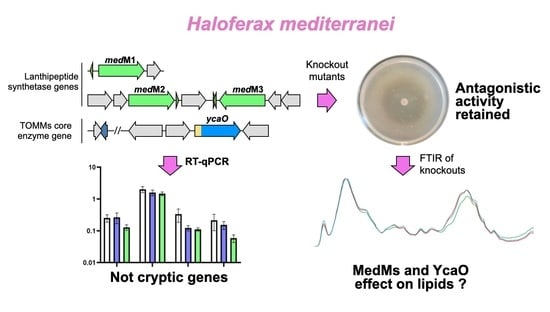
Towards the Understanding of the Function of Lanthipeptide and TOMM-Related Genes in Haloferax mediterranei
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Media
2.2. Generation of Knockout Mutants
2.3. Viable Cells Count and Anti-Haloarchaea Activity
2.4. Preparation of plates for RNA extraction and FTIR analysis
2.5. RNA Extraction and cDNA Synthesis
2.6. Transcriptional Analysis by qPCR
2.7. FTIR Analysis
2.8. Bioinformatic Analysis
3. Results and Discussion
3.1. Characterization of H. Mediterranei Growth and Antihaloarchaeal Activity on YPC Agar
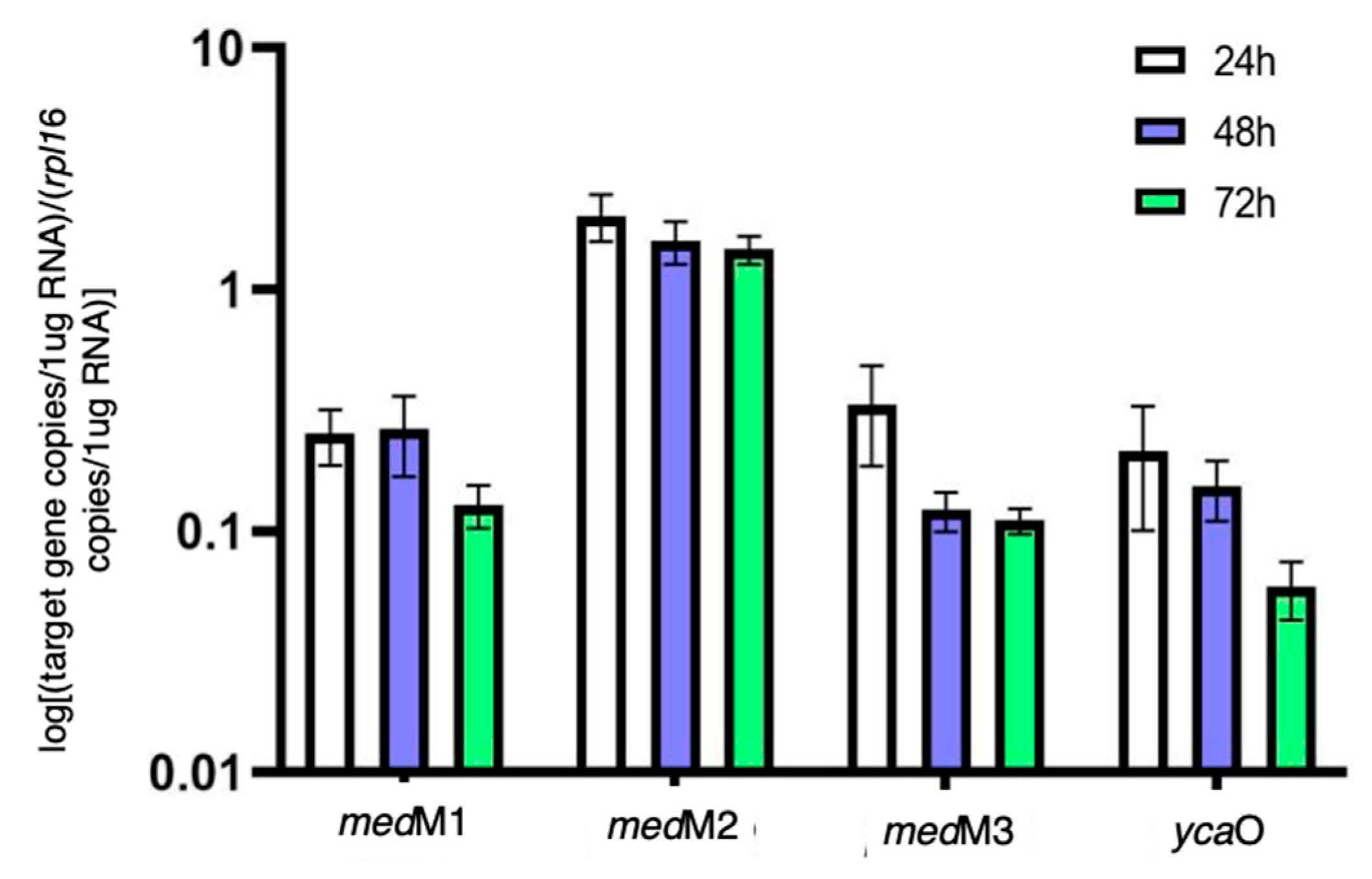
3.2. Transcriptional Analysis of the Biosynthetic Enzymes of RiPPs
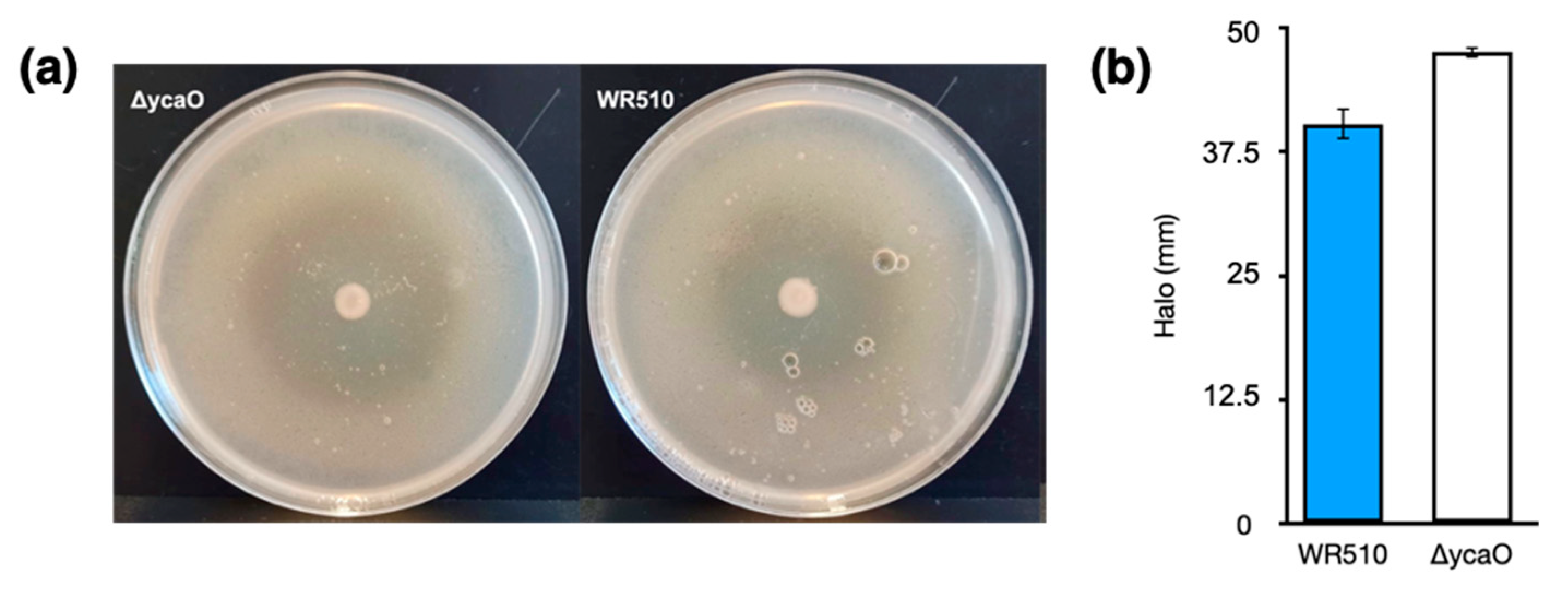
3.3. Generation of Haloazolisin Knockout Mutants and Their Antihaloarchaeal Activity
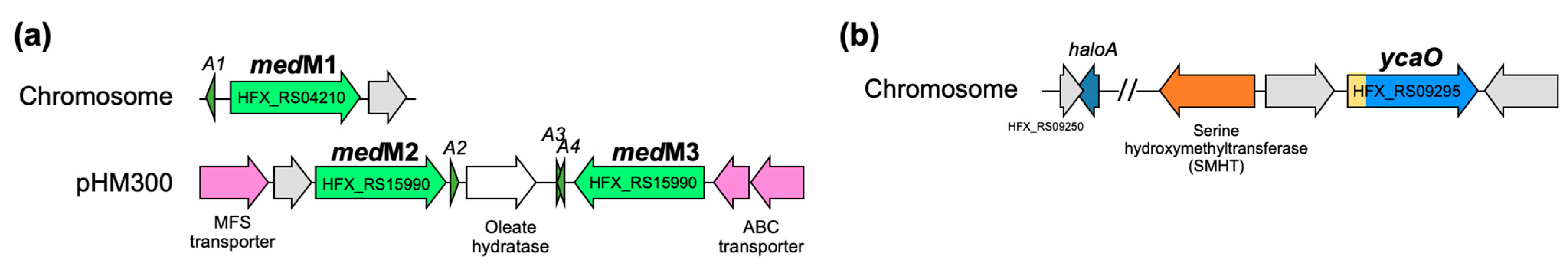
3.4. In Silico Analysis of H. Mediterranei Haloazolisin Genetic Environment
3.5. FTIR Analysis of H. Mediterranei Lacking RiPPs Core Biosynthetic Enzymes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Montalbán-López, M.; Scott, T.A.; Ramesh, S.; Rahman, I.R.; van Heel, A.J.; Viel, J.H.; Bandarian, V.; Dittmann, E.; Genilloud, O.; Goto, Y.; et al. New Developments in RiPP Discovery, Enzymology and Engineering. Nat. Prod. Rep. 2021, 38, 130–239. [Google Scholar] [CrossRef]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef]
- Zhong, Z.; He, B.; Li, J.; Li, Y.-X. Challenges and Advances in Genome Mining of Ribosomally Synthesized and Post-Translationally Modified Peptides (RiPPs). Synth. Syst. Biotechnol. 2020, 5, 155–172. [Google Scholar] [CrossRef]
- Cox, C.L.; Doroghazi, J.R.; Mitchell, D.A. The Genomic Landscape of Ribosomal Peptides Containing Thiazole and Oxazole Heterocycles. BMC Genomics 2015, 16, 778. [Google Scholar] [CrossRef]
- Castro, I.; Costa, H.; Turgeman-Grott, I.; Allers, T.; Mendo, S.; Caetano, T. The Lanthipeptide Biosynthetic Clusters of the Domain Archaea. Microbiol. Res. 2021, 253, 126884. [Google Scholar] [CrossRef]
- Repka, L.M.; Chekan, J.R.; Nair, S.K.; van der Donk, W.A. Mechanistic Understanding of Lanthipeptide Biosynthetic Enzymes. Chem. Rev. 2017, 117, 5457–5520. [Google Scholar] [CrossRef]
- Melby, J.O.; Nard, N.J.; Mitchell, D.A. Thiazole/Oxazole-Modified Microcins: Complex Natural Products from Ribosomal Templates. Curr. Opin. Chem. Biol. 2011, 15, 369–378. [Google Scholar] [CrossRef]
- Travin, D.Y.; Bikmetov, D.; Severinov, K. Translation-Targeting RiPPs and Where to Find Them. Front. Genet. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Bitan-Banin, G.; Ortenberg, R.; Mevarech, M. Development of a Gene Knockout System for the Halophilic Archaeon Haloferax Volcanii by Use of the PyrE Gene. J. Bacteriol. 2003, 185, 772–778. [Google Scholar] [CrossRef]
- Allers, T.; Ngo, H.-P.; Mevarech, M.; Lloyd, R.G. Development of Additional Selectable Markers for the Halophilic Archaeon Haloferax Volcanii Based on the LeuB and TrpA Genes. Appl. Environ. Microbiol. 2004, 70, 943–953. [Google Scholar] [CrossRef]
- Thomas, P.; Sekhar, A.C.; Upreti, R.; Mujawar, M.M.; Pasha, S.S. Optimization of Single Plate-Serial Dilution Spotting (SP-SDS) with Sample Anchoring as an Assured Method for Bacterial and Yeast Cfu Enumeration and Single Colony Isolation from Diverse Samples. Biotechnol. Rep. 2015, 8, 45–55. [Google Scholar] [CrossRef]
- Hwang, S.; Chavarria, N.E.; Hackley, R.K.; Schmid, A.K.; Maupin-Furlow, J.A. Gene Expression of Haloferax Volcanii on Intermediate and Abundant Sources of Fixed Nitrogen. Int. J. Mol. Sci. 2019, 20, 4784. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & Clustermap.Js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Zallot, R.; Oberg, N.; Gerlt, J.A. The EFI Web Resource for Genomic Enzymology Tools: Leveraging Protein, Genome, and Metagenome Databases to Discover Novel Enzymes and Metabolic Pathways. Biochemistry 2019, 58, 4169–4182. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Mining and Unearthing Hidden Biosynthetic Potential. Nat. Commun. 2021, 12, 3864. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Bedre, R.; Thapa, S.S.; Sabrin, A.; Wang, G.; Dassanayake, M.; Grove, A. Global Awakening of Cryptic Biosynthetic Gene Clusters in Burkholderia Thailandensis. ACS Chem. Biol. 2017, 12, 3012–3021. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Sun, S.; Wang, R.; Feng, H.; Xiang, H. Haloferax Mediterranei Halolysin R4 Confers Antagonistic and Defensive Capabilities. Appl. Environ. Microbiol. 2021, 87, e02889-20. [Google Scholar] [CrossRef]
- Caetano, T.; van der Donk, W.; Mendo, S. Bacteroidetes Can Be a Rich Source of Novel Lanthipeptides: The Case Study of Pedobacter Lusitanus. Microbiol. Res. 2020, 235, 126441. [Google Scholar] [CrossRef]
- Figueiredo, G.; Gomes, M.; Covas, C.; Mendo, S.; Caetano, T. The Unexplored Wealth of Microbial Secondary Metabolites: The Sphingobacteriaceae Case Study. Microb. Ecol. 2022, 83, 470–481. [Google Scholar] [CrossRef]
- Burkhart, B.J.; Schwalen, C.J.; Mann, G.; Naismith, J.H.; Mitchell, D.A. YcaO-Dependent Posttranslational Amide Activation: Biosynthesis, Structure, and Function. Chem. Rev. 2017, 117, 5389–5456. [Google Scholar] [CrossRef]
- Strader, M.B.; Costantino, N.; Elkins, C.A.; Chen, C.Y.; Patel, I.; Makusky, A.J.; Choy, J.S.; Court, D.L.; Markey, S.P.; Kowalak, J.A. A Proteomic and Transcriptomic Approach Reveals New Insight into β-Methylthiolation of Escherichia Coli Ribosomal Protein S12. Mol. Cell. Proteomics 2011, 10, M110.005199. [Google Scholar] [CrossRef] [PubMed]
- Mahanta, N.; Liu, A.; Dong, S.; Nair, S.K.; Mitchell, D.A. Enzymatic Reconstitution of Ribosomal Peptide Backbone Thioamidation. Proc. Natl. Acad. Sci. USA 2018, 115, 3030–3035. [Google Scholar] [CrossRef]
- Preisner, O.; Lopes, J.A.; Guiomar, R.; Machado, J.; Menezes, J.C. Fourier Transform Infrared (FT-IR) Spectroscopy in Bacteriology: Towards a Reference Method for Bacteria Discrimination. Anal. Bioanal. Chem. 2007, 387, 1739–1748. [Google Scholar] [CrossRef] [PubMed]
- Naumann, D. Infrared Spectroscopy in Microbiology. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Chichester, UK, 2006. [Google Scholar]
- Sivakesava, S.; Irudayaraj, J.; DebRoy, C. Differentitation of Microorganisms by FTIR-ATR and NIR Spectroscopy. Trans. ASAE 2004, 47, 951–957. [Google Scholar] [CrossRef]
- Mariey, L.; Signolle, J.P.; Amiel, C.; Travert, J. Discrimination, Classification, Identification of Microorganisms Using FTIR Spectroscopy and Chemometrics. Vib. Spectrosc. 2001, 26, 151–159. [Google Scholar] [CrossRef]
- Erukhimovitch, V.; Pavlov, V.; Talyshinsky, M.; Souprun, Y.; Huleihel, M. FTIR Microscopy as a Method for Identification of Bacterial and Fungal Infections. J. Pharm. Biomed. Anal. 2005, 37, 1105–1108. [Google Scholar] [CrossRef] [PubMed]
- Moreirinha, C.; Trindade, J.; Saraiva, J.A.; Almeida, A.; Delgadillo, I. MIR Spectroscopy as Alternative Method for Further Confirmation of Foodborne Pathogens Salmonella Spp. and Listeria Monocytogenes. J. Food Sci. Technol. 2018, 55, 3971–3978. [Google Scholar] [CrossRef] [PubMed]
- Puzey, K.A.; Gardner, P.J.; Petrova, V.K.; Donnelly, C.W.; Petrucci, G.A. Automated Species and Strain Identification of Bacteria in Complex Matrices Using FTIR Spectroscopy; Fountain, A.W., III, Gardner, P.J., Eds.; SPIE: Bellingham, WA, USA, 2008; p. 695412. [Google Scholar]
- Alaettin Guven, K.G. FT-IR for Rapid Discrimination of Halophilic Archaea and Bacteria. J. Phylogen. Evol. Biol. 2015, 3. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; ur Rehman, D.I. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Jones, R.N.; Angell, C.L.; Ito, T.; Smith, R.J.D. The Carnonyl Stretching Bands in the Infrared Spectra of the Unsaturated Lactones. Can. J. Chem. 1959, 37, 2007–2022. [Google Scholar] [CrossRef]




| Time | Strain | Wavenumber (cm−1) | Functional Group Assignments |
|---|---|---|---|
| Day 1 | ΔmedM1M2M3 | 1150 | Phosphodiester bonds |
| 1280 | Amides; Nucleic acids | ||
| 1450 | Lipids, including fatty acids; Amide III | ||
| 1550 | Amide II | ||
| 1650 | Amide I | ||
| Day 6 | ΔycaO | 1137–1145 | Phosphate and/or oligosaccharides |
| 1180–1190 | Amides and/or deoxyribose | ||
| 1650 | Amide I | ||
| 1740–1750 | C=O from lipids | ||
| Day 14 | WR510 | 1150 | Phosphodiester bonds |
| ΔycaO | 1430–1450 | Polysaccharides and/or lipids, including fatty acids; Amide III | |
| 1740–1750 | C=O from lipids; C=C from lipids including fatty acids | ||
| ΔmedM1M2M3 | 1050 | Phosphate; Oligosaccharides and/or C-O-C from nucleic acids and phospholipids | |
| 1373–1380 | C-N from guanines and/or cytosines; C-O and/or C-H and/or N-H | ||
| 1534 | Amide II | ||
| 1550 | Amide II | ||
| 1650 | Amide I | ||
| 1740–1750 | C=O from lipids |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, T.; Cassin, E.; Moreirinha, C.; Mendo, S.; Caetano, T.S. Towards the Understanding of the Function of Lanthipeptide and TOMM-Related Genes in Haloferax mediterranei. Biology 2023, 12, 236. https://doi.org/10.3390/biology12020236
Costa T, Cassin E, Moreirinha C, Mendo S, Caetano TS. Towards the Understanding of the Function of Lanthipeptide and TOMM-Related Genes in Haloferax mediterranei. Biology. 2023; 12(2):236. https://doi.org/10.3390/biology12020236
Chicago/Turabian StyleCosta, Thales, Elena Cassin, Catarina Moreirinha, Sónia Mendo, and Tânia Sousa Caetano. 2023. "Towards the Understanding of the Function of Lanthipeptide and TOMM-Related Genes in Haloferax mediterranei" Biology 12, no. 2: 236. https://doi.org/10.3390/biology12020236
APA StyleCosta, T., Cassin, E., Moreirinha, C., Mendo, S., & Caetano, T. S. (2023). Towards the Understanding of the Function of Lanthipeptide and TOMM-Related Genes in Haloferax mediterranei. Biology, 12(2), 236. https://doi.org/10.3390/biology12020236