CrossRef]
- Tang, J.; Zhou, H.; Yao, D.; Riaz, S.; You, D.; Klepacz-Smółka, A.; Daroch, M. Comparative Genomic Analysis Revealed Distinct Molecular Components and Organization of CO2-Concentrating Mechanism in Thermophilic Cyanobacteria. Front. Microbiol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Du, L.-M.; Li, M.; Yao, D.; Jiang, Y.; Waleron, M.; Waleron, K.; Daroch, M. Characterization of a Novel Hot-Spring Cyanobacterium Leptodesmis sichuanensis sp. Nov. and Genomic Insights of Molecular Adaptations Into Its Habitat. Front. Microbiol. 2022, 12, 3752–3775. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.I.; Bryant, D.A.; Casamatta, D.; Thomas-Keprta, K.L.; Sarkisova, S.A.; Shen, G.; Graham, J.E.; Boyd, E.S.; Peters, J.W.; Garrison, D.H.; et al. Polyphasic Characterization of a Thermotolerant Siderophilic Filamentous Cyanobacterium That Produces Intracellular Iron Deposits. Appl. Environ. Microbiol. 2010, 76, 6664–6672. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Shah, M.R.; Yao, D.; Jiang, Y.; Du, L.; Zhao, K.; Li, L.; Li, M.; Waleron, M.M.; Waleron, M.; et al. Polyphasic Identification and Genomic Insights of Leptothermofonsia sichuanensis gen. sp. nov., a Novel Thermophilic Cyanobacteria Within Leptolyngbyaceae. Front. Microbiol. 2022, 13, 765105. [Google Scholar] [CrossRef]
- Yoon, K.-S.; Nguyen, N.T.; Tran, K.T.; Tsuji, K.; Ogo, S. Nitrogen Fixation Genes and Nitrogenase Activity of the Non-Heterocystous Cyanobacterium Thermoleptolyngbya sp. O-77. Microbes Environ. 2017, 32, 324–329. [Google Scholar] [CrossRef]
- Tang, J.; Li, L.; Li, M.; Du, L.; Shah, M.R.; Waleron, M.M.; Waleron, M.; Waleron, K.F.; Daroch, M. Description, Taxonomy, and Comparative Genomics of a Novel species, Thermoleptolyngbya sichuanensis sp. nov., Isolated From Hot Springs of Ganzi, Sichuan, China. Front. Microbiol. 2021, 12, 696102. [Google Scholar] [CrossRef]
- Olsen, M.T.; Nowack, S.; Wood, J.; Becraft, E.D.; LaButti, K.; Lipzen, A.; Martin, J.; Schackwitz, W.S.; Rusch, D.B.; Cohan, F.M.; et al. The molecular dimension of microbial species: 3. Comparative genomics of Synechococcus strains with different light responses and in situ diel transcription patterns of associated putative ecotypes in the Mushroom Spring microbial mat. Front. Microbiol. 2015, 6, 604. [Google Scholar] [CrossRef]
- Bhaya, D.; Grossman, A.R.; Steunou, A.-S.; Khuri, N.; Cohan, F.M.; Hamamura, N.; Melendrez, M.; Bateson, M.M.; Ward, D.M.; Heidelberg, J. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J. 2007, 1, 703–713. [Google Scholar] [CrossRef]
- Cheng, Y.-I.; Chou, L.; Chiu, Y.-F.; Hsueh, H.-T.; Kuo, C.-H.; Chu, H.-A. Comparative Genomic Analysis of a Novel Strain of Taiwan Hot-Spring Cyanobacterium Thermosynechococcus sp. CL-1. Front. Microbiol. 2020, 11, 82. [Google Scholar] [CrossRef]
- Leu, J.-Y.; Lin, T.-H.; Selvamani, M.J.P.; Chen, H.-C.; Liang, J.-Z.; Pan, K.-M. Characterization of a novel thermophilic cyanobacterial strain from Taian hot springs in Taiwan for high CO2 mitigation and C-phycocyanin extraction. Process. Biochem. 2013, 48, 41–48. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kaneko, T.; Sato, S.; Ikeuchi, M.; Katoh, H.; Sasamoto, S.; Watanabe, A.; Iriguchi, M.; Kawashima, K.; Kimura, T.; et al. Complete Genome Structure of the Thermophilic Cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 2002, 9, 123–130. [Google Scholar] [CrossRef]
- Liang, Y.; Tang, J.; Luo, Y.; Kaczmarek, M.B.; Li, X.; Daroch, M. Thermosynechococcus as a thermophilic photosynthetic microbial cell factory for CO2 utilisation. Bioresour. Technol. 2019, 278, 255–265. [Google Scholar] [CrossRef]
- Liang, Y.; Kaczmarek, M.B.; Kasprzak, A.K.; Tang, J.; Shah, M.R.; Jin, P.; Klepacz-Smółka, A.; Cheng, J.; Ledakowicz, S.; Daroch, M. Thermosynechococcaceae as a source of thermostable C-phycocyanins: Properties and molecular insights. Algal Res. 2018, 35, 223–235. [Google Scholar] [CrossRef]
- Barakat, M.; Ortet, P.; E Whitworth, D. P2RP: A web-based framework for the identification and analysis of regulatory proteins in prokaryotic genomes. BMC Genom. 2013, 14, 269. [Google Scholar] [CrossRef]
- Borland, S.; Oudart, A.; Prigent-Combaret, C.; Brochier-Armanet, C.; Wisniewski-Dyé, F. Genome-wide survey of two-component signal transduction systems in the plant growth-promoting bacterium Azospirillum. BMC Genom. 2015, 16, 833. [Google Scholar] [CrossRef]
- Williams, R.H.; Whitworth, D.E. The genetic organisation of prokaryotic two-component system signalling pathways. BMC Genom. 2010, 11, 720. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Tang, J.; Yao, D.; Zhou, H.; Du, L.; Daroch, M. Reevaluation of Parasynechococcus-like Strains and Genomic Analysis of Their Microsatellites and Compound Microsatellites. Plants 2022, 11, 1060. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ 2016, 4, e1900v1901. [Google Scholar] [CrossRef]
- Mizuno, T. Compilation of All Genes Encoding Two-component Phosphotransfer Signal Transducers in the Genome of Escherichia coli. DNA Res. 1997, 4, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Hirao, K.; Oshima, T.; Aiba, H.; Utsumi, R.; Ishihama, A. Functional Characterization in Vitro of All Two-component Signal Transduction Systems from Escherichia coli. J. Biol. Chem. 2005, 280, 1448–1456. [Google Scholar] [CrossRef]
- Yao, D.; Cheng, L.; Du, L.; Li, M.; Daroch, M.; Tang, J. Genome-Wide Investigation and Analysis of Microsatellites and Compound Microsatellites in Leptolyngbya-like Species, Cyanobacteria. Life 2021, 11, 1258. [Google Scholar] [CrossRef]
- Wuichet, K.; Cantwell, B.J.; Zhulin, I.B. Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 2010, 13, 219–225. [Google Scholar] [CrossRef]
- Mascher, T. Bacterial (intramembrane-sensing) histidine kinases: Signal transfer rather than stimulus perception. Trends Microbiol. 2014, 22, 559–565. [Google Scholar] [CrossRef]
- Cock, P.; Whitworth, D.E. Evolution of Prokaryotic Two-Component System Signaling Pathways: Gene Fusions and Fissions. Mol. Biol. Evol. 2007, 24, 2355–2357. [Google Scholar] [CrossRef]
- Cheung, J.; Hendrickson, W.A. Sensor domains of two-component regulatory systems. Curr. Opin. Microbiol. 2010, 13, 116–123. [Google Scholar] [CrossRef]
- Jung, K.; Fried, L.; Behr, S.; Heermann, R. Histidine kinases and response regulators in networks. Curr. Opin. Microbiol. 2012, 15, 118–124. [Google Scholar] [CrossRef]
- Möglich, A.; Ayers, R.A.; Moffat, K. Structure and Signaling Mechanism of Per-ARNT-Sim Domains. Structure 2009, 17, 1282–1294. [Google Scholar] [CrossRef]
- Ho, Y.J.; Burden, L.M.; Hurley, J.H. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000, 19, 5288–5299. [Google Scholar] [CrossRef]
- Gao, R.; Stock, A.M. Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr. Opin. Microbiol. 2010, 13, 160–167. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Nikolskaya, A.N.; Koonin, E.V. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 2001, 203, 11–21. [Google Scholar] [CrossRef]
- Lamparter, T.; Mittmann, F.; Gärtner, W.; Börner, T.; Hartmann, E.; Hughes, J. Characterization of recombinant phytochrome from the cyanobacterium Synechocystis. Proc. Natl. Acad. Sci. USA 1997, 94, 11792–11797. [Google Scholar] [CrossRef]
- Sharrock, R.A. The phytochrome red/far-red photoreceptor superfamily. Genome Biol. 2008, 9, 230. [Google Scholar] [CrossRef]
- Gan, F.; Zhang, S.; Rockwell, N.C.; Martin, S.S.; Lagarias, J.C.; Bryant, D.A. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 2014, 345, 1312–1317. [Google Scholar] [CrossRef]
- Narikawa, R.; Ishizuka, T.; Muraki, N.; Shiba, T.; Kurisu, G.; Ikeuchi, M. Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. USA 2012, 110, 918–923. [Google Scholar] [CrossRef]
- Anders, K.; Essen, L.-O. The family of phytochrome-like photoreceptors: Diverse, complex and multi-colored, but very useful. Curr. Opin. Struct. Biol. 2015, 35, 7–16. [Google Scholar] [CrossRef]
- Fushimi, K.; Narikawa, R. Cyanobacteriochromes: Photoreceptors covering the entire UV-to-visible spectrum. Curr. Opin. Struct. Biol. 2019, 57, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Zorina, A.; Stepanchenko, N.; Novikova, G.V.; Sinetova, M.; Panichkin, V.B.; Moshkov, I.E.; Zinchenko, V.V.; Shestakov, S.V.; Suzuki, I.; Murata, N.; et al. Eukaryotic-like Ser/Thr Protein Kinases SpkC/F/K Are Involved in Phosphorylation of GroES in the Cyanobacterium Synechocystis. DNA Res. 2011, 18, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Salvado, B.; Vilaprinyo, E.; Sorribas, A.; Alves, R. A survey of HK, HPt, and RR domains and their organization in two-component systems and phosphorelay proteins of organisms with fully sequenced genomes. Peerj 2015, 3, e1183. [Google Scholar] [CrossRef] [PubMed]
- Lassak, J.; Bubendorfer, S.; Thormann, K.M. Domain Analysis of ArcS, the Hybrid Sensor Kinase of the Shewanella oneidensis MR-1 Arc Two-Component System, Reveals Functional Differentiation of Its Two Receiver Domains. J. Bacteriol. 2012, 195, 482–492. [Google Scholar] [CrossRef]
- Galperin, M.Y. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 2010, 13, 150–159. [Google Scholar] [CrossRef]
- Jenal, U.; Galperin, M.Y. Single domain response regulators: Molecular switches with emerging roles in cell organization and dynamics. Curr. Opin. Microbiol. 2009, 12, 152–160. [Google Scholar] [CrossRef]
- Whitworth, D.E.; Cock, P.J.A. Evolution of prokaryotic two-component systems: Insights from comparative genomics. Amino Acids 2009, 37, 459–466. [Google Scholar] [CrossRef]
- Kato, H.; Watanabe, S.; Nimura-Matsune, K.; Chibazakura, T.; Tozawa, Y.; Yoshikawa, H. Exploration of a Possible Partnership among Orphan Two-Component System Proteins in Cyanobacterium Synechococcus elongatus PCC 7942. Biosci. Biotechnol. Biochem. 2012, 76, 1484–1491. [Google Scholar] [CrossRef]
- Ibrahim, I.M.; Puthiyaveetil, S.; Allen, J.F. A Two-Component Regulatory System in Transcriptional Control of Photosystem Stoichiometry: Redox-Dependent and Sodium Ion-Dependent Phosphoryl Transfer from Cyanobacterial Histidine Kinase Hik2 to Response Regulators Rre1 and RppA. Front. Plant Sci. 2016, 7, 137. [Google Scholar] [CrossRef]
- Bairagi, N.; Watanabe, S.; Nimura-Matsune, K.; Tanaka, K.; Tsurumaki, T.; Nakanishi, S.; Tanaka, K. Conserved Two-component Hik2–Rre1 Signaling Is Activated Under Temperature Upshift and Plastoquinone-reducing Conditions in the Cyanobacterium Synechococcus elongatus PCC 7942. Plant Cell Physiol. 2021, 63, 176–188. [Google Scholar] [CrossRef]
- van Waasbergen, L.G.; Dolganov, N.; Grossman, A.R. nblS, a Gene Involved in Controlling Photosynthesis-Related Gene Expression during High Light and Nutrient Stress in Synechococcus elongatus PCC 7942. J. Bacteriol. 2002, 184, 2481–2490. [Google Scholar] [CrossRef]
- Kato, H.; Chibazakura, T.; Yoshikawa, H. NblR Is a Novel One-Component Response Regulator in the Cyanobacterium Synechococcus elongatus PCC 7942. Biosci. Biotechnol. Biochem. 2008, 72, 1072–1079. [Google Scholar] [CrossRef]
- Suzuki, S.; Ferjani, A.; Suzuki, I.; Murata, N. The SphS-SphR Two Component System Is the Exclusive Sensor for the Induction of Gene Expression in Response to Phosphate Limitation in Synechocystis. J. Biol. Chem. 2004, 279, 13234–13240. [Google Scholar] [CrossRef]
- Azuma, M.; Osanai, T.; Hirai, M.Y.; Tanaka, K. A Response Regulator Rre37 and an RNA Polymerase Sigma Factor SigE Represent Two Parallel Pathways to Activate Sugar Catabolism in a Cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2011, 52, 404–412. [Google Scholar] [CrossRef]
| Species | Genome Size (M) | No. of CDS | HK | RR | PP | ||||
|---|---|---|---|---|---|---|---|---|---|
| Classic | Hybrid | Unorthodox | CheA | HisKa | Hpt | ||||
| Leptodesmis sichuanensis A121 | 5.35 | 4917 | 42 | 23 | 1 | 2 | 61 | 1 | 1 |
| Leptolyngbya sp. JSC-1 | 7.87 | 7423 | 69 | 49 | 1 | 6 | 111 | 3 | 0 |
| Leptothermofonsia sichuanensis E412 | 6.43 | 5495 | 60 | 30 | 2 | 3 | 79 | 0 | 1 |
| Thermoleptolyngbya sichuanensis A183 | 5.53 | 4541 | 51 | 15 | 1 | 2 | 46 | 0 | 0 |
| Thermoleptolyngbya sp. O-77 | 5.48 | 4865 | 53 | 14 | 0 | 2 | 49 | 2 | 0 |
| Thermostichus sp. 60AY4M2 | 3.16 | 2622 | 16 | 1 | 1 | 2 | 29 | 2 | 0 |
| Thermostichus sp. 63AY4M2 | 3.09 | 2577 | 15 | 0 | 2 | 2 | 30 | 2 | 0 |
| Thermostichus sp. 65AY6A5 | 2.98 | 2597 | 17 | 1 | 1 | 2 | 28 | 2 | 0 |
| Thermostichus sp. 65AY6Li | 2.93 | 2632 | 15 | 1 | 2 | 2 | 29 | 2 | 0 |
| Thermostichus sp. JA-2-3Ba | 3.05 | 2862 | 16 | 1 | 2 | 2 | 28 | 2 | 0 |
| Thermostichus sp. JA-3-3Ab | 2.93 | 2760 | 15 | 1 | 2 | 2 | 29 | 2 | 0 |
| Thermosynechococcus lividus PCC 6715 | 2.66 | 2465 | 8 | 3 | 0 | 2 | 32 | 2 | 2 |
| Thermosynechococcus sp. CL-1 | 2.65 | 2549 | 13 | 4 | 0 | 2 | 26 | 1 | 1 |
| Thermosynechococcus sp. TA-1 | 2.66 | 2556 | 12 | 4 | 0 | 2 | 26 | 1 | 2 |
| Thermosynechococcus vestitus BP-1 | 2.59 | 2475 | 10 | 4 | 0 | 3 | 23 | 1 | 1 |
| Thermosynechococcus vestitus E542 | 2.65 | 2543 | 10 | 3 | 0 | 3 | 26 | 1 | 1 |
| Thermosynechococcus vulcanus NIES-2134 | 2.57 | 2496 | 10 | 4 | 0 | 2 | 24 | 1 | 1 |
| Species | Orphan | Pair | Triad | Tetrad | Pentad | Total |
|---|---|---|---|---|---|---|
| Leptodesmis A121 | 92 | 12 | 5 | 131 | ||
| Leptolyngbya JSC-1 | 137 | 35 | 9 | 1 | 239 | |
| Leptothermofonsia E412 | 120 | 17 | 4 | 1 | 1 | 175 |
| Thermoleptolyngbya A183 | 91 | 8 | 1 | 1 | 115 | |
| Thermoleptolyngbya O-77 | 84 | 14 | 1 | 1 | 120 | |
| Thermostichus 60AY4M2 | 46 | 1 | 1 | 51 | ||
| Thermostichus 63AY4M2 | 46 | 1 | 1 | 51 | ||
| Thermostichus 65AY6A5 | 46 | 1 | 1 | 51 | ||
| Thermostichus 65AY6Li | 44 | 2 | 1 | 51 | ||
| Thermostichus JA-2-3Ba | 39 | 3 | 2 | 51 | ||
| Thermostichus JA-3-3Ab | 42 | 3 | 1 | 51 | ||
| Thermosynechococcus PCC 6715 | 42 | 2 | 1 | 49 | ||
| Thermosynechococcus CL-1 | 33 | 7 | 47 | |||
| Thermosynechococcus TA-1 | 35 | 6 | 47 | |||
| Thermosynechococcus BP-1 | 32 | 5 | 42 | |||
| Thermosynechococcus E542 | 38 | 3 | 44 | |||
| Thermosynechococcus NIES-2134 | 36 | 3 | 42 | |||
| Cylindrospermum stagnale PCC 7417 | 107 | 27 | 3 | 1 | 174 | |
| Microcystis aeruginosa NIES-843 | 41 | 2 | 45 | |||
| Nostoc sp. PCC 7524 | 109 | 25 | 4 | 171 | ||
| Parasynechococcus sp. WH7803 | 19 | 2 | 1 | 26 | ||
| Synechocystis sp. PCC 6803 | 70 | 10 | 90 |
| A121 | JSC-1 | E412 | A183 | O-77 | 60AY4M2 | 63AY4M2 | 65AY6A5 | 65AY6Li | JA-2-3Ba | JA-3-3Ab | PCC 6715 | CL-1 | TA-1 | BP-1 | E542 | NIES-2134 | Putative Signals Detected | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PAS/PAC | 33/0 (32/1) | 31/0 (61/1) | 47/0 (24/0) | 52/0 (17/0) | 57/0 (6/0) | 22/1 (0/0) | 17/1 (0/0) | 22/1 (0/0) | 17/1 (0/0) | 16/0 (0/0) | 16/2 (0/0) | 3/0 (3/0) | 4/0 (2/1) | 4/0 (2/1) | 4/0 (2/1) | 3/0 (1/0) | 4/0 (2/1) | Small molecules, ions, gases, light, and redox State sensing |
| HAMP | 7(2) | 18(20) | 12(3) | 8(2) | 9(2) | 1(0) | 1(0) | 1(0) | 1(0) | 1(0) | 1(0) | 1(0) | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) | Signal transduction |
| GAF | 20(13) | 22(20) | 30(9) | 31(9) | 29(8) | 16(0) | 14(0) | 16(0) | 14(0) | 9(0) | 14(0) | 4(4) | 5(5) | 5(5) | 4(5) | 5(2) | 4(5) | Redox/oxygen sensing; cGMP binding |
| PHY | 2(0) | 4(0) | 2(0) | 0(0) | 0(0) | 1(0) | 1(0) | 1(0) | 1(0) | 0(0) | 1(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | Tetrapyrroles; light-sensing |
| CHASE | 3(2) | 7(1) | 1(3) | 3(0) | 3(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 2(0) | 2(0) | 0(0) | 1(0) | 0(0) | Small molecules recognition |
| MASE | 0(0) | 1(0) | 1(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | Membrane associated sensor |
| Cache | 2(1) | 6(7) | 1(2) | 1(1) | 2(1) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(1) | 0(1) | 0(1) | 0(1) | 0(1) | Small molecules recognition |
| cNMP_binding | 0(0) | 1(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | Cyclic nucleotide monophosphate-binding |
| S_TKc | 1(0) | 2(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | Serine/Threonine kinase catalytic domain |
| NIT | 0(0) | 0(0) | 0(1) | 0(1) | 0(1) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | Nitrate and nitrite responsive |
| FHA | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 1(0) | 1(0) | 1(0) | 1(0) | 1(0) | Phosphoserine/threonine binding |
| CBS | 3(0) | 0(5) | 0(2) | 1(0) | 1(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 1(0) | 0(0) | Adenosine nucleotides binding |
| Pkinase | 0(0) | 0(1) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | ATP binding |
   Sensing Transmit Receiver | A121 | JSC-1 | E412 | A183 | O-77 | 60AY4M2 | 63AY4M2 | 65AY6A5 | 65AY6Li | JA-2-3Ba | JA-3-3Ab | PCC 6715 | CL-1 | TA-1 | BP-1 | E542 | NIES-2134 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | 12 | 16 | 21 | 12 | 14 | 4 | 4 | 5 | 4 | 7 | 4 | 3 | 5 | 4 | 5 | 3 | 5 |
 | 12 | 27 | 14 | 17 | 16 | 3 | 3 | 3 | 3 | 2 | 3 | 3 | 5 | 5 | 2 | 4 | 2 |
 | 7 | 16 | 12 | 9 | 10 | 2 | 2 | 2 | 2 | 4 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
 | 5 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 2 | 2 | 2 | 2 |
 | 7 | 9 | 7 | 4 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
 | 3 | 0 | 4 | 3 | 2 | 3 | 3 | 3 | 3 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
 | 1 | 5 | 4 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
 | 1 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
 | 0 | 1 | 2 | 1 | 3 | 2 | 1 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 |
| Output Domain | RR Family | A121 | JSC-1 | E412 | A183 | O-77 | 60AY4M2 | 63AY4M2 | 65AY6A5 | 65AY6Li | JA-2-3Ba | JA-3-3Ab | PCC 6715 | CL-1 | TA-1 | BP-1 | E542 | NIES-2134 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stand-alone REC | CheY | 25 | 47 | 31 | 14 | 12 | 9 | 9 | 9 | 8 | 6 | 7 | 10 | 5 | 5 | 5 | 7 | 6 |
| DNA-binding | NarL | 12 | 12 | 8 | 8 | 10 | 6 | 6 | 6 | 6 | 6 | 6 | 3 | 4 | 4 | 4 | 4 | 4 |
| OmpR | 14 | 27 | 24 | 13 | 13 | 7 | 8 | 7 | 8 | 8 | 8 | 7 | 11 | 10 | 7 | 7 | 7 | |
| Chemotaxis | CheB | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| c-di-GMP signaling | RpfG | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| PleD | 1 | 3 | 4 | 3 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | |
| PleD-VieA | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | |
| Ser/Thr phosphorylation | RsbU | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pyridine nucleotide-disulphide oxidoreductase | TrxB | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Carbohydrate utilization | YesN | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Adenylate and Guanylate cyclase catalytic domain | CyC-C | 1 | 1 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unknown | Unclassified | 5 | 13 | 4 | 3 | 6 | 3 | 3 | 2 | 3 | 3 | 4 | 7 | 4 | 4 | 5 | 6 | 5 |
| Total | 61 | 111 | 79 | 46 | 49 | 29 | 30 | 28 | 29 | 28 | 29 | 32 | 26 | 26 | 23 | 26 | 24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.; Yao, D.; Zhou, H.; Wang, M.; Daroch, M. Distinct Molecular Patterns of Two-Component Signal Transduction Systems in Thermophilic Cyanobacteria as Revealed by Genomic Identification. Biology 2023, 12, 271. https://doi.org/10.3390/biology12020271
Tang J, Yao D, Zhou H, Wang M, Daroch M. Distinct Molecular Patterns of Two-Component Signal Transduction Systems in Thermophilic Cyanobacteria as Revealed by Genomic Identification. Biology. 2023; 12(2):271. https://doi.org/10.3390/biology12020271
Chicago/Turabian StyleTang, Jie, Dan Yao, Huizhen Zhou, Mingcheng Wang, and Maurycy Daroch. 2023. "Distinct Molecular Patterns of Two-Component Signal Transduction Systems in Thermophilic Cyanobacteria as Revealed by Genomic Identification" Biology 12, no. 2: 271. https://doi.org/10.3390/biology12020271
APA StyleTang, J., Yao, D., Zhou, H., Wang, M., & Daroch, M. (2023). Distinct Molecular Patterns of Two-Component Signal Transduction Systems in Thermophilic Cyanobacteria as Revealed by Genomic Identification. Biology, 12(2), 271. https://doi.org/10.3390/biology12020271









