RETRACTED: The Emerging Role of Epigenetics in Metabolism and Endocrinology
Simple Summary
Abstract
1. Introduction
2. Types of Epigenetic Modifications
2.1. DNA Methylation and the Role of DNA Methyltransferases (DNMTs)
2.2. Histone Modifications
2.3. Non-Coding RNA
3. Epigenetics and Metabolism
4. Epigenetics and Cancer Metabolism
4.1. Changes in DNA Methylation in Cancer
4.2. Changes in Histone Modifications in Cancer
4.3. miRNAs Modifications in Cancer
5. Epigenetics and Endocrine System
5.1. Epigenetics and Steroid Hormones
5.2. Epigenetics and Thyroid Hormones
5.3. Epigenetics and Peptide Hormones
6. Epigenetics and Endocrine Disruptors
6.1. Bisphenol-A (BPA)
6.2. Diethylstilbestrol (DES)
6.3. Dichlorodiphenyltrichloroethane (DDT)
6.4. Phthalates
6.5. Phytoestrogens
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2-HG | 2-hydroxyglutarate | HDMs | Histone demethylases |
| ACSS1/ACSS2/ACLY | acetyl-CoA synthetase short-chain family member/ATP citrate lyase | HMTs | Histone methyltransferases |
| ADP | Adenine-diphosphate | IDH/GLUD | Isocitrate dehydrogenase/glutamate dehydrogenase |
| AhR | Aryl hydrocarbon receptor | JAK-STAT | Janus kinase/signal transducers and activators of transcription |
| α-KG | α-ketoglutarate | JmjC | Jumonji C |
| AMPK | AMP-activated protein kinase | KDMs | Lysine demethylases |
| AR | Androgen receptor | KMT/PRMT | Protein arginine methyltransferase |
| ATP | Adenine-triphosphate | LH | Luteinizing hormone |
| BPA | Bisphenol A | lncRNAs | Long non-coding RNAs |
| CAR | Constitutive androstane receptor | LSD | Lysine-specific demethylase |
| CH3 | Methyl group | MAT | Methionine adenosyltransferase |
| circRNAs | Circular RNAs | MCF-7 | Michigan Cancer Foundation-7 |
| CoA | Co-enzyme A | MEOHP | Mono-(2-ethyl-5-oxohexyl) phthalate |
| DBP | Di-n-butyl phthalate | miRNAs | MicroRNAs |
| DDT | Dichlorodiphenyltrichloroethane | NAD+ | Nicotinamide adenine dinucleotide |
| DDT | Dichlorodiphenyltrichloroethane | ncRNAs | Non-coding RNA |
| DEHP | Di-2-ethyl-hexyl phthalate | NF-kB | Nuclear factor kappa B |
| DES | Diethylstilbestrol | NMAT | Nicotinamide mononucleotide adenyltransferase |
| DES | Diethylstilbestrol | NRs | Nuclear receptors |
| DMP | Dimethyl-phthalate | O-GlcNAc | O-linked N-Acetylglucosamine |
| DMR | DNA methylation regions | OGT/OGA | O-GlcNAc transferase/O-GlcNAcase |
| DNA | Deoxyribonucleic acid | PCBs | Polychlorinated biphenyls |
| DNMTs | DNA methyltransferases | pNEN | Pancreatic neuroendocrine neoplasms |
| EDC | Endocrine disruptors | PPARγ | Peroxisome proliferator-activated receptor gamma |
| EGFR/Akt/NF-kB | Epidermal growth factor receptor/serine/threonine kinase/nuclear factor kappa B | PR | Progesterone receptor |
| ER | Estrogen receptors | PTMs | Post-translational modifications |
| ERR | Estrogen-related receptor | PXR | Pregnane X receptor |
| FADH | Flavine adenine dinucleotides | RNAs | Ribose nucleic acid |
| FSH | Follicle-stimulating hormone | RXR | Retinoid X receptor |
| GnRH | Gonadotropin-releasing hormone | RXR | Retinoid X receptor |
| GPER | G protein-coupled estrogen receptor | SAH | S-adenosylhomocysteine |
| GR | Glucocorticoid receptor | SAM | S-adenosyl-L-methionine |
| GR | Glucocorticoid receptor | siRNAs | Short-interfering RNAs |
| HATs | Histone acetyltransferases | SIRT/PARP | Sirtuins/poly-ADP ribose polymerase |
| HDACs | Histone deacetylases | T2DM | Type 2 diabetes mellites |
| TCA | Tri-carboxylic cycle | tRNA | Transfer RNA |
| TH | Thyroid hormone | TSH | Thyroid-stimulating hormone |
| THRB | Thyroid hormone receptor beta | UDP-GlcNAc | Uridine diphospho-N-acetylglucosamine |
| TR | Thyroid hormone receptor |
References
- Hanna, C.W.; Demond, H.; Kelsey, G. Epigenetic Regulation in Development: Is the Mouse a Good Model for the Human? Hum. Reprod. Update 2018, 24, 556–576. [Google Scholar] [CrossRef] [PubMed]
- Arechederra, M.; Recalde, M.; Gárate-rascón, M.; Fernández-barrena, M.G.; Ávila, M.A.; Berasain, C. Epigenetic Biomarkers for the Diagnosis and Treatment of Liver Disease. Cancers 2021, 13, 1265. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Heard, E. Advances in Epigenetics Link Genetics to the Environment and Disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Flavahan, W.A.; Gaskell, E.; Bernstein, B.E. Epigenetic Plasticity and the Hallmarks of Cancer. Science 2017, 357, eaal2380. [Google Scholar] [CrossRef] [PubMed]
- Plunk, E.C.; Richards, S.M. Epigenetic Modifications Due to Environment, Ageing, Nutrition, and Endocrine Disrupting Chemicals and Their Effects on the Endocrine System. Int. J. Endocrinol. 2020, 2020, 9251980. [Google Scholar] [CrossRef]
- Mittelstaedt, N.N.; Becker, A.L.; de Freitas, D.N.; Zanin, R.F.; Stein, R.T.; de Souza, A.P.D. Dna Methylation and Immune Memory Response. Cells 2021, 10, 2943. [Google Scholar] [CrossRef]
- Parveen, N.; Dhawan, S. DNA Methylation Patterning and the Regulation of Beta Cell Homeostasis. Front. Endocrinol. 2021, 12, 651258. [Google Scholar] [CrossRef]
- Storck, W.K.; May, A.M.; Westbrook, T.C.; Duan, Z.; Morrissey, C.; Yates, J.A.; Alumkal, J.J. The Role of Epigenetic Change in Therapy-Induced Neuroendocrine Prostate Cancer Lineage Plasticity. Front. Endocrinol. 2022, 13, 926585. [Google Scholar] [CrossRef]
- Liu, P.; Yang, F.; Zhang, L.; Hu, Y.; Chen, B.; Wang, J.; Su, L.; Wu, M.; Chen, W. Emerging Role of Different DNA Methyltransferases in the Pathogenesis of Cancer. Front. Pharmacol. 2022, 13, 958146. [Google Scholar] [CrossRef]
- Pan, H.-M.; Lang, W.-Y.; Yao, L.-J.; Wang, Y.; Li, X.-L. ShRNA-Interfering LSD1 Inhibits Proliferation and Invasion of Gastric Cancer Cells via VEGF-C/PI3K/AKT Signaling Pathway. World J. Gastrointest. Oncol. 2019, 11, 622–633. [Google Scholar] [CrossRef]
- Sun, P.; Huang, T.; Huang, C.; Wang, Y.; Tang, D. Role of Histone Modification in the Occurrence and Development of Osteoporosis. Front. Endocrinol. 2022, 13, 964103. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Keating, S.T.; El-Osta, A. Metaboloepigenetics in Cancer, Immunity, and Cardiovascular Disease. Cardiovasc. Res. 2022, 1–14. [Google Scholar] [CrossRef]
- Akil, A.S.A.S.; Jerman, L.F.; Yassin, E.; Padmajeya, S.S.; Al-Kurbi, A.; Fakhro, K.A. Reading between the (Genetic) Lines: How Epigenetics Is Unlocking Novel Therapies for Type 1 Diabetes. Cells 2020, 9, 2403. [Google Scholar] [CrossRef]
- Wang, Z.; Long, H.; Chang, C.; Zhao, M.; Lu, Q. Crosstalk between Metabolism and Epigenetic Modifications in Autoimmune Diseases: A Comprehensive Overview. Cell. Mol. Life Sci. 2018, 75, 3353–3369. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Bultman, S.J. Metaboloepigenetics: Interrelationships between Energy Metabolism and Epigenetic Control of Gene Expression. J. Cell. Physiol. 2012, 227, 3169–3177. [Google Scholar] [CrossRef]
- Chang, M.; Yang, C.; Bao, X.; Wang, R. Genetic and Epigenetic Causes of Pituitary Adenomas. Front. Endocrinol. 2021, 11, 1–17. [Google Scholar] [CrossRef]
- Wang, J.; Xiao, M.; Wang, J.; Wang, S.; Zhang, J.; Guo, Y.; Tang, Y.; Gu, J. NRF2-Related Epigenetic Modifications in Cardiac and Vascular Complications of Diabetes Mellitus. Front. Endocrinol. 2021, 12, 598005. [Google Scholar] [CrossRef]
- Tiffon, C. The Impact of Nutrition and Environmental Epigenetics on Human Health and Disease. Int. J. Mol. Sci. 2018, 19, 3425. [Google Scholar] [CrossRef]
- Montjean, D.; Neyroud, A.S.; Yefimova, M.G.; Benkhalifa, M.; Cabry, R.; Ravel, C. Impact of Endocrine Disruptors upon Non-Genetic Inheritance. Int. J. Mol. Sci. 2022, 23, 3350. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Milagro, F.I.; Riezu-Boj, J.I.; Martinez, J.A. Epigenetic Signatures Underlying Inflammation: An Interplay of Nutrition, Physical Activity, Metabolic Diseases, and Environmental Factors for Personalized Nutrition. Inflamm. Res. 2021, 70, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Eggermann, T.; Elbracht, M.; Kurth, I.; Juul, A.; Juul, A.; Johannsen, T.H.; Johannsen, T.H.; Netchine, I.; Mastorakos, G.; Johannsson, G.; et al. Genetic Testing in Inherited Endocrine Disorders: Joint Position Paper of the European Reference Network on Rare Endocrine Conditions (Endo-ERN). Orphanet J. Rare Dis. 2020, 15, 144. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhou, J.; Chen, H.; Li, J.; Zhang, C.; Jiang, X.; Ni, C. The Immunomodulatory Effects of Endocrine Therapy in Breast Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, L.; Wang, Y.; Huang, G.; Li, X.; Xie, Z.; Zhou, Z. Insights Into the Role of DNA Methylation in Immune Cell Development and Autoimmune Disease. Front. Cell Dev. Biol. 2021, 9, 19. [Google Scholar] [CrossRef]
- Zhang, X.; Ho, S.-M. Epigenetics Meets Endocrinology. J. Mol. Endocrinol. 2011, 46, R11–R32. [Google Scholar] [CrossRef]
- Morris, M.J.; Monteggia, L.M. Role of DNA Methylation and the DNA Methyltransferases in Learning and Memory. Dialogues Clin. Neurosci. 2014, 16, 359–371. [Google Scholar] [CrossRef]
- Bollati, V.; Galimberti, D.; Pergoli, L.; Dalla Valle, E.; Barretta, F.; Cortini, F.; Scarpini, E.; Bertazzi, P.A.; Baccarelli, A. DNA Methylation in Repetitive Elements and Alzheimer Disease. Brain. Behav. Immun. 2011, 25, 1078–1083. [Google Scholar] [CrossRef]
- Klose, R.J.; Bird, A.P. Genomic DNA Methylation: The Mark and Its Mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef]
- Edwards, J.R.; Yarychkivska, O.; Boulard, M.; Bestor, T.H. DNA Methylation and DNA Methyltransferases. Epigenet. Chromatin 2017, 10, 23. [Google Scholar] [CrossRef]
- Jin, B.; Robertson, K.D. DNA Methyltransferases, DNA Damage Repair, and Cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar] [CrossRef]
- Lyko, F. The DNA Methyltransferase Family: A Versatile Toolkit for Epigenetic Regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Cristalli, C.; Manara, M.C.; Valente, S.; Pellegrini, E.; Bavelloni, A.; De Feo, A.; Blalock, W.; Di Bello, E.; Piñeyro, D.; Merkel, A.; et al. Novel Targeting of DNA Methyltransferase Activity Inhibits Ewing Sarcoma Cell Proliferation and Enhances Tumor Cell Sensitivity to DNA Damaging Drugs by Activating the DNA Damage Response. Front. Endocrinol. 2022, 13, 876602. [Google Scholar] [CrossRef]
- Tuorto, F.; Herbst, F.; Alerasool, N.; Bender, S.; Popp, O.; Federico, G.; Reitter, S.; Liebers, R.; Stoecklin, G.; Gröne, H.; et al. The TRNA Methyltransferase Dnmt2 Is Required for Accurate Polypeptide Synthesis during Haematopoiesis. EMBO J. 2015, 34, 2350–2362. [Google Scholar] [CrossRef]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and Biological Role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2017, 14, 1108–1123. [Google Scholar] [CrossRef]
- Jurkowska, R.Z.; Anspach, N.; Urbanke, C.; Jia, D.; Reinhardt, R.; Nellen, W.; Cheng, X.; Jeltsch, A. Formation of Nucleoprotein Filaments by Mammalian DNA Methyltransferase Dnmt3a in Complex with Regulator Dnmt3L. Nucleic Acids Res. 2008, 36, 6656–6663. [Google Scholar] [CrossRef]
- Laufer, B.I.; Gomez, J.A.; Jianu, J.M.; LaSalle, J.M. Stable DNMT3L Overexpression in SH-SY5Y Neurons Recreates a Facet of the Genome-Wide Down Syndrome DNA Methylation Signature. Epigenet. Chromatin 2021, 14, 13. [Google Scholar] [CrossRef]
- Emperle, M.; Bangalore, D.M.; Adam, S.; Kunert, S.; Heil, H.S.; Heinze, K.G.; Bashtrykov, P.; Tessmer, I.; Jeltsch, A. Structural and Biochemical Insight into the Mechanism of Dual CpG Site Binding and Methylation by the DNMT3A DNA Methyltransferase. Nucleic Acids Res. 2021, 49, 8294–8308. [Google Scholar] [CrossRef]
- Araujo, F.D.; Croteau, S.; Slack, A.D.; Milutinovic, S.; Bigey, P.; Price, G.B.; Zannis-Hajopoulos, M.; Szyf, M. The DNMT1 Target Recognition Domain Resides in the N Terminus. J. Biol. Chem. 2001, 276, 6930–6936. [Google Scholar] [CrossRef]
- Xie, S.; Wang, Z.; Okano, M.; Nogami, M.; Li, Y.; He, W.W.; Okumura, K.; Li, E. Cloning, Expression and Chromosome Locations of the Human DNMT3 Gene Family. Gene 1999, 236, 87–95. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, C.; Wu, C.; Cui, W.; Wang, L. DNA Methyltransferases in Cancer: Biology, Paradox, Aberrations, and Targeted Therapy. Cancers 2020, 12, 2123. [Google Scholar] [CrossRef]
- Ashapkin, V.V.; Kutueva, L.I.; Vanyushin, B.F. Dnmt2 Is the Most Evolutionary Conserved and Enigmatic Cytosine DNA Methyltransferase in Eukaryotes. Russ. J. Genet. 2016, 52, 237–248. [Google Scholar] [CrossRef]
- Dong, A.; Yoder, J.A.; Zhang, X.; Zhou, L.; Bestor, T.H.; Cheng, X. Structure of Human DNMT2, an Enigmatic DNA Methyltransferase Homolog That Displays Denaturant-Resistant Binding to DNA. Nucleic Acids Res. 2001, 29, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.J.; Botuyan, M.V.; Wu, Y.; Ward, C.J.; Nicholson, G.A.; Hammans, S.; Hojo, K.; Yamanishi, H.; Karpf, A.R.; Wallace, D.C.; et al. Mutations in DNMT1 Cause Hereditary Sensory Neuropathy with Dementia and Hearing Loss. Nat. Genet. 2011, 43, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Omura, N.; Hong, S.-M.; Goggins, M. Pancreatic Cancer DNMT1 Expression and Sensitivity to DNMT1 Inhibitors. Cancer Biol. Ther. 2010, 9, 321–329. [Google Scholar] [CrossRef]
- Emperle, M.; Adam, S.; Kunert, S.; Dukatz, M.; Baude, A.; Plass, C.; Rathert, P.; Bashtrykov, P.; Jeltsch, A. Mutations of R882 Change Flanking Sequence Preferences of the DNA Methyltransferase DNMT3A and Cellular Methylation Patterns. Nucleic Acids Res. 2019, 47, 11355–11367. [Google Scholar] [CrossRef]
- Man, X.; Li, Q.; Wang, B.; Zhang, H.; Zhang, S.; Li, Z. DNMT3A and DNMT3B in Breast Tumorigenesis and Potential Therapy. Front. Cell Dev. Biol. 2022, 10, 916725. [Google Scholar] [CrossRef]
- Hansen, R.S.; Wijmenga, C.; Luo, P.; Stanek, A.M.; Canfield, T.K.; Weemaes, C.M.R.; Gartler, S.M. The DNMT3B DNA Methyltransferase Gene Is Mutated in the ICF Immunodeficiency Syndrome. Proc. Natl. Acad. Sci. USA 1999, 96, 14412–14417. [Google Scholar] [CrossRef]
- Linnekamp, J.F.; Butter, R.; Spijker, R.; Medema, J.P.; van Laarhoven, H.W.M. Clinical and Biological Effects of Demethylating Agents on Solid Tumours—A Systematic Review. Cancer Treat. Rev. 2017, 54, 10–23. [Google Scholar] [CrossRef]
- Kamachi, K.; Ureshino, H.; Watanabe, T.; Yoshida, N.; Yamamoto, Y.; Kurahashi, Y.; Fukuda-Kurahashi, Y.; Hayashi, Y.; Hirai, H.; Yamashita, S.; et al. Targeting DNMT1 by Demethylating Agent OR-2100 Increases Tyrosine Kinase Inhibitors-Sensitivity and Depletes Leukemic Stem Cells in Chronic Myeloid Leukemia. Cancer Lett. 2022, 526, 273–283. [Google Scholar] [CrossRef]
- Hu, C.; Liu, X.; Zeng, Y.; Liu, J.; Wu, F. DNA Methyltransferase Inhibitors Combination Therapy for the Treatment of Solid Tumor: Mechanism and Clinical Application. Clin. Epigenet. 2021, 13, 166. [Google Scholar] [CrossRef]
- Wu, H.; Chang, C.; Lu, Q. The Epigenetics of Lupus Erythematosus BT-Epigenetics in Allergy and Autoimmunity; Chang, C., Lu, Q., Eds.; Springer: Singapore, 2020; pp. 185–207. ISBN 978-981-15-3449-2. [Google Scholar]
- Wardowska, A. The Epigenetic Face of Lupus: Focus on Antigen-Presenting Cells. Int. Immunopharmacol. 2020, 81, 106262. [Google Scholar] [CrossRef]
- Seal, R.L.; Denny, P.; Bruford, E.A.; Gribkova, A.K.; Landsman, D.; Marzluff, W.F.; McAndrews, M.; Panchenko, A.R.; Shaytan, A.K.; Talbert, P.B. A Standardized Nomenclature for Mammalian Histone Genes. Epigenet. Chromatin 2022, 15, 34. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef]
- Kaimala, S.; Kumar, C.A.; Allouh, M.Z.; Ansari, S.A.; Emerald, B.S. Epigenetic Modifications in Pancreas Development, Diabetes, and Therapeutics. Med. Res. Rev. 2022, 42, 1343–1371. [Google Scholar] [CrossRef]
- Mo, H.; Renna, C. Biomarker-Driven Targeted Therapies in Solid Tumor Malignancies. J. Hematol. Oncol. Pharm. 2021, 11, 84–91. [Google Scholar]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, Erasing and Reading Histone Lysine Methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y. The Diverse Functions of Histone Lysine Methylation. Nat. Rev. Mol. Cell Biol. 2005, 6, 838–849. [Google Scholar] [CrossRef]
- Thakur, C.; Chen, F. Connections between Metabolism and Epigenetics in Cancers. Semin. Cancer Biol. 2019, 57, 52–58. [Google Scholar] [CrossRef]
- Shahbazian, M.D.; Grunstein, M. Functions of Site-Specific Histone Acetylation and Deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef]
- Gujral, P.; Mahajan, V.; Lissaman, A.C.; Ponnampalam, A.P. Histone Acetylation and the Role of Histone Deacetylases in Normal Cyclic Endometrium. Reprod. Biol. Endocrinol. 2020, 18, 84. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Paderu, P.; Lee, A.; Eirekat, S.; Healey, K.; Chen, L.; Perlin, D.S.; Zhao, Y. Histone Acetylation Regulator Gcn5 Mediates Drug Resistance and Virulence of Candida Glabrata. Microbiol. Spectr. 2022, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiao, L.; Chen, X. Histone Acetylation and Methylation Underlie Oligodendroglial and Myelin Susceptibility in Schizophrenia. Front. Cell. Neurosci. 2022, 16, 823708. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Sanzón, E.; Prado-Garcia, H.; Romero-Garcia, S.; Nuñez-Corona, D.; Ortiz-Quintero, B.; Luna-Rivero, C.; Martínez-Cruz, V.; Carlos-Reyes, Á. Histone Deacetylases Modulate Resistance to the Therapy in Lung Cancer. Front. Genet. 2022, 13, 960263. [Google Scholar] [CrossRef] [PubMed]
- Nitsch, S.; Zorro Shahidian, L.; Schneider, R. Histone Acylations and Chromatin Dynamics: Concepts, Challenges, and Links to Metabolism. EMBO Rep. 2021, 22, e52774. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Joo, H.Y.; Jones, A.; Yang, C.; Zhai, L.; Smith IV, A.D.; Zhang, Z.; Chandrasekharan, M.B.; Sun, Z.W.; Renfrow, M.B.; Wang, Y.; et al. Regulation of Histone H2A and H2B Deubiquitination and Xenopus Development by USP12 and USP46. J. Biol. Chem. 2011, 286, 7190–7201. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, L.; Zhang, X.; Cui, W.; Liu, Y.; Sun, Q.; He, Q.; Zhao, S.; Zhang, G.; Wang, Y.; et al. Epigenetic Regulation of Ferroptosis by H2B Monoubiquitination and P53. EMBO Rep. 2019, 20, e47563. [Google Scholar] [CrossRef]
- Chen, Z.; Djekidel, M.N.; Zhang, Y. Distinct Dynamics and Functions of H2AK119ub1 and H3K27me3 in Mouse Preimplantation Embryos. Nat. Genet. 2021, 53, 551–563. [Google Scholar] [CrossRef]
- Cerutti, H.; Casas-Mollano, J.A. Histone H3 Phosphorylation: Universal Code or Lineage Specific Dialects? Epigenetics 2009, 4, 71–75. [Google Scholar] [CrossRef]
- Mazziotta, C.; Lanzillotti, C.; Gafà, R.; Touzé, A.; Durand, M.-A.; Martini, F.; Rotondo, J.C. The Role of Histone Post-Translational Modifications in Merkel Cell Carcinoma. Front. Oncol. 2022, 12, 832047. [Google Scholar] [CrossRef]
- Armache, A.; Yang, S.; Martínez de Paz, A.; Robbins, L.E.; Durmaz, C.; Cheong, J.Q.; Ravishankar, A.; Daman, A.W.; Ahimovic, D.J.; Klevorn, T.; et al. Histone H3.3 Phosphorylation Amplifies Stimulation-Induced Transcription. Nature 2020, 583, 852–857. [Google Scholar] [CrossRef]
- Basnet, H.; Su, X.B.; Tan, Y.; Meisenhelder, J.; Merkurjev, D.; Ohgi, K.A.; Hunter, T.; Pillus, L.; Rosenfeld, M.G. Tyrosine Phosphorylation of Histone H2A by CK2 Regulates Transcriptional Elongation. Nature 2014, 516, 267–271. [Google Scholar] [CrossRef]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone Phosphorylation: A Chromatin Modification Involved in Diverse Nuclear Events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef]
- Xia, F.; Wang, Y.; Xue, M.; Zhu, L.; Jia, D.; Shi, Y.; Gao, Y.; Li, L.; Li, Y.; Chen, S.; et al. LncRNA KCNQ1OT1: Molecular Mechanisms and Pathogenic Roles in Human Diseases. Genes Dis. 2022, 9, 1556–1565. [Google Scholar] [CrossRef]
- Gao, X.; Liu, L.; Min, X.; Jia, S.; Zhao, M. Non-Coding RNAs in CD4+ T Cells: New Insights Into the Pathogenesis of Systemic Lupus Erythematosus. Front. Immunol. 2020, 11, 568. [Google Scholar] [CrossRef]
- Tsai, C.Y.; Shen, C.Y.; Liu, C.W.; Hsieh, S.C.; Liao, H.T.; Li, K.J.; Lu, C.S.; Lee, H.T.; Lin, C.S.; Wu, C.H.; et al. Aberrant Non-Coding Rna Expression in Patients with Systemic Lupus Erythematosus: Consequences for Immune Dysfunctions and Tissue Damage. Biomolecules 2020, 10, 1641. [Google Scholar] [CrossRef]
- Bure, I.V.; Nemtsova, M.V.; Kuznetsova, E.B. Histone Modifications and Non-Coding RNAs: Mutual Epigenetic Regulation and Role in Pathogenesis. Int. J. Mol. Sci. 2022, 23, 5801. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, X.; Ye, L.; Guo, G.; Li, X.; Chen, C.; Sun, L.; Li, B.; Chen, N.; Xue, X. B Cell-Related Circulating MicroRNAs With the Potential Value of Biomarkers in the Differential Diagnosis, and Distinguishment Between the Disease Activity and Lupus Nephritis for Systemic Lupus Erythematosus. Front. Immunol. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Taufiqul Arif, K.M.; Elliot, E.K.; Haupt, L.M.; Griffiths, L.R. Regulatory Mechanisms of Epigenetic Mirna Relationships in Human Cancer and Potential as Therapeutic Targets. Cancers 2020, 12, 2922. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, Y.; Zhou, X. The Roles of MicroRNAs in Epigenetic Regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Hu, J.; Yin, W. Systematic Identification of Non-Coding RNAs. Adv. Exp. Med. Biol. 2018, 1094, 9–18. [Google Scholar] [CrossRef] [PubMed]
- de Brot, S.; Rutland, C.S.; Mongan, N.P.; James, V. Epigenetic Control of MicroRNA Expression and Cancer; Elsevier Inc.: Amsterdam, The Netherlands, 2018; ISBN 9780128110225. [Google Scholar]
- Ma, F.; Liu, X.; Li, D.; Wang, P.; Li, N.; Lu, L.; Cao, X. MicroRNA-466l Upregulates IL-10 Expression in TLR-Triggered Macrophages by Antagonizing RNA-Binding Protein Tristetraprolin-Mediated IL-10 MRNA Degradation. J. Immunol. 2010, 184, 6053–6059. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-MicroRNAs with a Role in Cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Wang, M.; Xia, X.; Chu, W.; Xia, L.; Meng, T.; Liu, L.; Liu, Y. Roles of MiR-186 and PTTG1 in Colorectal Neuroendocrine Tumors. Int. J. Clin. Exp. Med. 2015, 8, 22149–22157. [Google Scholar]
- Thorns, C.; Schurmann, C.; Gebauer, N.; Wallaschofski, H.; Kümpers, C.; Bernard, V.; Feller, A.C.; Keck, T.; Habermann, J.K.; Begum, N.; et al. Global MicroRNA Profiling of Pancreatic Neuroendocrine Neoplasias. Anticancer Res. 2014, 34, 2249–2254. [Google Scholar]
- Malczewska, A.; Kidd, M.; Matar, S.; Kos-Kudla, B.; Modlin, I.M. A Comprehensive Assessment of the Role of MiRNAs as Biomarkers in Gastroenteropancreatic Neuroendocrine Tumors. Neuroendocrinology 2018, 107, 73–90. [Google Scholar] [CrossRef]
- Tello-Flores, V.A.; Beltrán-Anaya, F.O.; Ramírez-Vargas, M.A.; Esteban-Casales, B.E.; Navarro-Tito, N.; Alarcón-Romero, L.D.C.; Luciano-Villa, C.A.; Ramírez, M.; Del Moral-Hernández, Ó.; Flores-Alfaro, E. Role of Long Non-Coding Rnas and the Molecular Mechanisms Involved in Insulin Resistance. Int. J. Mol. Sci. 2021, 22, 7256. [Google Scholar] [CrossRef]
- Formichi, C.; Nigi, L.; Grieco, G.E.; Maccora, C.; Fignani, D.; Brusco, N.; Licata, G.; Sebastiani, G.; Dotta, F. Non-coding Rnas: Novel Players in Insulin Resistance and Related Diseases. Int. J. Mol. Sci. 2021, 22, 7716. [Google Scholar] [CrossRef]
- Jones, A.; Danielson, K.M.; Benton, M.C.; Ziegler, O.; Shah, R.; Stubbs, R.S.; Das, S.; Macartney-Coxson, D. MiRNA Signatures of Insulin Resistance in Obesity. Obesity 2017, 25, 1734–1744. [Google Scholar] [CrossRef]
- Butz, H. Circulating Noncoding RNAs in Pituitary Neuroendocrine Tumors-Two Sides of the Same Coin. Int. J. Mol. Sci. 2022, 23, 5122. [Google Scholar] [CrossRef]
- Butz, H.; Patócs, A. MicroRNAs in Endocrine Tumors. EJIFCC 2019, 30, 146–164. [Google Scholar]
- Peng, C.; Wang, Y.-L. Editorial: MicroRNAs as New Players in Endocrinology. Front. Endocrinol. 2018, 9, 459. [Google Scholar] [CrossRef]
- Powell, E.E.; Wong, V.W.S.; Rinella, M. Non-Alcoholic Fatty Liver Disease. Lancet 2021, 397, 2212–2224. [Google Scholar] [CrossRef]
- Wang, G.; Han, J.J. Connections between Metabolism and Epigenetic Modifications in Cancer. Med. Rev. 2021, 1, 199–221. [Google Scholar] [CrossRef]
- Saggese, P.; Sellitto, A.; Martinez, C.A.; Giurato, G.; Nassa, G.; Rizzo, F.; Tarallo, R.; Scafoglio, C. Metabolic Regulation of Epigenetic Modifications and Cell Differentiation in Cancer. Cancers 2020, 12, 3788. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Craig, B. Thompson Emerging Metabolic Hallmarks of Cancer. Physiol. Behav. 2018, 176, 139–148. [Google Scholar] [CrossRef]
- Sebastian, C.; Vong, J.S.L.; Mayekar, M.K.; Tummala, K.S.; Singh, I. Editorial: Metabolism and Epigenetics. Front. Genet. 2022, 13, 4–6. [Google Scholar] [CrossRef]
- Janke, R.; Dodson, A.E.; Rine, J. Metabolism and Epigenetics. Annu. Rev. Cell Dev. Biol. 2015, 31, 473–496. [Google Scholar] [CrossRef]
- World Health Organization. Noncommunicable Diseases. Available online: https://www.who.int/news-room/fact-sheets/detail/noncommunicable-diseases (accessed on 2 November 2022).
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, Z.; Qin, Y. Connections between Metabolism and Epigenetics: Mechanisms and Novel Anti-Cancer Strategy. Front. Pharmacol. 2022, 13, 935536. [Google Scholar] [CrossRef] [PubMed]
- Keating, S.T.; El-Osta, A. Epigenetics and Metabolism. Circ. Res. 2015, 116, 715–736. [Google Scholar] [CrossRef] [PubMed]
- Pranzini, E.; Pardella, E.; Paoli, P.; Fendt, S.M.; Taddei, M.L. Metabolic Reprogramming in Anticancer Drug Resistance: A Focus on Amino Acids. Trends Cancer 2021, 7, 682–699. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Ramesh, V.; Locasale, J.W. The Evolving Metabolic Landscape of Chromatin Biology and Epigenetics. Nat. Rev. Genet. 2020, 21, 737–753. [Google Scholar] [CrossRef]
- Zhu, J.; Thompson, C.B. Metabolic Regulation of Cell Growth and Proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef]
- Campbell, S.L.; Wellen, K.E. Metabolic Signaling to the Nucleus in Cancer. Mol. Cell 2018, 71, 398–408. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic Reprogramming and Cancer Progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in Cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Kim, J.-A.; Yeom, Y. Il Metabolic Signaling to Epigenetic Alterations in Cancer. Biomol. Ther. 2018, 26, 69–80. [Google Scholar] [CrossRef]
- Wang, Y.-P.; Lei, Q.-Y. Metabolic Recoding of Epigenetics in Cancer. Cancer Commun. 2018, 38, 25. [Google Scholar] [CrossRef]
- Sanderson, S.M.; Gao, X.; Dai, Z.; Locasale, J.W. Methionine Metabolism in Health and Cancer: A Nexus of Diet and Precision Medicine. Nat. Rev. Cancer 2019, 19, 625–637. [Google Scholar] [CrossRef]
- Ravanel, S.; Gakière, B.; Job, D.; Douce, R. The Specific Features of Methionine Biosynthesis and Metabolism in Plants. Proc. Natl. Acad. Sci. USA 1998, 95, 7805–7812. [Google Scholar] [CrossRef]
- Zhang, N. Role of Methionine on Epigenetic Modification of DNA Methylation and Gene Expression in Animals. Anim. Nutr. 2018, 4, 11–16. [Google Scholar] [CrossRef]
- Mattocks, D.A.L.; Mentch, S.J.; Shneyder, J.; Ables, G.P.; Sun, D.; Richie, J.P.; Locasale, J.W.; Nichenametla, S.N. Short Term Methionine Restriction Increases Hepatic Global DNA Methylation in Adult but Not Young Male C57BL/6J Mice. Exp. Gerontol. 2017, 88, 1–8. [Google Scholar] [CrossRef]
- Usui, G.; Matsusaka, K.; Mano, Y.; Urabe, M.; Funata, S.; Fukayama, M.; Ushiku, T.; Kaneda, A. DNA Methylation and Genetic Aberrations in Gastric Cancer. Digestion 2021, 102, 25–32. [Google Scholar] [CrossRef]
- Easwaran, H.; Tsai, H.-C.; Baylin, S.B. Cancer Epigenetics: Tumor Heterogeneity, Plasticity of Stem-like States, and Drug Resistance. Mol. Cell 2014, 54, 716–727. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA Hypomethylation in Cancer Cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef]
- Frigola, J.; Solé, X.; Paz, M.F.; Moreno, V.; Esteller, M.; Capellà, G.; Peinado, M.A. Differential DNA Hypermethylation and Hypomethylation Signatures in Colorectal Cancer. Hum. Mol. Genet. 2005, 14, 319–326. [Google Scholar] [CrossRef]
- Rauluseviciute, I.; Drabløs, F.; Rye, M.B. DNA Hypermethylation Associated with Upregulated Gene Expression in Prostate Cancer Demonstrates the Diversity of Epigenetic Regulation. BMC Med. Genomics 2020, 13, 6. [Google Scholar] [CrossRef]
- Rodriguez, J.; Frigola, J.; Vendrell, E.; Risques, R.-A.; Fraga, M.F.; Morales, C.; Moreno, V.; Esteller, M.; Capellà, G.; Ribas, M.; et al. Chromosomal Instability Correlates with Genome-Wide DNA Demethylation in Human Primary Colorectal Cancers. Cancer Res. 2006, 66, 8462–9468. [Google Scholar] [CrossRef] [PubMed]
- Eden, A.; Gaudet, F.; Waghmare, A.; Jaenisch, R. Chromosomal Instability and Tumors Promoted by DNA Hypomethylation. Science 2003, 300, 455. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The Fundamental Role of Epigenetic Events in Cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef]
- Long, C.; Yin, B.; Lu, Q.; Zhou, X.; Hu, J.; Yang, Y.; Yu, F.; Yuan, Y. Promoter Hypermethylation of the RUNX3 Gene in Esophageal Squamous Cell Carcinoma. Cancer Investig. 2007, 25, 685–690. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA Hypermethylation in Disease: Mechanisms and Clinical Relevance. Epigenetics 2019, 14, 1141–1163. [Google Scholar] [CrossRef]
- Breiling, A.; Lyko, F. Epigenetic Regulatory Functions of DNA Modifications: 5-Methylcytosine and Beyond. Epigenet. Chromatin 2015, 8, 24. [Google Scholar] [CrossRef]
- McCabe, M.T.; Davis, J.N.; Day, M.L. Regulation of DNA Methyltransferase 1 by the PRb/E2F1 Pathway. Cancer Res. 2005, 65, 3624–3632. [Google Scholar] [CrossRef]
- Wu, X.-Y.; Chen, H.-C.; Li, W.-W.; Yan, J.-D.; Lv, R.-Y. DNMT1 Promotes Cell Proliferation via Methylating HMLH1 and HMSH2 Promoters in EGFR-Mutated Non-Small Cell Lung Cancer. J. Biochem. 2020, 168, 151–157. [Google Scholar] [CrossRef]
- Lu, G.-H.; Zhao, H.-M.; Liu, Z.-Y.; Cao, Q.; Shao, R.-D.; Sun, G. LncRNA SAMD12-AS1 Promotes the Progression of Gastric Cancer via DNMT1/P53 Axis. Arch. Med. Res. 2021, 52, 683–691. [Google Scholar] [CrossRef]
- Liu, H.; Song, Y.; Qiu, H.; Liu, Y.; Luo, K.; Yi, Y.; Jiang, G.; Lu, M.; Zhang, Z.; Yin, J.; et al. Downregulation of FOXO3a by DNMT1 Promotes Breast Cancer Stem Cell Properties and Tumorigenesis. Cell Death Differ. 2020, 27, 966–983. [Google Scholar] [CrossRef]
- Yao, Y.; Liu, C.; Wang, B.; Guan, X.; Fang, L.; Zhan, F.; Sun, H.; Li, H.; Lou, C.; Yan, F.; et al. HOXB9 Blocks Cell Cycle Progression to Inhibit Pancreatic Cancer Cell Proliferation through the DNMT1/RBL2/c-Myc Axis. Cancer Lett. 2022, 533, 215595. [Google Scholar] [CrossRef]
- Wong, K.K. DNMT1 as a Therapeutic Target in Pancreatic Cancer: Mechanisms and Clinical Implications. Cell. Oncol. 2020, 43, 779–792. [Google Scholar] [CrossRef]
- Lee, E.; Wang, J.; Yumoto, K.; Jung, Y.; Cackowski, F.C.; Decker, A.M.; Li, Y.; Franceschi, R.T.; Pienta, K.J.; Taichman, R.S. DNMT1 Regulates Epithelial-Mesenchymal Transition and Cancer Stem Cells, Which Promotes Prostate Cancer Metastasis. Neoplasia 2016, 18, 553–566. [Google Scholar] [CrossRef]
- Luo, Y.; Xie, C.; Brocker, C.N.; Fan, J.; Wu, X.; Feng, L.; Wang, Q.; Zhao, J.; Lu, D.; Tandon, M.; et al. Intestinal PPARα Protects Against Colon Carcinogenesis via Regulation of Methyltransferases DNMT1 and PRMT6. Gastroenterology 2019, 157, 744–759.e4. [Google Scholar] [CrossRef]
- Leonard, S.; Pereira, M.; Fox, R.; Gordon, N.; Yap, J.; Kehoe, S.; Luesley, D.; Woodman, C.; Ganesan, R. Over-Expression of DNMT3A Predicts the Risk of Recurrent Vulvar Squamous Cell Carcinomas. Gynecol. Oncol. 2016, 143, 414–420. [Google Scholar] [CrossRef]
- Sun, W.; Ma, G.; Zhang, L.; Wang, P.; Zhang, N.; Wu, Z.; Dong, Y.; Cai, F.; Chen, L.; Liu, H.; et al. DNMT3A-Mediated Silence in ADAMTS9 Expression Is Restored by RNF180 to Inhibit Viability and Motility in Gastric Cancer Cells. Cell Death Dis. 2021, 12, 428. [Google Scholar] [CrossRef]
- Husni, R.E.; Shiba-Ishii, A.; Iiyama, S.; Shiozawa, T.; Kim, Y.; Nakagawa, T.; Sato, T.; Kano, J.; Minami, Y.; Noguchi, M. DNMT3a Expression Pattern and Its Prognostic Value in Lung Adenocarcinoma. Lung Cancer 2016, 97, 59–65. [Google Scholar] [CrossRef]
- Miao, J.; Zhao, C.; Tang, K.; Xiong, X.; Wu, F.; Xue, W.; Duan, B.; Zhang, H.; Jing, X.; Li, W.; et al. TDG Suppresses the Migration and Invasion of Human Colon Cancer Cells via the DNMT3A/TIMP2 Axis. Int. J. Biol. Sci. 2022, 18, 2527–2539. [Google Scholar] [CrossRef]
- Gui, T.; Liu, M.; Yao, B.; Jiang, H.; Yang, D.; Li, Q.; Zeng, X.; Wang, Y.; Cao, J.; Deng, Y.; et al. TCF3 Is Epigenetically Silenced by EZH2 and DNMT3B and Functions as a Tumor Suppressor in Endometrial Cancer. Cell Death Differ. 2021, 28, 3316–3328. [Google Scholar] [CrossRef]
- Ibrahim, M.L.; Klement, J.D.; Lu, C.; Redd, P.S.; Xiao, W.; Yang, D.; Browning, D.D.; Savage, N.M.; Buckhaults, P.J.; Morse, H.C.; et al. Myeloid-Derived Suppressor Cells Produce IL-10 to Elicit DNMT3b-Dependent IRF8 Silencing to Promote Colitis-Associated Colon Tumorigenesis. Cell Rep. 2018, 25, 3036–3046.e6. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Tu, G.; Yang, G.; Wang, X.; Kang, L.; Yang, L.; Zeng, H.; Wan, X.; Qiao, Y.; Cui, X.; et al. Autocrine TGF-Β1/MiR-200s/MiR-221/DNMT3B Regulatory Loop Maintains CAF Status to Fuel Breast Cancer Cell Proliferation. Cancer Lett. 2019, 452, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, M.; Wang, Y. The Roles of Histone Modifications in Tumorigenesis and Associated Inhibitors in Cancer Therapy. J. Natl. Cancer Cent. 2022, 2, 277–290. [Google Scholar] [CrossRef]
- Zhao, Z.; Shilatifard, A. Epigenetic Modifications of Histones in Cancer. Genome Biol. 2019, 20, 245. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone Methylation: A Dynamic Mark in Health, Disease and Inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Herz, H.-M.; Garruss, A.; Shilatifard, A. SET for Life: Biochemical Activities and Biological Functions of SET Domain-Containing Proteins. Trends Biochem. Sci. 2013, 38, 621–639. [Google Scholar] [CrossRef]
- Barghout, S.H.; Machado, R.A.C.; Barsyte-Lovejoy, D. Chemical Biology and Pharmacology of Histone Lysine Methylation Inhibitors. Biochim. Biophys. Acta Gene Regul. Mech. 2022, 1865, 194840. [Google Scholar] [CrossRef]
- Husmann, D.; Gozani, O. Histone Lysine Methyltransferases in Biology and Disease. Nat. Struct. Mol. Biol. 2019, 26, 880–889. [Google Scholar] [CrossRef]
- Mohan, M.; Herz, H.-M.; Shilatifard, A. SnapShot: Histone Lysine Methylase Complexes. Cell 2012, 149, 498–498.e1. [Google Scholar] [CrossRef]
- Dorna, D.; Paluszczak, J. The Emerging Significance of Histone Lysine Demethylases as Prognostic Markers and Therapeutic Targets in Head and Neck Cancers. Cells 2022, 11, 1023. [Google Scholar] [CrossRef]
- McAllister, T.E.; England, K.S.; Hopkinson, R.J.; Brennan, P.E.; Kawamura, A.; Schofield, C.J. Recent Progress in Histone Demethylase Inhibitors. J. Med. Chem. 2016, 59, 1308–1329. [Google Scholar] [CrossRef]
- Thinnes, C.C.; England, K.S.; Kawamura, A.; Chowdhury, R.; Schofield, C.J.; Hopkinson, R.J. Targeting Histone Lysine Demethylases-Progress, Challenges, and the Future. Biochim. Biophys. Acta Gene Regul. Mech. 2014, 1839, 1416–1432. [Google Scholar] [CrossRef]
- Feng, J.; Meng, X. Histone Modification and Histone Modification-Targeted Anti-Cancer Drugs in Breast Cancer: Fundamentals and Beyond. Front. Pharmacol. 2022, 13, 946811. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the Histone Code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Haws, S.A.; Yu, D.; Ye, C.; Wille, C.K.; Nguyen, L.C.; Krautkramer, K.A.; Tomasiewicz, J.L.; Yang, S.E.; Miller, B.R.; Liu, W.H.; et al. Methyl-Metabolite Depletion Elicits Adaptive Responses to Support Heterochromatin Stability and Epigenetic Persistence. Mol. Cell 2020, 78, 210–223.e8. [Google Scholar] [CrossRef]
- Dai, Z.; Mentch, S.J.; Gao, X.; Nichenametla, S.N.; Locasale, J.W. Methionine Metabolism Influences Genomic Architecture and Gene Expression through H3K4me3 Peak Width. Nat. Commun. 2018, 9, 1955. [Google Scholar] [CrossRef]
- Mentch, S.J.; Mehrmohamadi, M.; Huang, L.; Liu, X.; Gupta, D.; Mattocks, D.; Gómez Padilla, P.; Ables, G.; Bamman, M.M.; Thalacker-Mercer, A.E.; et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015, 22, 861–873. [Google Scholar] [CrossRef]
- Sperber, H.; Mathieu, J.; Wang, Y.; Ferreccio, A.; Hesson, J.; Xu, Z.; Fischer, K.A.; Devi, A.; Detraux, D.; Gu, H.; et al. The Metabolome Regulates the Epigenetic Landscape during Naive-to-Primed Human Embryonic Stem Cell Transition. Nat. Cell Biol. 2015, 17, 1523–1535. [Google Scholar] [CrossRef]
- Shyh-Chang, N.; Locasale, J.W.; Lyssiotis, C.A.; Zheng, Y.; Teo, R.Y.; Ratanasirintrawoot, S.; Zhang, J.; Onder, T.; Unternaehrer, J.J.; Zhu, H.; et al. Influence of Threonine Metabolism on S -Adenosylmethionine and Histone Methylation. Science 2013, 339, 222–226. [Google Scholar] [CrossRef]
- Shiraki, N.; Shiraki, Y.; Tsuyama, T.; Obata, F.; Miura, M.; Nagae, G.; Aburatani, H.; Kume, K.; Endo, F.; Kume, S. Methionine Metabolism Regulates Maintenance and Differentiation of Human Pluripotent Stem Cells. Cell Metab. 2014, 19, 780–794. [Google Scholar] [CrossRef]
- Lakshmikuttyamma, A.; Scott, S.A.; DeCoteau, J.F.; Geyer, C.R. Reexpression of Epigenetically Silenced AML Tumor Suppressor Genes by SUV39H1 Inhibition. Oncogene 2010, 29, 576–588. [Google Scholar] [CrossRef] [PubMed]
- McLean, C.M.; Karemaker, I.D.; van Leeuwen, F. The Emerging Roles of DOT1L in Leukemia and Normal Development. Leukemia 2014, 28, 2131–2138. [Google Scholar] [CrossRef] [PubMed]
- Reynoird, N.; Mazur, P.K.; Stellfeld, T.; Flores, N.M.; Lofgren, S.M.; Carlson, S.M.; Brambilla, E.; Hainaut, P.; Kaznowska, E.B.; Arrowsmith, C.H.; et al. Coordination of Stress Signals by the Lysine Methyltransferase SMYD2 Promotes Pancreatic Cancer. Genes Dev. 2016, 30, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-S.; Lee, M.-H.; Lee, M.-O. Histone Methyltransferases Regulate the Transcriptional Expression of ERα and the Proliferation of Tamoxifen-Resistant Breast Cancer Cells. Breast Cancer Res. Treat. 2020, 180, 45–54. [Google Scholar] [CrossRef]
- Fenizia, C.; Bottino, C.; Corbetta, S.; Fittipaldi, R.; Floris, P.; Gaudenzi, G.; Carra, S.; Cotelli, F.; Vitale, G.; Caretti, G. SMYD3 Promotes the Epithelial–Mesenchymal Transition in Breast Cancer. Nucleic Acids Res. 2019, 47, 1278–1293. [Google Scholar] [CrossRef]
- Tajima, K.; Matsuda, S.; Yae, T.; Drapkin, B.J.; Morris, R.; Boukhali, M.; Niederhoffer, K.; Comaills, V.; Dubash, T.; Nieman, L.; et al. SETD1A Protects from Senescence through Regulation of the Mitotic Gene Expression Program. Nat. Commun. 2019, 10, 2854. [Google Scholar] [CrossRef]
- Gala, K.; Li, Q.; Sinha, A.; Razavi, P.; Dorso, M.; Sanchez-Vega, F.; Chung, Y.R.; Hendrickson, R.; Hsieh, J.J.; Berger, M.; et al. KMT2C Mediates the Estrogen Dependence of Breast Cancer through Regulation of ERα Enhancer Function. Oncogene 2018, 37, 4692–4710. [Google Scholar] [CrossRef]
- Su, C.-H.; Lin, I.-H.; Tzeng, T.-Y.; Hsieh, W.-T.; Hsu, M.-T. Regulation of IL-20 Expression by Estradiol through KMT2B-Mediated Epigenetic Modification. PLoS ONE 2016, 11, e0166090. [Google Scholar] [CrossRef]
- Jin, Y.; Park, S.; Park, S.-Y.; Lee, C.-Y.; Eum, D.-Y.; Shim, J.-W.; Choi, S.-H.; Choi, Y.-J.; Park, S.-J.; Heo, K. G9a Knockdown Suppresses Cancer Aggressiveness by Facilitating Smad Protein Phosphorylation through Increasing BMP5 Expression in Luminal A Type Breast Cancer. Int. J. Mol. Sci. 2022, 23, 589. [Google Scholar] [CrossRef]
- Casciello, F.; Al-Ejeh, F.; Miranda, M.; Kelly, G.; Baxter, E.; Windloch, K.; Gannon, F.; Lee, J.S. G9a-Mediated Repression of CDH10 in Hypoxia Enhances Breast Tumour Cell Motility and Associates with Poor Survival Outcome. Theranostics 2020, 10, 4515–4529. [Google Scholar] [CrossRef]
- Siouda, M.; Dujardin, A.D.; Barbollat-Boutrand, L.; Mendoza-Parra, M.A.; Gibert, B.; Ouzounova, M.; Bouaoud, J.; Tonon, L.; Robert, M.; Foy, J.-P.; et al. CDYL2 Epigenetically Regulates MIR124 to Control NF-ΚB/STAT3-Dependent Breast Cancer Cell Plasticity. iScience 2020, 23, 101141. [Google Scholar] [CrossRef]
- Crawford, N.T.; McIntyre, A.J.; McCormick, A.; D’Costa, Z.C.; Buckley, N.E.; Mullan, P.B. TBX2 Interacts with Heterochromatin Protein 1 to Recruit a Novel Repression Complex to EGR1-Targeted Promoters to Drive the Proliferation of Breast Cancer Cells. Oncogene 2019, 38, 5971–5986. [Google Scholar] [CrossRef]
- Zhang, J.; Yao, D.; Jiang, Y.; Huang, J.; Yang, S.; Wang, J. Synthesis and Biological Evaluation of Benzimidazole Derivatives as the G9a Histone Methyltransferase Inhibitors That Induce Autophagy and Apoptosis of Breast Cancer Cells. Bioorg. Chem. 2017, 72, 168–181. [Google Scholar] [CrossRef]
- Yi, C.; Li, G.; Wang, W.; Sun, Y.; Zhang, Y.; Zhong, C.; Stovall, D.B.; Li, D.; Shi, J.; Sui, G. Disruption of YY1-EZH2 Interaction Using Synthetic Peptides Inhibits Breast Cancer Development. Cancers 2021, 13, 2402. [Google Scholar] [CrossRef]
- Dong, H.; Liu, Q.; Zhao, T.; Yao, F.; Xu, Y.; Chen, B.; Wu, Y.; Zheng, X.; Jin, F.; Li, J.; et al. Long Non-Coding RNA LOXL1-AS1 Drives Breast Cancer Invasion and Metastasis by Antagonizing MiR-708-5p Expression and Activity. Mol. Ther. Nucleic Acids 2020, 19, 696–705. [Google Scholar] [CrossRef]
- Yomtoubian, S.; Lee, S.B.; Verma, A.; Izzo, F.; Markowitz, G.; Choi, H.; Cerchietti, L.; Vahdat, L.; Brown, K.A.; Andreopoulou, E.; et al. Inhibition of EZH2 Catalytic Activity Selectively Targets a Metastatic Subpopulation in Triple-Negative Breast Cancer. Cell Rep. 2020, 30, 755–770.e6. [Google Scholar] [CrossRef]
- Zeng, Y.; Qiu, R.; Yang, Y.; Gao, T.; Zheng, Y.; Huang, W.; Gao, J.; Zhang, K.; Liu, R.; Wang, S.; et al. Regulation of EZH2 by SMYD2-Mediated Lysine Methylation Is Implicated in Tumorigenesis. Cell Rep. 2019, 29, 1482–1498.e4. [Google Scholar] [CrossRef]
- Gong, C.; Yao, S.; Gomes, A.R.; Man, E.P.S.; Lee, H.J.; Gong, G.; Chang, S.; Kim, S.-B.; Fujino, K.; Kim, S.-W.; et al. BRCA1 Positively Regulates FOXO3 Expression by Restricting FOXO3 Gene Methylation and Epigenetic Silencing through Targeting EZH2 in Breast Cancer. Oncogenesis 2016, 5, e214. [Google Scholar] [CrossRef]
- Esteller, M. Cancer Epigenomics: DNA Methylomes and Histone-Modification Maps. Nat. Rev. Genet. 2007, 8, 286–298. [Google Scholar] [CrossRef]
- Van Den Broeck, A.; Brambilla, E.; Moro-Sibilot, D.; Lantuejoul, S.; Brambilla, C.; Eymin, B.; Khochbin, S.; Gazzeri, S. Loss of Histone H4K20 Trimethylation Occurs in Preneoplasia and Influences Prognosis of Non–Small Cell Lung Cancer. Clin. Cancer Res. 2008, 14, 7237–7245. [Google Scholar] [CrossRef]
- Barlési, F.; Giaccone, G.; Gallegos-Ruiz, M.I.; Loundou, A.; Span, S.W.; Lefesvre, P.; Kruyt, F.A.E.; Rodriguez, J.A. Global Histone Modifications Predict Prognosis of Resected Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2007, 25, 4358–4364. [Google Scholar] [CrossRef] [PubMed]
- Seligson, D.B.; Horvath, S.; McBrian, M.A.; Mah, V.; Yu, H.; Tze, S.; Wang, Q.; Chia, D.; Goodglick, L.; Kurdistani, S.K. Global Levels of Histone Modifications Predict Prognosis in Different Cancers. Am. J. Pathol. 2009, 174, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Yamakawa, M.; Kimura, S.; Usuba, O.; Toyono, M. Expression of Acetylated and Dimethylated Histone H3 in Colorectal Cancer. Dig. Surg. 2013, 30, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Karczmarski, J.; Rubel, T.; Paziewska, A.; Mikula, M.; Bujko, M.; Kober, P.; Dadlez, M.; Ostrowski, J. Histone H3 Lysine 27 Acetylation Is Altered in Colon Cancer. Clin. Proteomics 2014, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.J.; Zhou, X.; Zheng, J.H.; Lu, M.D.; Nie, J.Y.; Yang, X.J.; Zheng, Z.Q. Histone Acetyltransferases and Deacetylases: Molecular and Clinical Implications to Gastrointestinal Carcinogenesis. Acta Biochim. Biophys. Sin. 2012, 44, 80–91. [Google Scholar] [CrossRef]
- Bardhan, K.; Paschall, A.V.; Yang, D.; Chen, M.R.; Simon, P.S.; Bhutia, Y.D.; Martin, P.M.; Thangaraju, M.; Browning, D.D.; Ganapathy, V.; et al. IFNγ Induces DNA Methylation-Silenced GPR109A Expression via PSTAT1/P300 and H3K18 Acetylation in Colon Cancer. Cancer Immunol. Res. 2015, 3, 795–805. [Google Scholar] [CrossRef]
- Tamagawa, H.; Oshima, T.; Shiozawa, M.; Morinaga, S.; Nakamura, Y.; Yoshihara, M.; Sakuma, Y.; Kameda, Y.; Akaike, M.; Masuda, M.; et al. The Global Histone Modification Pattern Correlates with Overall Survival in Metachronous Liver Metastasis of Colorectal Cancer. Oncol. Rep. 2012, 27, 637–642. [Google Scholar] [CrossRef][Green Version]
- Ashktorab, H.; Belgrave, K.; Hosseinkhah, F.; Brim, H.; Nouraie, M.; Takkikto, M.; Hewitt, S.; Lee, E.L.; Dashwood, R.H.; Smoot, D. Global Histone H4 Acetylation and HDAC2 Expression in Colon Adenoma and Carcinoma. Dig. Dis. Sci. 2009, 54, 2109–2117. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The Role of MicroRNAs in Human Cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Parikh, A.; Lee, C.; Joseph, P.; Marchini, S.; Baccarini, A.; Kolev, V.; Romualdi, C.; Fruscio, R.; Shah, H.; Wang, F.; et al. MicroRNA-181a Has a Critical Role in Ovarian Cancer Progression through the Regulation of the Epithelial–Mesenchymal Transition. Nat. Commun. 2014, 5, 2977. [Google Scholar] [CrossRef]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 Family Reverts Aberrant Methylation in Lung Cancer by Targeting DNA Methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.A.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b Induces Global DNA Hypomethylation and Tumor Suppressor Gene Reexpression in Acute Myeloid Leukemia by Targeting Directly DNMT3A and 3B and Indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar] [CrossRef]
- Ng, E.K.O.; Tsang, W.P.; Ng, S.S.M.; Jin, H.C.; Yu, J.; Li, J.J.; Röcken, C.; Ebert, M.P.A.; Kwok, T.T.; Sung, J.J.Y. MicroRNA-143 Targets DNA Methyltransferases 3A in Colorectal Cancer. Br. J. Cancer 2009, 101, 699–706. [Google Scholar] [CrossRef]
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.-T.; Xu, Q.; Li, W.; Lu, B.; et al. A Regulatory Circuit of MiR-148a/152 and DNMT1 in Modulating Cell Transformation and Tumor Angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2013, 5, 3–13. [Google Scholar] [CrossRef]
- Zhu, A.; Xia, J.; Zuo, J.; Jin, S.; Zhou, H.; Yao, L.; Huang, H.; Han, Z. MicroRNA-148a Is Silenced by Hypermethylation and Interacts with DNA Methyltransferase 1 in Gastric Cancer. Med. Oncol. 2012, 29, 2701–2709. [Google Scholar] [CrossRef]
- Le, F.; Luo, P.; Yang, Q.O.; Zhong, X.M. MiR-181a Promotes Growth of Thyroid Cancer Cells by Targeting Tumor Suppressor RB1. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5638–5647. [Google Scholar] [CrossRef]
- Armstrong, C.M.; Liu, C.; Lou, W.; Lombard, A.P.; Evans, C.P.; Gao, A.C. MicroRNA-181a Promotes Docetaxel Resistance in Prostate Cancer Cells. Prostate 2017, 77, 1020–1028. [Google Scholar] [CrossRef]
- Patel, N.; Garikapati, K.R.; Ramaiah, M.J.; Polavarapu, K.K.; Bhadra, U.; Bhadra, M.P. MiR-15a/MiR-16 Induces Mitochondrial Dependent Apoptosis in Breast Cancer Cells by Suppressing Oncogene BMI1. Life Sci. 2016, 164, 60–70. [Google Scholar] [CrossRef]
- Frixa, T.; Donzelli, S.; Blandino, G. Oncogenic MicroRNAs: Key Players in Malignant Transformation. Cancers 2015, 7, 2466–2485. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. MicroRNAs as Oncogenes and Tumor Suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A MicroRNA Polycistron as a Potential Human Oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Kaller, M.; Hünten, S.; Siemens, H.; Hermeking, H. Analysis of the P53/MicroRNA Network in Cancer. Adv. Exp. Med. Biol. 2022, 1385, 187–228. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, X.; Lim, L.P.; De Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A MicroRNA Component of the P53 Tumour Suppressor Network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef]
- Cao, L.; Yang, X.; Chen, Y.; Zhang, D.; Jiang, X.-F.; Xue, P. Exosomal MiR-21 Regulates the TETs/PTENp1/PTEN Pathway to Promote Hepatocellular Carcinoma Growth. Mol. Cancer 2019, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Henson, R.; Wehbe–Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Balatti, V.; Croce, C.M. BCL2 and MiR-15/16: From Gene Discovery to Treatment. Cell Death Differ. 2018, 25, 21–26. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. MiR-15 and MiR-16 Induce Apoptosis by Targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Oshi, M.; Huyser, M.R.; Yan, L.; Fukada, M.; Matsuhashi, N.; Futamura, M.; Akao, Y.; Yoshida, K.; Takabe, K. Low Expression of MiR-29a Is Associated with Aggressive Biology and Worse Survival in Gastric Cancer. Sci. Rep. 2021, 11, 14134. [Google Scholar] [CrossRef]
- Roderburg, C.; Urban, G.-W.; Bettermann, K.; Vucur, M.; Zimmermann, H.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.; Castoldi, M.; et al. Micro-RNA Profiling Reveals a Role for MiR-29 in Human and Murine Liver Fibrosis. Hepatology 2011, 53, 209–218. [Google Scholar] [CrossRef]
- Van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of MicroRNAs after Myocardial Infarction Reveals a Role of MiR-29 in Cardiac Fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef]
- Boon, R.A.; Seeger, T.; Heydt, S.; Fischer, A.; Hergenreider, E.; Horrevoets, A.J.G.; Vinciguerra, M.; Rosenthal, N.; Sciacca, S.; Pilato, M.; et al. MicroRNA-29 in Aortic Dilation: Implications for Aneurysm Formation. Circ. Res. 2011, 109, 1115–1119. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, G.; Wu, J.-H.; Jiang, C.-P. Diverse Roles of MiR-29 in Cancer (Review). Oncol. Rep. 2014, 31, 1509–1516. [Google Scholar] [CrossRef]
- Watamura, S.E. Endocrine System. In Encyclopedia of Infant and Early Childhood Development; Elsevier: Amsterdam, The Netherlands, 2008; pp. 450–459. ISBN 9781496320711. [Google Scholar]
- Manotas, M.C.; González, D.M.; Céspedes, C.; Forero, C.; Rojas Moreno, A.P. Genetic and Epigenetic Control of Puberty. Sex. Dev. 2022, 16, 1–10. [Google Scholar] [CrossRef]
- Gluckman, P.D.; Hanson, M.A.; Buklijas, T.; Low, F.M.; Beedle, A.S. Epigenetic Mechanisms That Underpin Metabolic and Cardiovascular Diseases. Nat. Rev. Endocrinol. 2009, 5, 401–408. [Google Scholar] [CrossRef]
- Roy, A.; Palli, S.R. Epigenetic Modifications Acetylation and Deacetylation Play Important Roles in Juvenile Hormone Action. BMC Genom. 2018, 19, 934. [Google Scholar] [CrossRef]
- Lomniczi, A.; Ojeda, S.R. The Emerging Role of Epigenetics in the Regulation of Female Puberty. Endocr. Dev. 2016, 29, 1–16. [Google Scholar] [CrossRef]
- Kong, S.; Peng, Y.; Chen, W.; Ma, X.; Wei, Y.; Zhao, Y.; Li, R.; Qiao, J.; Yan, L. Epigenetic Consequences of Hormonal Interactions between Opposite-sex Twin Fetuses. Clin. Transl. Med. 2020, 10, e234. [Google Scholar] [CrossRef]
- Shepherd, R.; Bretherton, I.; Pang, K.; Mansell, T.; Czajko, A.; Kim, B.; Vlahos, A.; Zajac, J.D.; Saffery, R.; Cheung, A.; et al. Gender-Affirming Hormone Therapy Induces Specific DNA Methylation Changes in Blood. Clin. Epigenet. 2022, 14, 24. [Google Scholar] [CrossRef]
- Vazquez, M.J.; Daza-Dueñas, S.; Tena-Sempere, M. Emerging Roles of Epigenetics in the Control of Reproductive Function: Focus on Central Neuroendocrine Mechanisms. J. Endocr. Soc. 2021, 5, bvab152. [Google Scholar] [CrossRef]
- Holst, J.P.; Soldin, O.P.; Guo, T.; Soldin, S.J. Steroid Hormones: Relevance and Measurement in the Clinical Laboratory. Clin. Lab. Med. 2004, 24, 105–118. [Google Scholar] [CrossRef]
- Sever, R.; Glass, C.K. Signaling by Nuclear Receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a016709. [Google Scholar] [CrossRef]
- Nugent, B.M.; McCarthy, M.M. Epigenetic Underpinnings of Developmental Sex Differences in the Brain. Neuroendocrinology 2011, 93, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhang, Z.; Shen, W.J.; Azhar, S. Cellular Cholesterol Delivery, Intracellular Processing and Utilization for Biosynthesis of Steroid Hormones. Nutr. Metab. 2010, 7, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Kharwanlang, B.; Sharma, R. Glucocorticoid Hormones in Aging BT-Hormones in Ageing and Longevity; Rattan, S., Sharma, R., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 37–55. ISBN 978-3-319-63001-4. [Google Scholar]
- McGowan, P.O.; Sasaki, A.; D’Alessio, A.C.; Dymov, S.; Labonté, B.; Szyf, M.; Turecki, G.; Meaney, M.J. Epigenetic Regulation of the Glucocorticoid Receptor in Human Brain Associates with Childhood Abuse. Nat. Neurosci. 2009, 12, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Sheinkopf, S.J.; Righi, G.; Marsit, C.J.; Lester, B.M. Methylation of the Glucocorticoid Receptor (NR3C1) in Placenta Is Associated with Infant Cry Acoustics. Front. Behav. Neurosci. 2016, 10, 100. [Google Scholar] [CrossRef]
- Lv, J.; Ma, Q.; Dasgupta, C.; Xu, Z.; Zhang, L. Antenatal Hypoxia and Programming of Glucocorticoid Receptor Expression in the Adult Rat Heart. Front. Physiol. 2019, 10, 323. [Google Scholar] [CrossRef]
- Auger, C.J.; Coss, D.; Auger, A.P.; Forbes-Lorman, R.M. Epigenetic Control of Vasopressin Expression Is Maintained by Steroid Hormones in the Adult Male Rat Brain. Proc. Natl. Acad. Sci. USA 2011, 108, 4242–4247. [Google Scholar] [CrossRef]
- Zhou, Y.; Gu, B.; Brichant, G.; Singh, J.P.; Yang, H.; Chang, H.; Zhao, Y.; Cheng, C.; Liu, Z.W.; Alderman, M.H.; et al. The Steroid Hormone Estriol (E3) Regulates Epigenetic Programming of Fetal Mouse Brain and Reproductive Tract. BMC Biol. 2022, 20, 93. [Google Scholar] [CrossRef]
- Houshdaran, S.; Oke, A.B.; Fung, J.C.; Vo, K.C.; Nezhat, C.; Giudice, L.C. Steroid Hormones Regulate Genome-Wide Epigenetic Programming and Gene Transcription in Human Endometrial Cells with Marked Aberrancies in Endometriosis. PLoS Genet. 2020, 16, e1008601. [Google Scholar] [CrossRef]
- Fröhlich, E.; Wahl, R. Physiological Role and Use of Thyroid Hormone Metabolites-Potential Utility in COVID-19 Patients. Front. Endocrinol. 2021, 12, 587518. [Google Scholar] [CrossRef]
- Ren, B.; Zhu, Y. A New Perspective on Thyroid Hormones: Crosstalk with Reproductive Hormones in Females. Int. J. Mol. Sci. 2022, 23, 2708. [Google Scholar] [CrossRef]
- Pirahanchi, Y.; Jialal, I. Physiology, Thyroid Stimulating Hormone (TSH); StatPearls Publishing: Treasure Island, FL, USA, 2018. [Google Scholar]
- Liu, Y.-Y.; Milanesi, A.; Brent, G.A. Thyroid Hormones. In Hormonal Signaling in Biology and Medicine; Elsevier: Amsterdam, The Netherlands, 2020; Volume 1, pp. 487–506. ISBN 9780128138144. [Google Scholar]
- Köhrle, J. Local Activation and Inactivation of Thyroid Hormones: The Deiodinase Family. Mol. Cell. Endocrinol. 1999, 151, 103–119. [Google Scholar] [CrossRef]
- Panicker, V. Genetics of Thyroid Function and Disease. Clin. Biochem. Rev. 2011, 32, 165–175. [Google Scholar]
- Friesema, E.C.H.; Ganguly, S.; Abdalla, A.; Fox, J.E.M.; Halestrap, A.P.; Visser, T.J. Identification of Monocarboxylate Transporter 8 as a Specific Thyroid Hormone Transporter. J. Biol. Chem. 2003, 278, 40128–40135. [Google Scholar] [CrossRef]
- Pizzagalli, F.; Hagenbuch, B.; Stieger, B.; Klenk, U.; Folkers, G.; Meier, P.J. Identification of a Novel Human Organic Anion Transporting Polypeptide as a High Affinity Thyroxine Transporter. Mol. Endocrinol. 2002, 16, 2283–2296. [Google Scholar] [CrossRef]
- Kashyap, V.; Gudas, L.J. Epigenetic Regulatory Mechanisms Distinguish Retinoic Acid-Mediated Transcriptional Responses in Stem Cells and Fibroblasts. J. Biol. Chem. 2010, 285, 14534–14548. [Google Scholar] [CrossRef]
- Smith, J.A.; Fan, C.-Y.; Zou, C.; Bodenner, D.; Kokoska, M.S. Methylation Status of Genes in Papillary Thyroid Carcinoma. Arch. Otolaryngol. Neck Surg. 2007, 133, 1006. [Google Scholar] [CrossRef]
- Pozzi, S.; Rossetti, S.; Bistulfi, G.; Sacchi, N. RAR-Mediated Epigenetic Control of the Cytochrome P450 Cyp26a1 in Embryocarcinoma Cells. Oncogene 2006, 25, 1400–1407. [Google Scholar] [CrossRef]
- Shimi, G.; Pourvali, K.; Ghorbani, A.; Nooshin, S.; Zare Karizi, S.; Iranirad, R.; Zand, H. Alterations of DNA Methylation and Expression of Genes Related to Thyroid Hormone Metabolism in Colon Epithelium of Obese Patients. BMC Med. Genom. 2022, 15, 229. [Google Scholar] [CrossRef]
- Kim, S.; Cho, Y.H.; Won, S.; Ku, J.-L.; Moon, H.-B.; Park, J.; Choi, G.; Kim, S.; Choi, K. Maternal Exposures to Persistent Organic Pollutants Are Associated with DNA Methylation of Thyroid Hormone-Related Genes in Placenta Differently by Infant Sex. Environ. Int. 2019, 130, 104956. [Google Scholar] [CrossRef]
- Li, L.; Ying, Y.; Zhang, C.; Wang, W.; Li, Y.; Feng, Y.; Liang, J.; Song, H.; Wang, Y. Bisphenol A Exposure and Risk of Thyroid Nodules in Chinese Women: A Case-Control Study. Environ. Int. 2019, 126, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Pitto, L.; Gorini, F.; Bianchi, F.; Guzzolino, E. New Insights into Mechanisms of Endocrine-Disrupting Chemicals in Thyroid Diseases: The Epigenetic Way. Int. J. Environ. Res. Public Health 2020, 17, 7787. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Marinelli, B.; Apostoli, P.; Bonzini, M.; Nordio, F.; Hoxha, M.; Pegoraro, V.; Motta, V.; Tarantini, L.; Cantone, L.; et al. Exposure to Metal-Rich Particulate Matter Modifies the Expression of Candidate MicroRNAs in Peripheral Blood Leukocytes. Environ. Health Perspect. 2010, 118, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Wang, D.; Baccarelli, A. Environmental Chemicals and MicroRNAs. Mutat. Res. Mol. Mech. Mutagen. 2011, 714, 105–112. [Google Scholar] [CrossRef]
- Vrijens, K.; Bollati, V.; Nawrot, T.S. MicroRNAs as Potential Signatures of Environmental Exposure or Effect: A Systematic Review. Environ. Health Perspect. 2015, 123, 399–411. [Google Scholar] [CrossRef]
- Pacifico, F.; Crescenzi, E.; Mellone, S.; Iannetti, A.; Porrino, N.; Liguoro, D.; Moscato, F.; Grieco, M.; Formisano, S.; Leonardi, A. Nuclear Factor-Κb Contributes to Anaplastic Thyroid Carcinomas through up-Regulation of MiR-146a. J. Clin. Endocrinol. Metab. 2010, 95, 1421–1430. [Google Scholar] [CrossRef]
- Kołodziejski, P.A.; Pruszyńska-Oszmałek, E.; Wojciechowicz, T.; Sassek, M.; Leciejewska, N.; Jasaszwili, M.; Billert, M.; Małek, E.; Szczepankiewicz, D.; Misiewicz-Mielnik, M.; et al. The Role of Peptide Hormones Discovered in the 21st Century in the Regulation of Adipose Tissue Functions. Genes 2021, 12, 756. [Google Scholar] [CrossRef]
- Ahmed, S.A.H.; Ansari, S.A.; Mensah-Brown, E.P.K.; Emerald, B.S. The Role of DNA Methylation in the Pathogenesis of Type 2 Diabetes Mellitus. Clin. Epigenet. 2020, 12, 104. [Google Scholar] [CrossRef]
- Bansal, A.; Pinney, S.E. DNA Methylation and Its Role in the Pathogenesis of Diabetes. Pediatr. Diabetes 2017, 18, 167–177. [Google Scholar] [CrossRef]
- Tang, X.; Muniappan, L.; Tang, G.; Özcan, S. Identification of Glucose-Regulated MiRNAs from Pancreatic β Cells Reveals a Role for MiR-30d in Insulin Transcription. RNA 2009, 15, 287–293. [Google Scholar] [CrossRef]
- Madogwe, E.; Tanwar, D.K.; Taibi, M.; Schuermann, Y.; St-Yves, A.; Duggavathi, R. Global Analysis of FSH-regulated Gene Expression and Histone Modification in Mouse Granulosa Cells. Mol. Reprod. Dev. 2020, 87, 1082–1096. [Google Scholar] [CrossRef]
- Luján, S.; Caroppo, E.; Niederberger, C.; Arce, J.-C.; Sadler-Riggleman, I.; Beck, D.; Nilsson, E.; Skinner, M.K. Sperm DNA Methylation Epimutation Biomarkers for Male Infertility and FSH Therapeutic Responsiveness. Sci. Rep. 2019, 9, 16786. [Google Scholar] [CrossRef]
- Małodobra-Mazur, M.; Cierzniak, A.; Myszczyszyn, A.; Kaliszewski, K.; Dobosz, T. Histone Modifications Influence the Insulin-Signaling Genes and Are Related to Insulin Resistance in Human Adipocytes. Int. J. Biochem. Cell Biol. 2021, 137, 106031. [Google Scholar] [CrossRef]
- Emamgholipour, S.; Ebrahimi, R.; Bahiraee, A.; Niazpour, F.; Meshkani, R. Acetylation and Insulin Resistance: A Focus on Metabolic and Mitogenic Cascades of Insulin Signaling. Crit. Rev. Clin. Lab. Sci. 2020, 57, 196–214. [Google Scholar] [CrossRef]
- Castellano-Castillo, D.; Denechaud, P.-D.; Fajas, L.; Moreno-Indias, I.; Oliva-Olivera, W.; Tinahones, F.; Queipo-Ortuño, M.I.; Cardona, F. Human Adipose Tissue H3K4me3 Histone Mark in Adipogenic, Lipid Metabolism and Inflammatory Genes Is Positively Associated with BMI and HOMA-IR. PLoS ONE 2019, 14, e0215083. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Sun, Y.; Li, R.; Deng, J.; Deng, J. DNA Methylation and Histone Deacetylation Regulating Insulin Sensitivity Due to Chronic Cold Exposure. Cryobiology 2017, 74, 36–42. [Google Scholar] [CrossRef]
- La Merrill, M.A.; Vandenberg, L.N.; Smith, M.T.; Goodson, W.; Browne, P.; Patisaul, H.B.; Guyton, K.Z.; Kortenkamp, A.; Cogliano, V.J.; Woodruff, T.J.; et al. Consensus on the Key Characteristics of Endocrine-Disrupting Chemicals as a Basis for Hazard Identification. Nat. Rev. Endocrinol. 2020, 16, 45–57. [Google Scholar] [CrossRef]
- Axelstad, M.; Hass, U.; Scholze, M.; Christiansen, S.; Kortenkamp, A.; Boberg, J. EDC IMPACT: Reduced Sperm Counts in Rats Exposed to Human Relevant Mixtures of Endocrine Disrupters. Endocr. Connect. 2018, 7, 139–148. [Google Scholar] [CrossRef]
- Johansson, H.K.L.; Svingen, T.; Fowler, P.A.; Vinggaard, A.M.; Boberg, J. Environmental Influences on Ovarian Dysgenesis-Developmental Windows Sensitive to Chemical Exposures. Nat. Rev. Endocrinol. 2017, 13, 400–414. [Google Scholar] [CrossRef]
- Amano, I.; Takatsuru, Y.; Khairinisa, M.A.; Kokubo, M.; Haijima, A.; Koibuchi, N. Effects of Mild Perinatal Hypothyroidism on Cognitive Function of Adult Male Offspring. Endocrinology 2018, 159, 1910–1921. [Google Scholar] [CrossRef]
- Ghassabian, A.; Trasande, L. Disruption in Thyroid Signaling Pathway: A Mechanism for the Effect of Endocrine-Disrupting Chemicals on Child Neurodevelopment. Front. Endocrinol. 2018, 9, 204. [Google Scholar] [CrossRef] [PubMed]
- Zamora, A.N.; Marchlewicz, E.; Téllez-Rojo, M.M.; Burant, C.F.; Cantoral, A.; Song, P.X.K.; Mercado, A.; Dolinoy, D.C.; Peterson, K.E. Trimester Two Gestational Exposure to Bisphenol A and Adherence to Mediterranean Diet Are Associated with Adolescent Offspring Oxidative Stress and Metabolic Syndrome Risk in a Sex-Specific Manner. Front. Nutr. 2022, 9, 961082. [Google Scholar] [CrossRef] [PubMed]
- Abulehia, H.F.S.; Nor, N.S.M.; Sheikh Abdul Kadir, S.H. The Current Findings on the Impact of Prenatal BPA Exposure on Metabolic Parameters: In Vivo and Epidemiological Evidence. Nutrients 2022, 14, 2766. [Google Scholar] [CrossRef] [PubMed]
- Cano-Sancho, G.; Salmon, A.G.; La Merrill, M.A. Association between Exposure to p,p ′-DDT and Its Metabolite p,p ′-DDE with Obesity: Integrated Systematic Review and Meta-Analysis. Environ. Health Perspect. 2017, 125, 096002. [Google Scholar] [CrossRef]
- Alonso-Magdalena, P.; Vieira, E.; Soriano, S.; Menes, L.; Burks, D.; Quesada, I.; Nadal, A. Bisphenol A Exposure during Pregnancy Disrupts Glucose Homeostasis in Mothers and Adult Male Offspring. Environ. Health Perspect. 2010, 118, 1243–1250. [Google Scholar] [CrossRef]
- Lacouture, A.; Lafront, C.; Peillex, C.; Pelletier, M.; Audet-Walsh, É. Impacts of Endocrine-Disrupting Chemicals on Prostate Function and Cancer. Environ. Res. 2022, 204, 112085. [Google Scholar] [CrossRef]
- Wan, M.L.Y.; Co, V.A.; El-Nezami, H. Endocrine Disrupting Chemicals and Breast Cancer: A Systematic Review of Epidemiological Studies. Crit. Rev. Food Sci. Nutr. 2022, 62, 6549–6576. [Google Scholar] [CrossRef]
- Eve, L.; Fervers, B.; Le Romancer, M.; Etienne-Selloum, N. Exposure to Endocrine Disrupting Chemicals and Risk of Breast Cancer. Int. J. Mol. Sci. 2020, 21, 9139. [Google Scholar] [CrossRef]
- Buoso, E.; Masi, M.; Racchi, M.; Corsini, E. Endocrine-Disrupting Chemicals’ (EDCs) Effects on Tumour Microenvironment and Cancer Progression: Emerging Contribution of RACK1. Int. J. Mol. Sci. 2020, 21, 9229. [Google Scholar] [CrossRef]
- Nettore, I.C.; Franchini, F.; Palatucci, G.; Macchia, P.E.; Ungaro, P. Epigenetic Mechanisms of Endocrine-Disrupting Chemicals in Obesity. Biomedicines 2021, 9, 1716. [Google Scholar] [CrossRef]
- Lauretta, R.; Sansone, A.; Sansone, M.; Romanelli, F.; Appetecchia, M. Endocrine Disrupting Chemicals: Effects on Endocrine Glands. Front. Endocrinol. 2019, 10, 178. [Google Scholar] [CrossRef]
- Diamanti-Kandarakis, E.; Bourguignon, J.-P.; Giudice, L.C.; Hauser, R.; Prins, G.S.; Soto, A.M.; Zoeller, R.T.; Gore, A.C. Endocrine-Disrupting Chemicals: An Endocrine Society Scientific Statement. Endocr. Rev. 2009, 30, 293–342. [Google Scholar] [CrossRef]
- Balaguer, P.; Delfosse, V.; Grimaldi, M.; Bourguet, W. Structural and Functional Evidences for the Interactions between Nuclear Hormone Receptors and Endocrine Disruptors at Low Doses. Comptes Rendus Biol. 2017, 340, 414–420. [Google Scholar] [CrossRef]
- Monneret, C. What Is an Endocrine Disruptor? Comptes Rendus Biol. 2017, 340, 403–405. [Google Scholar] [CrossRef]
- Sweeney, M.F.; Hasan, N.; Soto, A.M.; Sonnenschein, C. Environmental Endocrine Disruptors: Effects on the Human Male Reproductive System. Rev. Endocr. Metab. Disord. 2015, 16, 341–357. [Google Scholar]
- Sikka, S.C.; Wang, R. Endocrine Disruptors and Estrogenic Effects on Male Reproductive Axis. Asian J. Androl. 2008, 10, 134–145. [Google Scholar] [CrossRef]
- Maqbool, F.; Mostafalou, S.; Bahadar, H.; Abdollahi, M. Review of Endocrine Disorders Associated with Environmental Toxicants and Possible Involved Mechanisms. Life Sci. 2016, 145, 265–273. [Google Scholar] [CrossRef]
- Murata, M.; Kang, J.-H. Bisphenol A (BPA) and Cell Signaling Pathways. Biotechnol. Adv. 2018, 36, 311–327. [Google Scholar] [CrossRef]
- Hart, R.J. The Impact of Prenatal Exposure to Bisphenol A on Male Reproductive Function. Front. Endocrinol. 2020, 11, 320. [Google Scholar] [CrossRef]
- Matuszczak, E.; Komarowska, M.D.; Debek, W.; Hermanowicz, A. The Impact of Bisphenol A on Fertility, Reproductive System, and Development: A Review of the Literature. Int. J. Endocrinol. 2019, 2019, 6–8. [Google Scholar] [CrossRef]
- Tomza-Marciniak, A.; Stępkowska, P.; Kuba, J.; Pilarczyk, B. Effect of Bisphenol A on Reproductive Processes: A Review of in Vitro, in Vivo and Epidemiological Studies. J. Appl. Toxicol. 2018, 38, 51–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qu, K.; Hai, Y.; Zhao, C. Bisphenol A (BPA) Binding on Full-length Architectures of Estrogen Receptor. J. Cell. Biochem. 2018, 119, 6784–6794. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-B.; He, Y.; Song, C.; Ke, X.; Fan, S.-J.; Peng, W.-J.; Tan, R.; Kawata, M.; Matsuda, K.-I.; Pan, B.-X.; et al. Bisphenol a Regulates the Estrogen Receptor Alpha Signaling in Developing Hippocampus of Male Rats through Estrogen Receptor. Hippocampus 2014, 24, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Gould, J.C.; Leonard, L.S.; Maness, S.C.; Wagner, B.L.; Conner, K.; Zacharewski, T.; Safe, S.; McDonnell, D.P.; Gaido, K.W. Bisphenol A Interacts with the Estrogen Receptor α in a Distinct Manner from Estradiol. Mol. Cell. Endocrinol. 1998, 142, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, P.; Romano, R.M.; Kizys, M.M.L.; Oliveira, K.C.; Kasamatsu, T.; Giannocco, G.; Chiamolera, M.I.; Dias-da-Silva, M.R.; Romano, M.A. Adult Exposure to Bisphenol A (BPA) in Wistar Rats Reduces Sperm Quality with Disruption of the Hypothalamic-Pituitary-Testicular Axis. Toxicology 2015, 329, 1–9. [Google Scholar] [CrossRef]
- Ye, L.; Zhao, B.; Hu, G.; Chu, Y.; Ge, R.-S. Inhibition of Human and Rat Testicular Steroidogenic Enzyme Activities by Bisphenol A. Toxicol. Lett. 2011, 207, 137–142. [Google Scholar] [CrossRef]
- Gao, Z.; Liu, S.; Tan, L.; Gao, X.; Fan, W.; Ding, C.; Li, M.; Tang, Z.; Shi, X.; Luo, Y.; et al. Testicular Toxicity of Bisphenol Compounds: Homeostasis Disruption of Cholesterol/Testosterone via PPARα Activation. Sci. Total Environ. 2022, 836, 155628. [Google Scholar] [CrossRef]
- Zhou, W.; Fang, F.; Zhu, W.; Chen, Z.J.; Du, Y.; Zhang, J. Bisphenol A and Ovarian Reserve among Infertile Women with Polycystic Ovarian Syndrome. Int. J. Environ. Res. Public Health 2017, 14, 18. [Google Scholar] [CrossRef]
- Hossein Rashidi, B.; Amanlou, M.; Behrouzi Lak, T.; Ghazizadeh, M.; Haghollahi, F.; Bagheri, M.; Eslami, B. The Association Between Bisphenol A and Polycystic Ovarian Syndrome: A Case-Control Study. Acta Med. Iran. 2017, 55, 759–764. [Google Scholar]
- Susiarjo, M.; Sasson, I.; Mesaros, C.; Bartolomei, M.S. Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse. PLoS Genet. 2013, 9, e1003401. [Google Scholar] [CrossRef]
- Anderson, O.S.; Nahar, M.S.; Faulk, C.; Jones, T.R.; Liao, C.; Kannan, K.; Weinhouse, C.; Rozek, L.S.; Dolinoy, D.C. Epigenetic Responses Following Maternal Dietary Exposure to Physiologically Relevant Levels of Bisphenol A. Environ. Mol. Mutagen. 2012, 53, 334–342. [Google Scholar] [CrossRef]
- Hatch, E.E.; Troisi, R.; Palmer, J.R.; Wise, L.A.; Titus, L.; Strohsnitter, W.C.; Ricker, W.; Hyer, M.; Hoover, R.N. Prenatal Diethylstilbestrol Exposure and Risk of Obesity in Adult Women. J. Dev. Orig. Health Dis. 2015, 6, 201–207. [Google Scholar] [CrossRef]
- Doherty, L.F.; Bromer, J.G.; Zhou, Y.; Aldad, T.S.; Taylor, H.S. In Utero Exposure to Diethylstilbestrol (DES) or Bisphenol-A (BPA) Increases EZH2 Expression in the Mammary Gland: An Epigenetic Mechanism Linking Endocrine Disruptors to Breast Cancer. Horm. Cancer 2010, 1, 146–155. [Google Scholar] [CrossRef]
- Harlid, S.; Xu, Z.; Panduri, V.; D’Aloisio, A.A.; DeRoo, L.A.; Sandler, D.P.; Taylor, J.A. In Utero Exposure to Diethylstilbestrol and Blood DNA Methylation in Women Ages 40–59 Years from the Sister Study. PLoS ONE 2015, 10, e0118757. [Google Scholar] [CrossRef]
- Jefferson, T.B.; Wang, T.; Jefferson, W.N.; Li, Y.; Hamilton, K.J.; Wade, P.A.; Williams, C.J.; Korach, K.S. Multiple Tissue-Specific Epigenetic Alterations Regulate Persistent Gene Expression Changes Following Developmental DES Exposure in Mouse Reproductive Tissues. Epigenetics 2022. [Google Scholar] [CrossRef]
- Li, Y.; Hamilton, K.J.; Wang, T.; Coons, L.A.; Jefferson, W.N.; Li, R.; Wang, Y.; Grimm, S.A.; Ramsey, J.T.; Liu, L.; et al. DNA Methylation and Transcriptome Aberrations Mediated by ERα in Mouse Seminal Vesicles Following Developmental DES Exposure. Proc. Natl. Acad. Sci. USA 2018, 115, E4189–E4198. [Google Scholar] [CrossRef]
- Feroe, A.; Broene, R.; Albuquerque, D.; Ruiz, P. Endocrine Disrupting Chemicals, Transgenerational Epigenetics and Metabolic Diseases. EC Endocrinol. Metab. Res. 2017, 21, 31–51. [Google Scholar]
- Timokhina, E.P.; Yaglov, V.V.; Nazimova, S.V. Dichlorodiphenyltrichloroethane and the Adrenal Gland: From Toxicity to Endocrine Disruption. Toxics 2021, 9, 243. [Google Scholar] [CrossRef]
- Tilghman, S.L.; Bratton, M.R.; Segar, H.C.; Martin, E.C.; Rhodes, L.V.; Li, M.; McLachlan, J.A.; Wiese, T.E.; Nephew, K.P.; Burow, M.E. Endocrine Disruptor Regulation of MicroRNA Expression in Breast Carcinoma Cells. PLoS ONE 2012, 7, e32754. [Google Scholar] [CrossRef]
- Skinner, M.K.; Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Haque, M.; Nilsson, E.E. Ancestral Dichlorodiphenyltrichloroethane (DDT) Exposure Promotes Epigenetic Transgenerational Inheritance of Obesity. BMC Med. 2013, 11, 228. [Google Scholar] [CrossRef]
- Nilsson, E.E.; Sadler-Riggleman, I.; Skinner, M.K. Environmentally Induced Epigenetic Transgenerational Inheritance of Disease. Environ. Epigenet. 2018, 4, dvy016. [Google Scholar] [CrossRef] [PubMed]
- Moreira, S.; Pereira, S.C.; Seco-Rovira, V.; Oliveira, P.F.; Alves, M.G.; Pereira, M. de L. Pesticides and Male Fertility: A Dangerous Crosstalk. Metabolites 2021, 11, 799. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, M.; Smith, A.D.; Swaminathan, H.; Sangngern, P.; Douglas, A.; Horsager, A.; Carrell, D.T.; Uren, P.J. Establishing a Stable, Repeatable Platform for Measuring Changes in Sperm DNA Methylation. Clin. Epigenet. 2018, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Zhao, P.; Liu, J.; Zhang, J.; Zhang, J.; Wang, Y.; Wu, L.; Song, M.; Wang, W. Novel Epigenomic Biomarkers of Male Infertility Identified by Methylation Patterns of CpG Sites Within Imprinting Control Regions of H19 and SNRPN Genes. OMICS J. Integr. Biol. 2018, 22, 354–364. [Google Scholar] [CrossRef]
- Jenkins, T.G.; Aston, K.I.; Meyer, T.D.; Hotaling, J.M.; Shamsi, M.B.; Johnstone, E.B.; Cox, K.J.; Stanford, J.B.; Porucznik, C.A.; Carrell, D.T. Decreased Fecundity and Sperm DNA Methylation Patterns. Fertil. Steril. 2016, 105, 51–57.e3. [Google Scholar] [CrossRef]
- Varshavsky, J.R.; Morello-Frosch, R.; Woodruff, T.J.; Zota, A.R. Dietary Sources of Cumulative Phthalates Exposure among the U.S. General Population in NHANES 2005–2014. Environ. Int. 2018, 115, 417–429. [Google Scholar] [CrossRef]
- Viñas, P.; Campillo, N.; Pastor-Belda, M.; Oller, A.; Hernández-Córdoba, M. Determination of Phthalate Esters in Cleaning and Personal Care Products by Dispersive Liquid–Liquid Microextraction and Liquid Chromatography–Tandem Mass Spectrometry. J. Chromatogr. A 2015, 1376, 18–25. [Google Scholar] [CrossRef]
- Wójtowicz, A.K.; Sitarz-Głownia, A.M.; Szczęsna, M.; Szychowski, K.A. The Action of Di-(2-Ethylhexyl) Phthalate (DEHP) in Mouse Cerebral Cells Involves an Impairment in Aryl Hydrocarbon Receptor (AhR) Signaling. Neurotox. Res. 2019, 35, 183–195. [Google Scholar] [CrossRef]
- Nassan, F.L.; Korevaar, T.I.M.; Coull, B.A.; Skakkebæk, N.E.; Krawetz, S.A.; Estill, M.; Hait, E.J.; Korzenik, J.R.; Ford, J.B.; De Poortere, R.A.; et al. Dibutyl-Phthalate Exposure from Mesalamine Medications and Serum Thyroid Hormones in Men. Int. J. Hyg. Environ. Health 2019, 222, 101–110. [Google Scholar] [CrossRef]
- Wójtowicz, A.K.; Szychowski, K.A.; Wnuk, A.; Kajta, M. Dibutyl Phthalate (DBP)-Induced Apoptosis and Neurotoxicity Are Mediated via the Aryl Hydrocarbon Receptor (AhR) but Not by Estrogen Receptor Alpha (ERα), Estrogen Receptor Beta (ERβ), or Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) in Mouse C. Neurotox. Res. 2017, 31, 77–89. [Google Scholar] [CrossRef]
- Boas, M.; Frederiksen, H.; Feldt-Rasmussen, U.; Skakkebæk, N.E.; Hegedüs, L.; Hilsted, L.; Juul, A.; Main, K.M. Childhood Exposure to Phthalates: Associations with Thyroid Function, Insulin-like Growth Factor I, and Growth. Environ. Health Perspect. 2010, 118, 1458–1464. [Google Scholar] [CrossRef]
- Meruvu, S.; Zhang, J.; Choudhury, M. Butyl Benzyl Phthalate Promotes Adipogenesis in 3T3-L1 Cells via the MiRNA-34a-5p Signaling Pathway in the Absence of Exogenous Adipogenic Stimuli. Chem. Res. Toxicol. 2021, 34, 2251–2260. [Google Scholar] [CrossRef]
- Zhang, J.; Choudhury, M. Benzyl Butyl Phthalate Induced Early LncRNA H19 Regulation in C3H10T1/2 Stem Cell Line. Chem. Res. Toxicol. 2021, 34, 54–62. [Google Scholar] [CrossRef]
- Deng, T.; Du, Y.; Wang, Y.; Teng, X.; Hua, X.; Yuan, X.; Yao, Y.; Guo, N.; Li, Y. The Associations of Urinary Phthalate Metabolites with the Intermediate and Pregnancy Outcomes of Women Receiving IVF/ICSI Treatments: A Prospective Single-Center Study. Ecotoxicol. Environ. Saf. 2020, 188, 109884. [Google Scholar] [CrossRef]
- Kabra, A.N.; Ji, M.-K.; Choi, J.; Kim, J.R.; Govindwar, S.P.; Jeon, B.-H. Toxicity of Atrazine and Its Bioaccumulation and Biodegradation in a Green Microalga, Chlamydomonas Mexicana. Environ. Sci. Pollut. Res. 2014, 21, 12270–12278. [Google Scholar] [CrossRef]
- Buser, M.C.; Murray, H.E.; Scinicariello, F. Age and Sex Differences in Childhood and Adulthood Obesity Association with Phthalates: Analyses of NHANES 2007-2010. Int. J. Hyg. Environ. Health 2014, 217, 687–694. [Google Scholar] [CrossRef]
- Trasande, L.; Spanier, A.J.; Sathyanarayana, S.; Attina, T.M.; Blustein, J. Urinary Phthalates and Increased Insulin Resistance in Adolescents. Pediatrics 2013, 132, e646–e655. [Google Scholar] [CrossRef]
- Lee, D.W.; Lim, H.M.; Lee, J.Y.; Min, K.B.; Shin, C.H.; Lee, Y.A.; Hong, Y.C. Prenatal Exposure to Phthalate and Decreased Body Mass Index of Children: A Systematic Review and Meta-Analysis. Sci. Rep. 2022, 12, 8961. [Google Scholar] [CrossRef]
- Dong, Y.; Gao, D.; Li, Y.; Yang, Z.; Wang, X.; Chen, M.; Wang, Z.; Song, Y.; Zou, Z.; Ma, J. Effect of Childhood Phthalates Exposure on the Risk of Overweight and Obesity: A Nested Case-Control Study in China. Environ. Int. 2022, 158, 106886. [Google Scholar] [CrossRef]
- Chuang, S.C.; Chen, H.C.; Sun, C.W.; Chen, Y.A.; Wang, Y.H.; Chiang, C.J.; Chen, C.C.; Wang, S.L.; Chen, C.J.; Hsiung, C.A. Phthalate Exposure and Prostate Cancer in a Population-Based Nested Case-Control Study. Environ. Res. 2020, 181, 108902. [Google Scholar] [CrossRef]
- Yang, P.J.; Hou, M.F.; Ou-Yang, F.; Hsieh, T.H.; Lee, Y.J.; Tsai, E.M.; Wang, T.N. Association between Recurrent Breast Cancer and Phthalate Exposure Modified by Hormone Receptors and Body Mass Index. Sci. Rep. 2022, 12, 2858. [Google Scholar] [CrossRef] [PubMed]
- Rochester, J.R.; Millam, J.R. Phytoestrogens and Avian Reproduction: Exploring the Evolution and Function of Phytoestrogens and Possible Role of Plant Compounds in the Breeding Ecology of Wild Birds. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2009, 154, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Adlercreutz, H. Phytoestrogens and Breast Cancer. J. Steroid Biochem. Mol. Biol. 2002, 83, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Rietjens, I.M.C.M.; Louisse, J.; Beekmann, K. The Potential Health Effects of Dietary Phytoestrogens. Br. J. Pharmacol. 2017, 174, 1263–1280. [Google Scholar] [CrossRef]
- Domínguez-López, I.; Yago-Aragón, M.; Salas-Huetos, A.; Tresserra-Rimbau, A.; Hurtado-Barroso, S. Effects of Dietary Phytoestrogens on Hormones throughout a Human Lifespan: A Review. Nutrients 2020, 12, 2456. [Google Scholar] [CrossRef]
- Jefferson, W.N. Adult Ovarian Function Can Be Affected by High Levels of Soy. J. Nutr. 2010, 140, 2322S–2325S. [Google Scholar] [CrossRef]
- Nicholls, J.; Lasley, B.L.; Nakajima, S.T.; Setchell, K.D.R.; Schneeman, B.O. Effects of Soy Consumption on Gonadotropin Secretion and Acute Pituitary Responses to Gonadotropin-Releasing Hormone in Women. J. Nutr. 2002, 132, 708–714. [Google Scholar] [CrossRef]
- Basu, P.; Maier, C. Phytoestrogens and Breast Cancer: In Vitro Anticancer Activities of Isoflavones, Lignans, Coumestans, Stilbenes and Their Analogs and Derivatives. Biomed. Pharmacother. 2018, 107, 1648–1666. [Google Scholar] [CrossRef]
- Nguyen, M.; Osipo, C. Targeting Breast Cancer Stem Cells Using Naturally Occurring Phytoestrogens. Int. J. Mol. Sci. 2022, 23, 6813. [Google Scholar] [CrossRef]
- Lecomte, S.; Demay, F.; Ferrière, F.; Pakdel, F. Phytochemicals Targeting Estrogen Receptors: Beneficial Rather Than Adverse Effects? Int. J. Mol. Sci. 2017, 18, 1381. [Google Scholar] [CrossRef]
- Omoto, Y.; Iwase, H. Clinical Significance of Estrogen Receptor β in Breast and Prostate Cancer from Biological Aspects. Cancer Sci. 2015, 106, 337–343. [Google Scholar] [CrossRef]
- Pons, D.G.; Nadal-Serrano, M.; Blanquer-Rossello, M.M.; Sastre-Serra, J.; Oliver, J.; Roca, P. Genistein Modulates Proliferation and Mitochondrial Functionality in Breast Cancer Cells Depending on ERalpha/ERbeta Ratio. J. Cell. Biochem. 2014, 115, 949–958. [Google Scholar] [CrossRef]
- Attia, D.M.A.; Ederveen, A.G.H. Opposing Roles of ERα and ERβ in the Genesis and Progression of Adenocarcinoma in the Rat Ventral Prostate. Prostate 2012, 72, 1013–1022. [Google Scholar] [CrossRef]
- Thomas, C.; Gustafsson, J.-Å. The Different Roles of ER Subtypes in Cancer Biology and Therapy. Nat. Rev. Cancer 2011, 11, 597–608. [Google Scholar] [CrossRef]
- Adjakly, M.; Bosviel, R.; Rabiau, N.; Boiteux, J.-P.; Bignon, Y.-J.; Guy, L.; Bernard-Gallon, D. DNA Methylation and Soy Phytoestrogens: Quantitative Study in DU-145 and PC-3 Human Prostate Cancer Cell Lines. Epigenomics 2011, 3, 795–803. [Google Scholar] [CrossRef]
- Vardi, A.; Bosviel, R.; Rabiau, N.; Adjakly, M.; Satih, S.; Dechelotte, P.; Boiteux, J.-P.; Fontana, L.; Bignon, Y.-J.; Guy, L.; et al. Soy Phytoestrogens Modify DNA Methylation of GSTP1, RASSF1A, EPH2 and BRCA1 Promoter in Prostate Cancer Cells. In Vivo 2010, 24, 393–400. [Google Scholar]
- Day, J.K.; Bauer, A.M.; DesBordes, C.; Zhuang, Y.; Kim, B.; Newton, L.G.; Nehra, V.; Forsee, K.M.; MacDonald, R.S.; Besch-Williford, C.; et al. Genistein Alters Methylation Patterns in Mice. J. Nutr. 2002, 132, 2419S–2423S. [Google Scholar] [CrossRef]
- Barnes, S. The Biochemistry, Chemistry and Physiology of the Isoflavones in Soybeans and Their Food Products. Lymphat. Res. Biol. 2010, 8, 89–98. [Google Scholar] [CrossRef]
- Yan, G.-R.; Xiao, C.-L.; He, G.-W.; Yin, X.-F.; Chen, N.-P.; Cao, Y.; He, Q.-Y. Global Phosphoproteomic Effects of Natural Tyrosine Kinase Inhibitor, Genistein, on Signaling Pathways. Proteomics 2010, 10, 976–986. [Google Scholar] [CrossRef]
- Javed, Z.; Khan, K.; Herrera-Bravo, J.; Naeem, S.; Iqbal, M.J.; Sadia, H.; Qadri, Q.R.; Raza, S.; Irshad, A.; Akbar, A.; et al. Genistein as a Regulator of Signaling Pathways and MicroRNAs in Different Types of Cancers. Cancer Cell Int. 2021, 21, 388. [Google Scholar] [CrossRef]
- Pandima Devi, K.; Rajavel, T.; Daglia, M.; Nabavi, S.F.; Bishayee, A.; Nabavi, S.M. Targeting MiRNAs by Polyphenols: Novel Therapeutic Strategy for Cancer. Semin. Cancer Biol. 2017, 46, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-J.; Hsu, Y.-L.; Huang, Y.-F.; Tsai, E.-M. Molecular Mechanisms of Anticancer Effects of Phytoestrogens in Breast Cancer. Curr. Protein Pept. Sci. 2018, 19, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xiong, H.; Zhao, Z.; Luo, M.; Ju, Y.; Yang, G.; Mei, Z. Genistein Suppresses Allergic Contact Dermatitis through Regulating the MAP2K2/ERK Pathway. Food Funct. 2021, 12, 4556–4569. [Google Scholar] [CrossRef] [PubMed]
- Mukund, V. Genistein: Its Role in Breast Cancer Growth and Metastasis. Curr. Drug Metab. 2020, 21, 6–10. [Google Scholar] [CrossRef]
- Cheng, W.X.; Huang, H.; Chen, J.H.; Zhang, T.T.; Zhu, G.Y.; Zheng, Z.T.; Lin, J.T.; Hu, Y.P.; Zhang, Y.; Bai, X.L.; et al. Genistein Inhibits Angiogenesis Developed during Rheumatoid Arthritis through the IL-6/JAK2/STAT3/VEGF Signalling Pathway. J. Orthop. Transl. 2020, 22, 92–100. [Google Scholar] [CrossRef]
- Malloy, K.M.; Wang, J.; Clark, L.H.; Fang, Z.; Sun, W.; Yin, Y.; Kong, W.; Zhou, C.X.; Bae-Jump, V.L. Novasoy and Genistein Inhibit Endometrial Cancer Cell Proliferation through Disruption of the AKT/MTOR and MAPK Signaling Pathways. Am. J. Transl. Res. 2018, 10, 784–795. [Google Scholar]
- Zhang, H.; Zhao, Z.; Pang, X.; Yang, J.; Yu, H.; Zhang, Y.; Zhou, H.; Zhao, J. Genistein Protects Against Ox-LDL-Induced Inflammation Through MicroRNA-155/SOCS1-Mediated Repression of NF-ĸB Signaling Pathway in HUVECs. Inflammation 2017, 40, 1450–1459. [Google Scholar] [CrossRef]
- Pan, H.; Zhou, W.; He, W.; Liu, X.; Ding, Q.; Ling, L.; Zha, X.; Wang, S. Genistein Inhibits MDA-MB-231 Triple-Negative Breast Cancer Cell Growth by Inhibiting NF-ΚB Activity via the Notch-1 Pathway. Int. J. Mol. Med. 2012, 30, 337–343. [Google Scholar] [CrossRef]
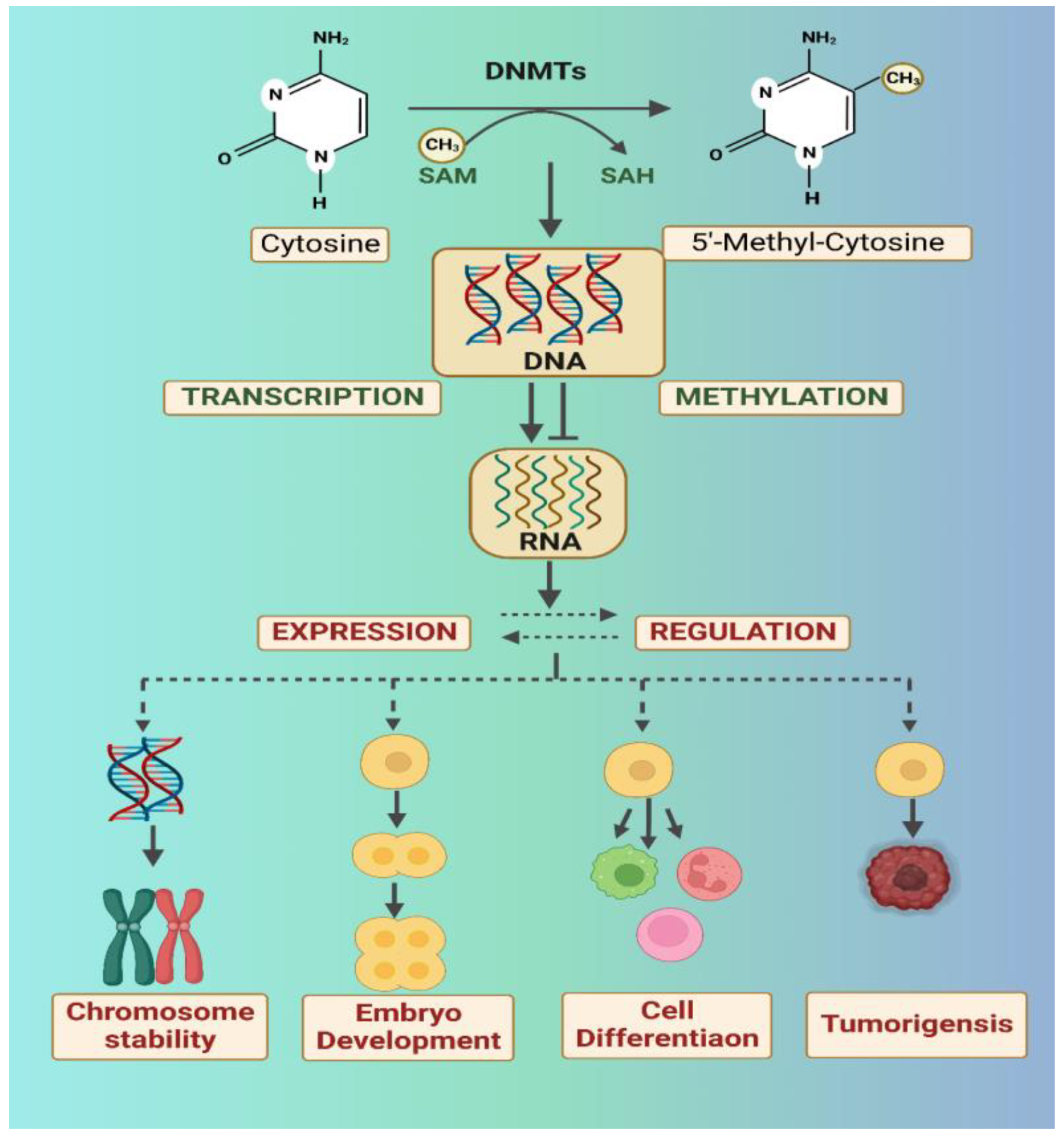
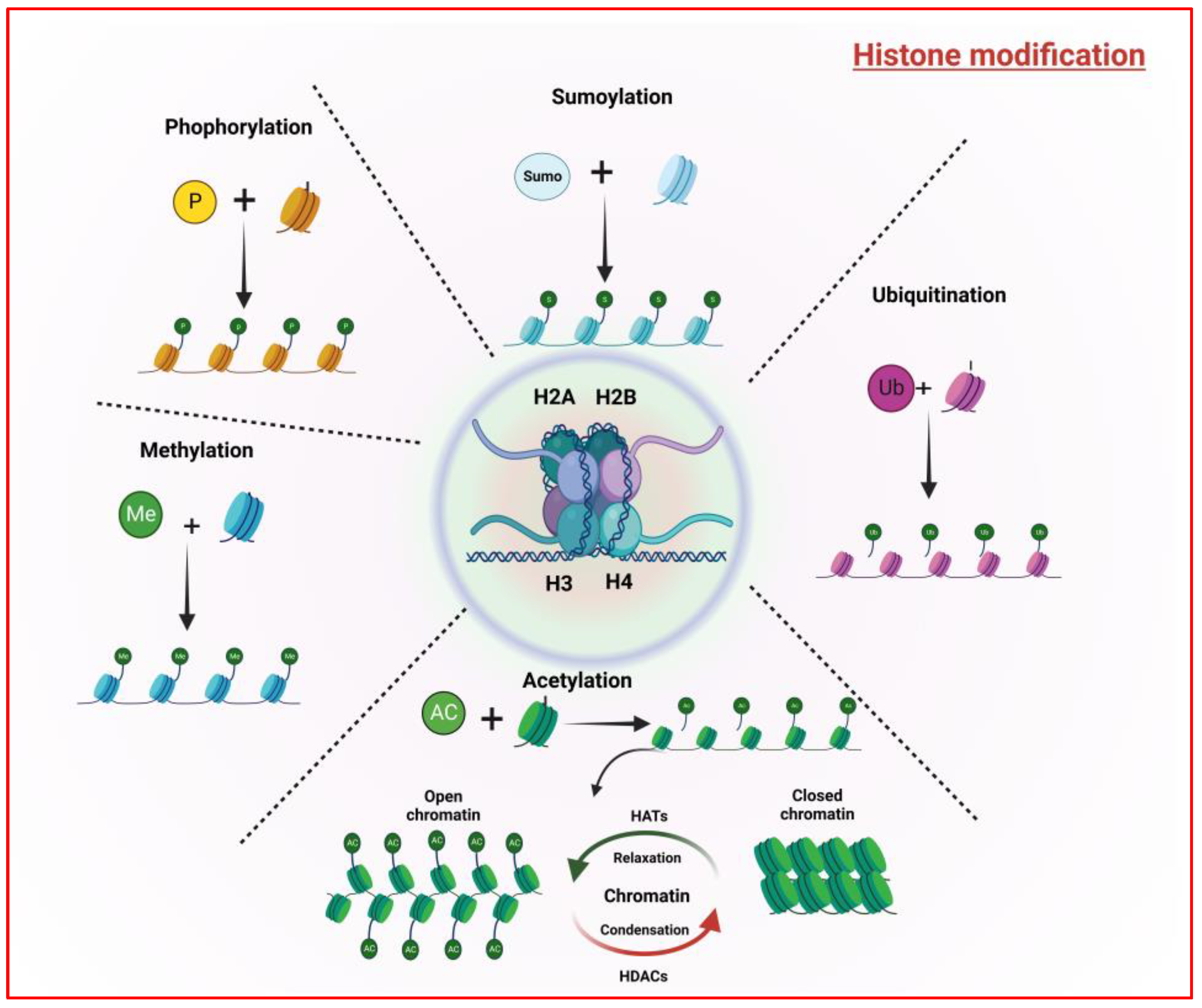
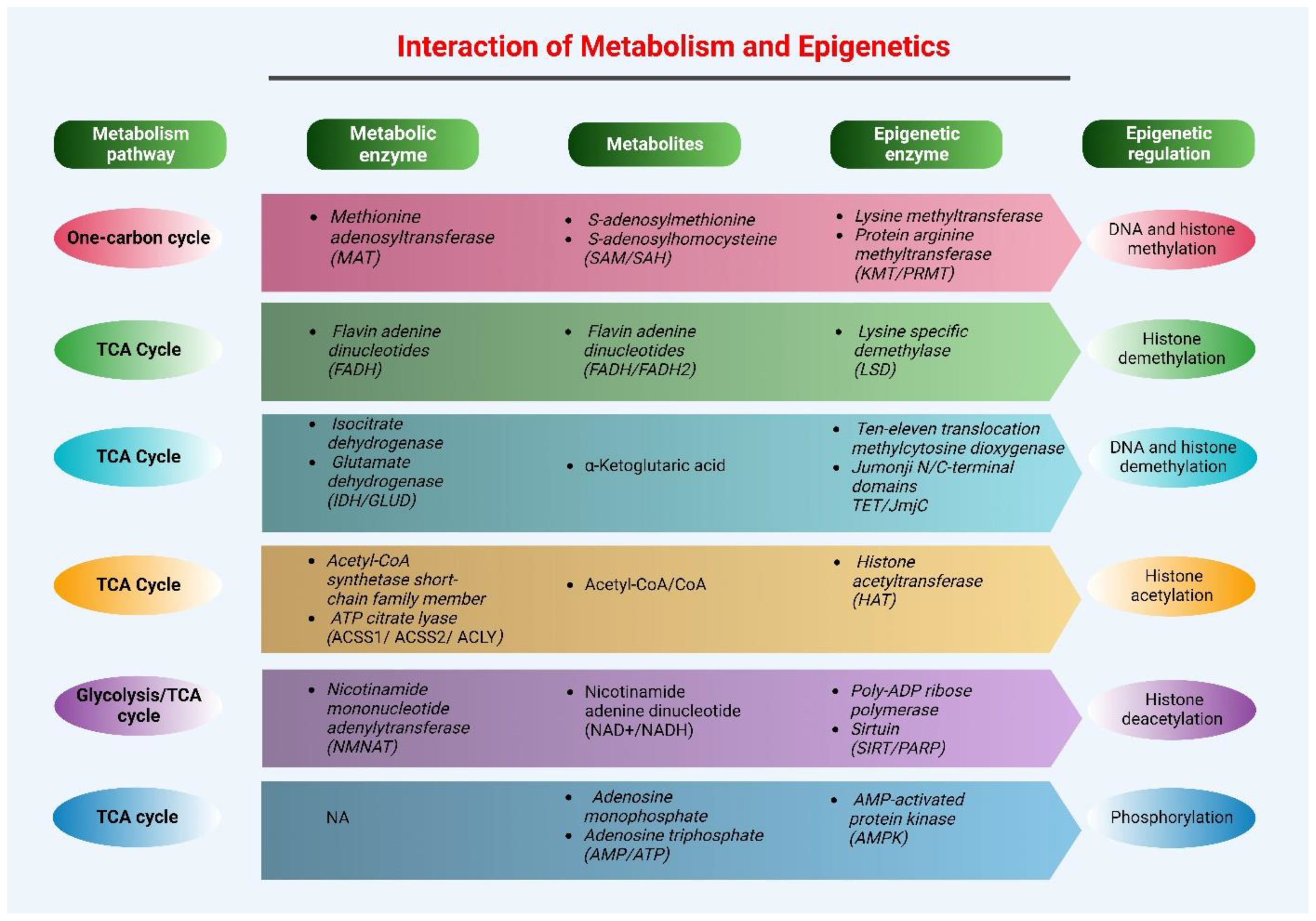

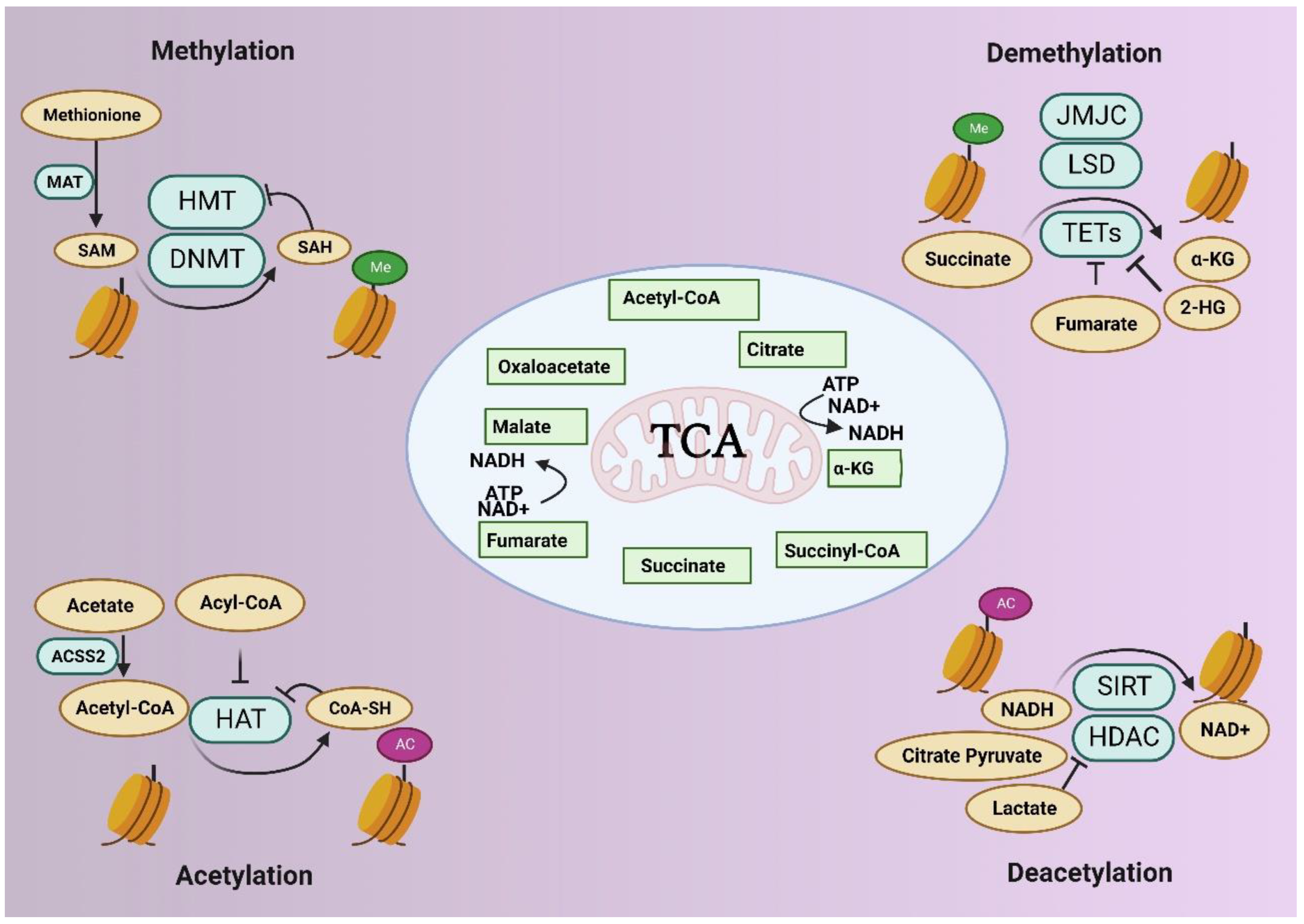
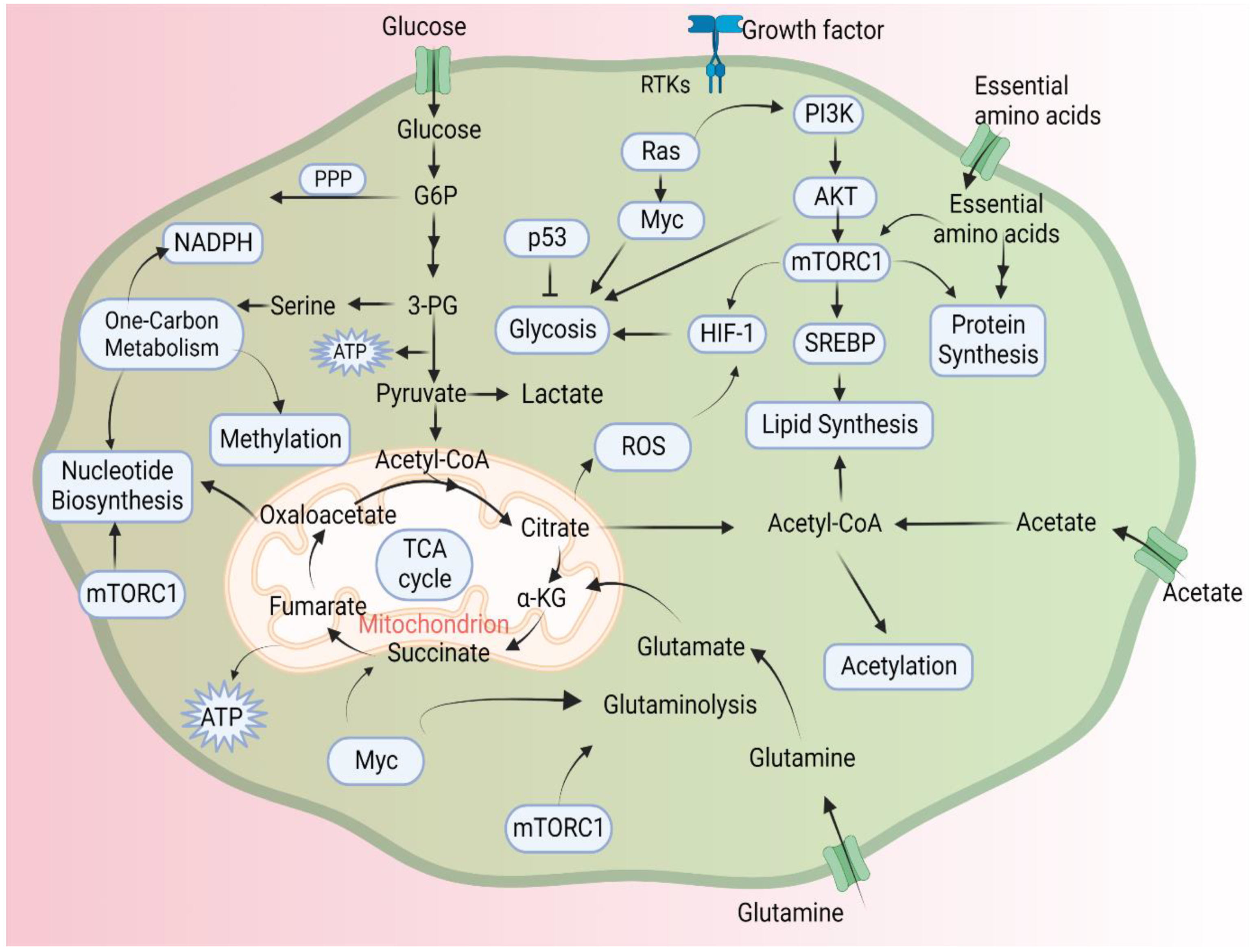
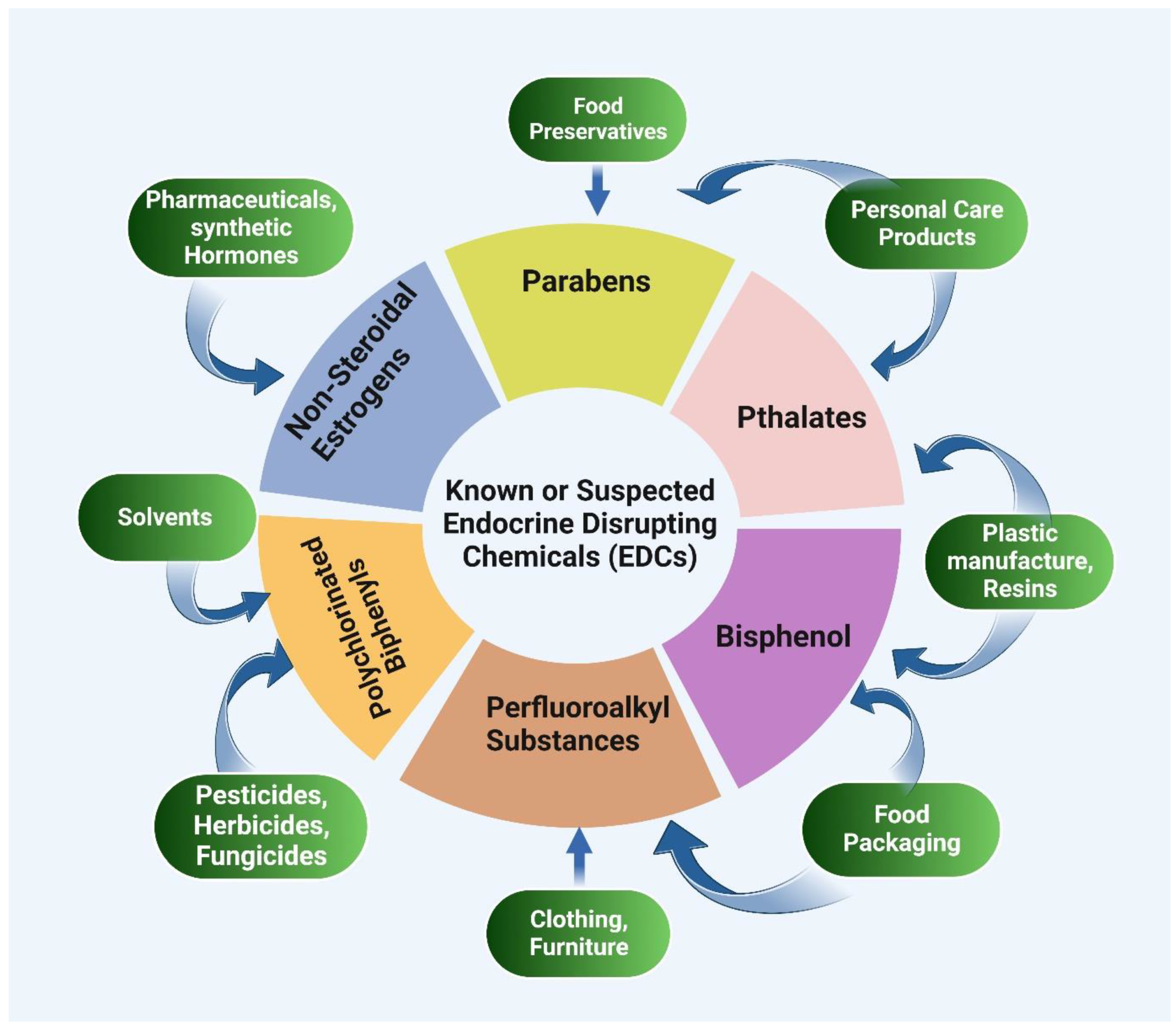
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sibuh, B.Z.; Quazi, S.; Panday, H.; Parashar, R.; Jha, N.K.; Mathur, R.; Jha, S.K.; Taneja, P.; Jha, A.K. RETRACTED: The Emerging Role of Epigenetics in Metabolism and Endocrinology. Biology 2023, 12, 256. https://doi.org/10.3390/biology12020256
Sibuh BZ, Quazi S, Panday H, Parashar R, Jha NK, Mathur R, Jha SK, Taneja P, Jha AK. RETRACTED: The Emerging Role of Epigenetics in Metabolism and Endocrinology. Biology. 2023; 12(2):256. https://doi.org/10.3390/biology12020256
Chicago/Turabian StyleSibuh, Belay Zeleke, Sameer Quazi, Hrithika Panday, Ritika Parashar, Niraj Kumar Jha, Runjhun Mathur, Saurabh Kumar Jha, Pankaj Taneja, and Abhimanyu Kumar Jha. 2023. "RETRACTED: The Emerging Role of Epigenetics in Metabolism and Endocrinology" Biology 12, no. 2: 256. https://doi.org/10.3390/biology12020256
APA StyleSibuh, B. Z., Quazi, S., Panday, H., Parashar, R., Jha, N. K., Mathur, R., Jha, S. K., Taneja, P., & Jha, A. K. (2023). RETRACTED: The Emerging Role of Epigenetics in Metabolism and Endocrinology. Biology, 12(2), 256. https://doi.org/10.3390/biology12020256






