Characterization of Enrichment Cultures of Anammox, Nitrifying and Denitrifying Bacteria Obtained from a Cold, Heavily Nitrogen-Polluted Aquifer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. High-Throughput Sequencing of 16S rRNA Genes
2.3. Fed-Batch Cultivation without Carrier Materials
2.4. Batch Cultivation with the Addition of Carrier Materials
2.5. Continuous Flow Cultivation of AnAOB
2.6. Analytical Methods
2.7. Fluorescence In Situ Hybridization (FISH)
2.8. Visualization and Analysis of the Microbial Biofilms
2.9. Quantitative Polymerase Chain Reaction (qPCR)
3. Results
3.1. Enrichment Cultures of N-Cycle Bacteria
3.1.1. N Consumption/Production Rates in Fed-Batch Cultures
3.1.2. Results of FISH Analysis of AnAOB Enrichment Culture
3.1.3. Microbial Diversity at the Family Level in Enrichment Cultures
3.2. Continuous Flow Cultivation of AnAOB
3.2.1. Anammox Performance with Various Carriers
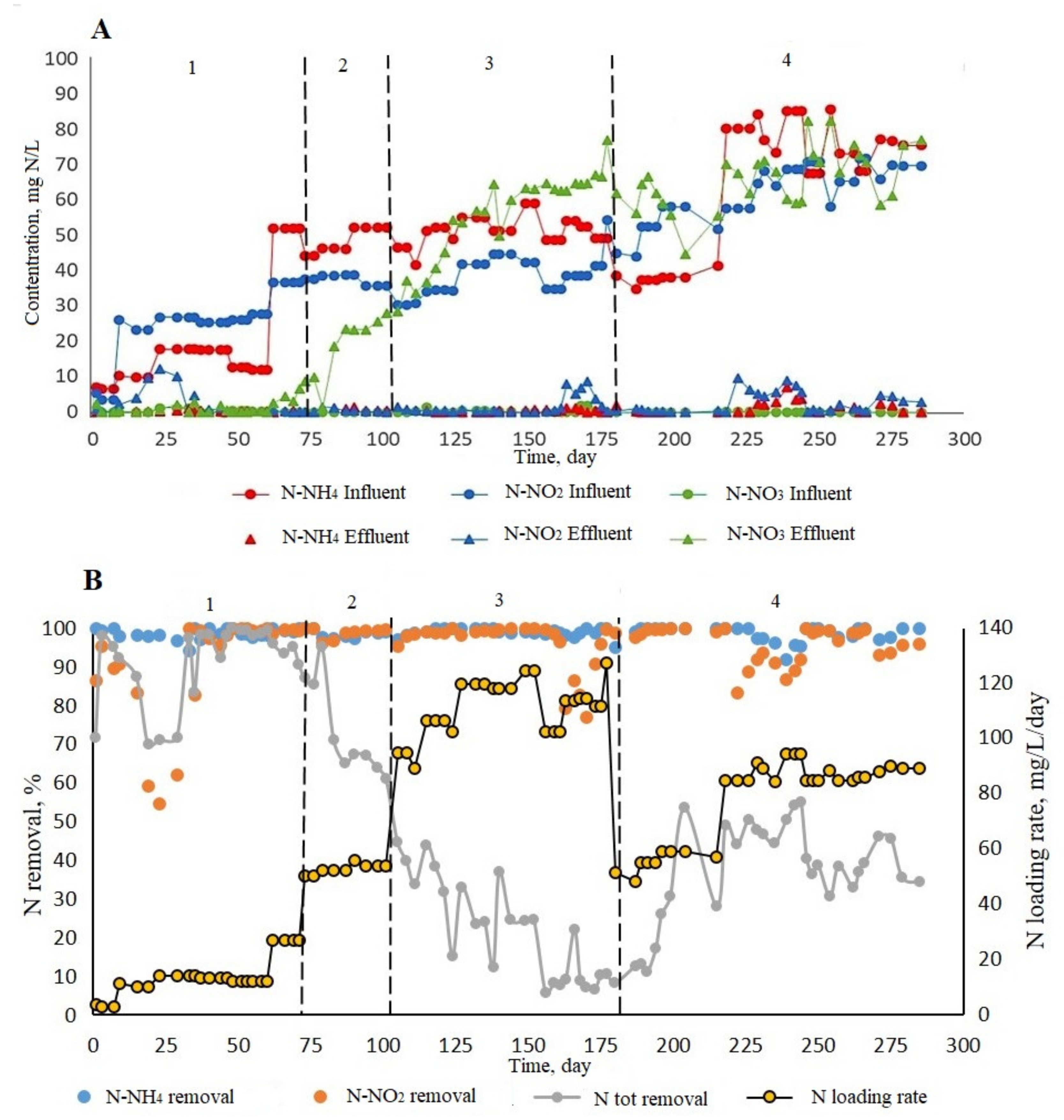
3.2.2. N Removal Performance in ABR
3.3. Key N Cycle Bacteria in the ABR Microbial Community
3.3.1. Fluorescence In Situ Hybridization
3.3.2. qPCR
3.3.3. Microbial Community in ABR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Safonov, A.V.; Babich, T.L.; Sokolova, D.S.; Grouzdev, D.S.; Tourova, T.P.; Poltaraus, A.B.; Zakharova, E.V.; Merkel, A.Y.; Novikov, A.P.; Nazina, T.N. Microbial Community and in situ Bioremediation of Groundwater by Nitrate Removal in the Zone of a Radioactive Waste Surface Repository. Front. Microbiol. 2018, 9, 1985. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Herrmann, M.; Thamdrup, B.; Schwab, V.F.; Geesink, P.; Trumbore, S.E.; Totsche, K.U.; Küsel, K. Nitrogen Loss from Pristine Carbonate-Rock Aquifers of the Hainich Critical Zone Exploratory (Germany) Is Primarily Driven by Chemolithoautotrophic Anammox Processes. Front. Microbial. 2017, 8, 1951. [Google Scholar] [CrossRef]
- Mosley, O.E.; Gios, E.; Close, M.; Weaver, L.; Daughney, C.; Handley, K.M. Nitrogen cycling and microbial cooperation in the terrestrial subsurface. ISME J. 2022, 16, 2561–2573. [Google Scholar] [CrossRef]
- Aguilar-Rangel, E.J.; Prado, B.L.; Vásquez-Murrieta, M.S.; Santos, P.E.-D.L.; Siebe, C.; Falcón, L.I.; Santillán, J.; Alcántara-Hernández, R.J. Temporal analysis of the microbial communities in a nitrate-contaminated aquifer and the co-occurrence of anammox, n-damo and nitrous-oxide reducing bacteria. J. Contam. Hydrol. 2020, 234, 103657. [Google Scholar] [CrossRef]
- Wegner, C.; Gaspar, M.; Geesink, P.; Herrmann, M.; Marz, M.; Küsel, K. Biogeochemical Regimes in Shallow Aquifers Reflect the Metabolic Coupling of the Elements Nitrogen, Sulfur, and Carbon. ASM J. Appl. Environ. Microbiol. 2019, 85, e02346-18. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Herrmann, M.; Blohm, A.; Hilke, I.; Frosch, T.; Trumbore, S.E.; Küsel, K. Thiosulfate- and hydrogen-driven autotrophic denitrification by a microbial consortium enriched from groundwater of an oligotrophic limestone aquifer. FEMS Microbiol. Ecol. 2018, 94, 10. [Google Scholar] [CrossRef]
- Boguslavsky, A.E.; Gaskova, O.L.; Naymushina, O.S.; Popova, N.M.; Safonov, A.V. Environmental monitoring of low-level radioactive waste disposal in electrochemical plant facilities in Zelenogorsk, Russia. Appl. Geochem. 2020, 119, 104598. [Google Scholar] [CrossRef]
- Nazina, T.; Babich, T.; Kostryukova, N.; Sokolova, D.; Abdullin, R.; Tourova, T.; Kadnikov, V.; Mardanov, A.; Ravin, N.; Grouzdev, D.; et al. Ultramicrobacteria from Nitrate- and Radionuclide-Contaminated Groundwater. Sustainability 2020, 12, 1239. [Google Scholar] [CrossRef]
- Nazina, T.N.; Safonov, A.V.; Kosareva, I.M.; Ivoilov, V.S.; Poltaraus, A.B.; Ershov, B.G. Microbiological processes in the Severnyi deep disposal site for liquid radioactive wastes. Microbiology 2010, 79, 528–537. [Google Scholar] [CrossRef]
- Safonov, A.V.; Boguslavsky, A.E.; Gaskova, O.L.; Boldyrev, K.A.; Shvartseva, O.S.; Khvashchevskaya, A.A.; Popova, N.M. Biogeochemical Modelling of Uranium Immobilization and Aquifer Remediation Strategies Near NCCP Sludge Storage Facilities. Appl. Sci. 2021, 11, 2875. [Google Scholar] [CrossRef]
- Reed, D.W.; Smith, J.M.; Francis, C.A.; Fujita, Y. Responses of Ammonia-Oxidizing Bacterial and Archaeal Populations to Organic Nitrogen Amendments in Low-Nutrient Groundwater. Appl. Environ. Microbiol. 2010, 76, 26. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Wang, S.; Ye, F.; Zhu, G. Global Distribution of Anaerobic Ammonia Oxidation (Anammox) Bacteria—Field Surveys in Wetland, Dryland, Groundwater Aquifer and Snow. Front. Microbiol. 2019, 10, 2583. [Google Scholar] [CrossRef]
- Strous, M.; Fuerst, J.A.; Kramer, E.H.M.; Logemann, S.; Muyzer, G.; van de Pas-Schoonen, K.T.; Webb, R.; Kuenen, J.G.; Jetten, M.S.M. Missing lithotroph identified as new planctomycete. Nature 1999, 400, 446–449. [Google Scholar] [CrossRef]
- Dalsgaard, T.; Thamdrup, B.; Canfield, D.E. Anaerobic ammonium oxidation (anammox) in the marine environment. Res. Microbiol. 2005, 156, 457–464. [Google Scholar] [CrossRef]
- Zhuab, A.; Chen, J.; Gao, L.; Shimizu, Y.; Liange, D.; Yi, M.; Cao, L. Combined microbial and isotopic signature approach to identify nitrate sources and transformation processes in groundwater. Chemosphere 2019, 228, 721–734. [Google Scholar] [CrossRef]
- Mosley, O.E.; Gios, E.; Weaver, L.; Close, M.; Daughney, C.; van der Raaij, R.; Martindale, H.; Handley, K.M. Metabolic Diversity and Aero-Tolerance in Anammox Bacteria from Geochemically Distinct Aquifers. mSystems 2022, 7, 125521. [Google Scholar] [CrossRef] [PubMed]
- Ludington, W.B.; Seher, T.D.; Applegate, O.; Li, X.; Kliegman, J.I.; Langelier, C.; Atwill, E.R.; Harter, T.; DeRisi, J.L. Assessing biosynthetic potential of agricultural groundwater through metagenomic sequencing: A diverse anammox community dominates nitrate-rich groundwater. PLoS ONE 2017, 12, e0174930. [Google Scholar] [CrossRef] [PubMed]
- Moore, T.A.; Xing, Y.; Lazenby, B.; Lynch, M.D.J.; Schiff, S.; Robertson, W.D.; Timlin, R.; Lanza, S.; Cathryn Ryan, M.; Aravena, R.; et al. Prevalence of Anaerobic Ammonium-Oxidizing Bacteria in Contaminated Groundwater. Environ. Sci. Technol. 2011, 45, 7217–7225. [Google Scholar] [CrossRef]
- Smith, R.L.; Böhlke, J.K.; Song, B.; Tobias, C.R. Role of Anaerobic Ammonium Oxidation (Anammox) in Nitrogen Removal from a Freshwater Aquifer. Environ. Sci. Technol. 2015, 49, 12169–12177. [Google Scholar] [CrossRef]
- Nakano, M.; Kamei, T.; Man Shakya, B.; Nakamura, T.; Tanaka, Y.; Haramoto, E.; Toyama, T.; Kazama, F. Distribution and Community Composition of Anammox Bacteria in Shallow Groundwater of the Kathmandu Valley, Nepal. Microbes Environ. 2021, 36, 1. [Google Scholar] [CrossRef]
- Wang, S.; Zhu, G.; Zhuang, L.; Li, Y.; Liu, L.; Lavik, G.; Berg, M.; Liu, S.; Long, X.-E.; Guo, J.; et al. Anaerobic ammonium oxidation is a major N-sink in aquifer systems around the world. ISME J 2020, 14, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Radny, D.; Huang, S.; Zhuang, L.; Zhao, S.; Berg, M.; Jetten, M.S.M.; Zhu, G. Nitrogen loss by anaerobic ammonium oxidation in unconfined aquifer soils. Sci. Rep. 2017, 7, 40173. [Google Scholar] [CrossRef]
- Xiong, Y.; Du, Y.; Deng, Y.; Ma, T.; Wang, Y. Feammox in alluvial-lacustrine aquifer system: Nitrogen/iron isotopic and biogeochemical evidences. Water Res. 2022, 222, 118867. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Jin, R.C. Summary of the preservation techniques and the evolution of the anammox bacteria characteristics during preservation. Appl. Microbiol. Biotechnol. 2017, 101, 4349–4362. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Yan, S.; Gong, Z.; Zhang, S. Enrichment and characterization of Anammox bacteria in a non-woven membrane reactor. Water Pract. Technol. 2022, 17, 798–807. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Q.; Shena, Y.; Yang, D. Enhancing the in-situ enrichment of anammox bacteria in aerobic granules to achieve high-rate CANON at low temperatures. Chemosphere. 2021, 278, 130395. [Google Scholar] [CrossRef]
- Engström, P.; Dalsgaard, T.; Hulth, S.; Aller, R.C. Anaerobic ammonium oxidation by nitrite (anammox): Implications for N2 production in coastal marine sediments. Geochim. Cosmochim. Acta 2005, 69, 2057–2065. [Google Scholar] [CrossRef]
- Canion, A.; Kostka, J.E.; Gihring, T.M.; Huettel, M.; van Beusekom, J.E.E.; Gao, H.; Lavik, G.; Kuypers, M.M.M. Temperature response of denitrification and anammox reveals the adaptation of microbial communities to in situ temperatures in permeable marine sediments that span 50° in latitude. Biogeosciences 2014, 11, 309–320. [Google Scholar] [CrossRef]
- Rysgaard, S.; Nøhr Glud, R.; Risgaard-Petersen, N.; Dalsgaard, T. Denitrification and anammox activity in Arctic marine sediments. Limnol. Oceanogr. 2004, 49, 1493–1502. [Google Scholar] [CrossRef]
- Thamdrup, B.; Dalsgaard, T. Production of N2 through Anaerobic Ammonium Oxidation Coupled to Nitrate Reduction in Marine Sediments. Appl. Environ. Microbiol. 2002, 68, 3. [Google Scholar] [CrossRef]
- Hu, Z.; Lotti, T.; de Kreuk, M.; Kleerebezem, R.; van Loosdrecht, M.; Kruit, J.; Jetten, M.S.M.; Kartal, B. ; Kruit, J.; Jetten, M.S.M.; Kartal, B. Nitrogen Removal by a Nitritation-Anammox Bioreactor at Low Temperature. Appl. Environ. Microbiol. 2013, 79, 8. [Google Scholar] [CrossRef] [PubMed]
- Lotti, T.; Kleerebezem, R.; van Loosdrecht, M.C.M. Effect of temperature change on anammox activity. Biotechnol. Bioeng. 2015, 112, 98–103. [Google Scholar] [CrossRef]
- Vishnyakova, A.; Popova, N.; Artemiev, G.; Botchkova, E.; Litti, Y.; Safonov, A. Effect of Mineral Carriers on Biofilm Formation and Nitrogen Removal Activity by an Indigenous Anammox Community from Cold Groundwater Ecosystem Alone and Bioaugmented with Biomass from a “Warm” Anammox Reactor. Biology 2022, 11, 1421. [Google Scholar] [CrossRef] [PubMed]
- Popova, N.; Vishnyakova, A.; Artemiev, G.; Sitanskaia, A.; Litti, Y.; Safonov, A. Biofilms of anammox bacteria on mineral carriers to establish a subterranean permeable barrier. Int. Environ, J. Sci. Technol. 2022, 2022, 1–12. [Google Scholar] [CrossRef]
- Safonov, A.V.; Andryushchenko, N.D.; Ivanov, P.V.; Boldyrev, K.A.; Babich, T.L.; German, K.E.; Zakharova, E.V. Biogenic Factors of Radionuclide Immobilization on Sandy Rocks of Upper Aquifers. Radiochemistry 2019, 61, 99–108. [Google Scholar] [CrossRef]
- Safonov, A.; Popova, N.; Boldyrev, K.; Lavrinovich, E.; Boeva, N.; Artemiev, G.; Kuzovkina, E.; Emelyanov, A.; Myasnikov, I.; Zakharova, E.; et al. The microbial impact on U, Pu, Np, and Am immobilization on aquifer sandy rocks, collected at the deep LRW injection site. J. Geochem. Explor. 2022, 240, 107052. [Google Scholar] [CrossRef]
- Safonov, A.; Lavrinovich, E.; Emel’yanov, A.; Boldyrev, K.; Kuryakov, V.; Rodygina, N.; Zakharova, E.; Novikov, A. Risk of colloidal and pseudo-colloidal transport of actinides in nitrate contaminated groundwater near a radioactive waste repository after bioremediation. Sci. Rep. 2022, 12, 4557. [Google Scholar] [CrossRef] [PubMed]
- Gohl, D.M.; MacLean, A.; Hauge, A.; Becker, A.; Walek, D.; Beckman, K.B. An optimized protocol for high-throughput ampliconbased microbiome profiling. Protoc. Exch. 2016, 2016, 1–15. [Google Scholar] [CrossRef]
- Hugerth, L.W.; Wefer, H.A.; Lundin, S.; Jakobsson, H.E.; Lindberg, M.; Rodin, S.; Engstrand, L.; Andersson, A.F. Dege Prime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies. Appl. Environ. Microbiol. 2014, 80, 5116–5123. [Google Scholar] [CrossRef]
- Merkel, A.Y.; Tarnovetskii, I.Y.; Podosokorskaya, O.A.; Toshchakov, S.V. Analysis of 16S rRNA Primer Systems for Profiling of Thermophilic Microbial Communities. Microbiology 2019, 88, 671–680. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. Dada2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef] [PubMed]
- Mironov, V.V.; Bochkova, E.A.; Gannesen, A.V.; Vanteeva, A.V.; Russkova, Y.I.; Nozhevnikova, A.N. Dynamics of Biological Processes during Composting of Anaerobically Digested Wastewater Sludge. Microbiology 2020, 89, 470–482. [Google Scholar] [CrossRef]
- Nozhevnikova, A.N.; Litti, Y.V.; Nekrasova, V.K.; Kulichevskaya, I.S.; Grigoryeva, N.V.; Kulikov, N.I.; Zubov, M.G. Anaerobic ammonium oxidation (Anammox) in immobilized activated sludge biofilms during the treatment of weak wastewater. Microbiology 2012, 81, 25–34. [Google Scholar] [CrossRef]
- van der Star, W.R.L.; Miclea, A.I.; van Dongen, U.G.J.M.; Muyzer, G.; Picioreanu, C.; van Loosdrecht, M.C.M. The membrane bioreactor: A novel tool to grow anammox bacteria as free cells. Biotechnol. Bioeng. 2008, 101, 286–294. [Google Scholar] [CrossRef]
- Li, X.; Xiao, Y.; Liao, D.; Zheng, W.; Yi, T.; Yang, Q.; Zeng, G. Granulation of simultaneous partial nitrification and anammox biomass in one single SBR system. Appl. Biochem. Biotechnol. 2011, 163, 1053–1065. [Google Scholar] [CrossRef]
- Kallistova, A.; Nikolaev, Y.; Grachev, V.; Beletsky, A.; Gruzdev, E.; Kadnikov, V.; Dorofeev, A.; Berestovskaya, J.; Pelevina, A.; Zekker, I.; et al. New Insight into the Interspecies Shift of Anammox Bacteria Ca. “Brocadia” and Ca. “Jettenia” in Reactors Fed with Formate and Folate. Front. Microbiol. 2022, 12, 802201. [Google Scholar] [CrossRef]
- Van Hulle, S.W.H.; Vandeweyer, H.J.P.; Meesschaert, B.D.; Vanrolleghem, P.A.; Dejans, P.; Dumoulin, A. Engineering aspects and practical application of Autotroph ic nitrogen removal from nitrogen rich streams. Chem. Eng. J. 2010, 162, 1–20. [Google Scholar] [CrossRef]
- Yakimov, A.V.; Zasukhin, D.S.; Vorobkalo, V.A.; Ponomareva, O.A.; Knyazeva, E.E.; Zaikovskii, V.I.; Kolozhvari, B.A.; Ivanova, I.I. Dealumination of Nanosized Zeolites Y. Pet. Chem. 2019, 59, 540–545. [Google Scholar] [CrossRef]
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; Stahl, D.A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990, 56, 6. [Google Scholar] [CrossRef]
- Botchkova, E.A.; Litti, Y.V.; Novikov, A.A.; Grouzdev, D.S.; Bochkareva, E.S.; Beskorovayny, A.V.; Kuznetsov, B.B.; Nozhevnikova, A.N. Description of “Candidatus Jettenia ecosi” sp. nov., a New Species of Anammox Bacteria. Microbiology 2018, 87, 766–776. [Google Scholar] [CrossRef]
- Vishnyakova, A.V.; Litti, Y.V.; Botchkova, E.A.; Ermoshin, A.A.; Nozhevnikova, A.N. Changes in Relative Abundance of Microbial Groups Involved in Nitrogen Removal in the Anammox‒Partial Nitrification Reactor System at Increase in Ammonium Nitrogen and COD Load. Microbiology 2020, 89, 205–211. [Google Scholar] [CrossRef]
- Zhang, L.; Zheng, P.; Tang, C.; Ren-cun, J. Anaerobic ammonium oxidation for treatment of ammonium-rich wastewaters. J. Zhejiang Univ. Sci. B 2008, 9, 416–426. [Google Scholar] [CrossRef]
- Mobarry, B.K.; Wagner, M.; Urbain, V.; Rittmann, B.E.; Stahl, D.A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 1996, 62, 2156–2162. [Google Scholar] [CrossRef]
- Maixner, F.; Noguera, D.R.; Anneser, B.; Stoecker, K.; Wegl, G.; Wagner, M.; Daims, H. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Appl. Microbiol. Int. 2006, 8, 1487–1495. [Google Scholar] [CrossRef]
- Wagner, M.; Rath, G.; Koops, H.-P.; Flood, J.; Amann, R. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci. Technol. 1996, 34, 237–244. [Google Scholar] [CrossRef]
- Daims, H.; Nielsen, J.L.; Nielsen, P.H.; Schleifer, K.-H.; Wagner, M. In Situ Characterization of Nitrospira-Like Nitrite-Oxidizing Bacteria Active in Wastewater Treatment Plants. Appl. Environ. Microbiol. 2001, 67, 5273–5284. [Google Scholar] [CrossRef]
- Humbert, S.; Zopfi, J.; Tarnawski, S.-E. Abundance of anammox bacteria in different wetland soils. Environ. Microbiol. Rep. 2012, 4, 484–490. [Google Scholar] [CrossRef]
- Hermansson, A.; Lindgren, P.E. Quantification of ammonia-oxidizing bacteria in arable soil by real-time PCR. Appl. Environ. Microbiol. 2001, 67, 972–976. [Google Scholar] [CrossRef] [PubMed]
- Braker, G.; Fesefeldt, A.; Witzel, K.-P. Development of PCR Primer Systems for Amplification of Nitrite Reductase Genes (nirK and nirS) To Detect Denitrifying Bacteria in Environmental Samples. Appl. Environ. Microbiol. 1998, 64, 3769–3775. [Google Scholar] [CrossRef] [PubMed]
- Nicol, G.W.; Hink, L.; Gubry-Rangin, C.; Prosser, J.I.; Lehtovirta-Morley, L.E. Genome Sequence of “Candidatus Nitrosocosmicus franklandus” C13, a Terrestrial Ammonia-Oxidizing Archaeon. Microbiol. Resour. Announc. 2019, 8, e00435-19. [Google Scholar] [CrossRef] [PubMed]
- Siegert, M.; Li, X.-F.; Yates, M.D.; Logan, B.E. The presence of hydrogenotrophic methanogens in the inoculum improves methane gas production in microbial electrolysis cells. Front. Microbiol. 2015, 5, 778. [Google Scholar] [CrossRef] [PubMed]
- Golyshina, O.V.; Pivovarova, T.A.; Karavaiko, G.I.; Kondrateva, T.F.; Moore, E.R.; Abraham, W.R.; Lunsdorf, H.; Timmis, K.N.; Yakimov, M.M.; Golyshin, P.N. Ferroplasma acidiphilum gen. nov., sp. nov., an acidophilic, autotrophic, ferrous-iron-oxidizing, cell-wall-lacking, mesophilic member of the Ferroplasmaceae fam. nov., comprising a distinct lineage of the Archaea. Int. J. Syst. Evol. Microbiol. 2000, 50, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, T.; Huang, K.; He, X.; Zhang, X.-X. Roles and correlations of functional bacteria and genes in the start-up of simultaneous anammox and denitrification system for enhanced nitrogen removal. Sci. Total Environ. 2019, 655, 1355–1363. [Google Scholar] [CrossRef]
- Green, S.J.; Prakash, O.; Jasrotia, P.; Overholt, W.A.; Cardenas, E.; Hubbard, D.; Tiedje, J.M.; Watson, D.B.; Schadt, C.W.; Brooks, S.C.; et al. Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl. Environ. Microbiol. 2012, 78, 1039–1047. [Google Scholar] [CrossRef]
- Joo, H.S.; Hirai, M.; Shoda, M. Improvement in ammonium removal efficiency in wastewater treatment by mixed culture of Alcaligenes faecalis No. 4 and L1. Biosci. J. Bioeng. 2007, 103, 66–73. [Google Scholar] [CrossRef]
- Gu, W.; Wang, L.; Liu, Y.; Lian, P.; Zhan, X.; Li, Y.; Huang, X. Anammox bacteria enrichment and denitrification in moving bed biofilm reactors packed with different buoyant carriers: Performances and mechanisms. Sci. Total Environ. 2020, 719, 137277. [Google Scholar] [CrossRef]
- Meng, F.; Su, G.; Hu, Y.; Lu, H.; Huang, L.; Chen, G. Improving nitrogen removal in an ANAMMOX reactor using a permeable reactive biobarrier. Water Res. 2014, 58, 82–91. [Google Scholar] [CrossRef]
- Connan, R.; Dabert, P.; Le Roux, S.; Chapleur, O.; Bridoux, G.; Vanotti, M.F.; Béline, B.; Magrí, A. Characterization of a combined batch-continuous procedure for the culture of anammox biomass. Ecol. Eng. 2017, 106, 231–241. [Google Scholar] [CrossRef]
- Chen, P.; Li, S.; Zheng, X.; Ren, T.; Wang, L. Heterotrophic Nitrification by Bacterium Shinella Zoogloeoides Sp. CPZ56. FEB Fresenius Environ. Bull. 2016, 25, 3017–3022. [Google Scholar]
- Adav, S.S.; Lee, D.-J.; Lai, J.-Y. Biological nitrification–denitrification with alternating oxic and anoxic operations using aerobic granules. Appl. Microbiol. Biotechnol. 2009, 84, 1181–1189. [Google Scholar] [CrossRef]
- Akaboci, T.R.; Gich, F.; Ruscalleda, M.; Balaguer, M.D.; Colprim, J. Assessment of operational conditions towards mainstream partial nitritation-anammox stability at moderate to low temperature: Reactor performance and bacterial community. Chem. Eng. J. 2018, 350, 192–200. [Google Scholar] [CrossRef]
- Lawson, C.; Wu, S.; Bhattacharjee, A.S.; Hamilton, J.J.; McMahon, K.D.; Goel, R.; Noguera, D.R. Metabolic network analysis reveals microbial community interactions in anammox granules. Nat. Commun. 2017, 8, 15416. [Google Scholar] [CrossRef] [PubMed]
- Becking, J.H. The Genus Beijerinckia. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006. [Google Scholar] [CrossRef]
- Bei, Q.; Peng, J.; Liesack, W. Shedding light on the functional role of the Ignavibacteria in Italian rice field soil: A meta-genomic/transcriptomic analysis. Soil Biol. Biochem. 2021, 163, 108444. [Google Scholar] [CrossRef]
- Liu, Y.; Niud, Q.; Wangce, S.; Jib, J.; Zhang, Y.; Yang, M.; Hojob, T.; Li, Y. Upgrading of the symbiosis of Nitrosomanas and anammox bacteria in a novel single-stage partial nitritation–anammox system: Nitrogen removal potential and Microbial characterization. Bioresour. Technol. 2017, 244, 1–463. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef]
- Zhao, X.; Zhong, J.; Wei, C.; Lin, C.-W.; Ding, T. Current Perspectives on Viable but Non-culturable State in Foodborne Pathogens. Front. Microbiol. 2017, 8, 580. [Google Scholar] [CrossRef]
- Shu, D.; He, Y.; Yue, H.; Gao, J.; Wang, Q.; Yang, S. Enhanced long-term nitrogen removal by organotrophic anammox bacteria under different C/N ratio constraints: Quantitative molecular mechanism and microbial community dynamics. RSC Adv. 2016, 6, 87593–87606. [Google Scholar] [CrossRef]
- Gonzalez-Martinez, A.; Osorio, F.; Rodriguez-Sanchez, A.; Martinez-Toledo, M.V.; Gonzalez-Lopez, J.; Lotti, T.; van Loosdrecht, M.C.M. Bacterial community structure of a lab-scale anammox membrane bioreactor. Biotechnol. Prog. 2015, 31, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Lv, L.; Kang, Q.; Gao, B.; Ni, S.; Chen, Y.; Xu, S. Microbial dynamics of biofilm and suspended flocs in anammox membrane bioreactor: The effect of non-woven fabric membrane. Bioresour. Technol. 2018, 247, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Peng, Y.; Fan, L.; Zhang, L.; Ni, B.-J.; Kartal, B.; Feng, X.; Jetten, M.S.M.; Yuan, Z. Metagenomic analysis of anammox communities. Environ. Microbiol. 2016, 18, 2979–2993. [Google Scholar] [CrossRef]
- Hink, L.; Lycus, P.; Gubry-Rangin, C.; Frostegård, Å.; Nicol, G.W.; Prosser, J.I.; Bakken, L.R. Kinetics of NH3-oxidation, NO-turnover, N2O-production and electron flow during oxygen depletion in model bacterial and archaeal ammonia oxidisers. Environ. Microbiol. 2017, 19, 4882–4896. [Google Scholar] [CrossRef]
- Li, W.; Zhuang, J.; Zhou, Y.; Meng, F.; Kang, D.; Zheng, P.; Shapleigh, J.P.; Liu, Y. Metagenomics reveals microbial community differences lead to differential nitrate production in anammox reactors with differing nitrogen loading rates. Water Res. 2020, 169, 115279. [Google Scholar] [CrossRef] [PubMed]
- Fan, N.-S.; Bai, Y.-H.; Wu, J.; Zhang, Q.; Fu, J.-J.; Zhou, W.-L.; Huang, B.-C.; Jin, R.-C. A two-stage anammox process for the advanced treatment of high-strength ammonium wastewater: Microbial community and nitrogen transformation. J. Clean. Prod. 2020, 261, 121148. [Google Scholar] [CrossRef]
- Orschler, L.; Agrawal, S.; Lackner, S. Targeted metagenomics reveals extensive diversity of the denitrifying community in partial nitritation anammox and activated sludge systems. Biotechnol. Bioeng. 2020, 118, 433–441. [Google Scholar] [CrossRef]
- Kim, H.; Ogram, A.; Bae, H. Nitrification, Anammox and Denitrificationalong a Nutrient Gradient in the Florida Everglades. Wetlands 2017, 37, 391–399. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, L.; Sun, S.; Li, J.; Jia, T.; Peng, Y. In situ enrichment of anammox bacteria in anoxic biofilms are possible due to the stable and long-term accumulation of nitrite during denitrification. Bioresour. Technol. 2020, 300, 122668. [Google Scholar] [CrossRef]
- Trojanowicz, K.; Plaza, E.; Trela, J. Pilot scale studies on nitritation-anammox process for mainstream wastewater at low temperature Water. Sci. Technol. 2016, 73, 761–768. [Google Scholar] [CrossRef]
- Sharif Shourjeh, M.; Kowal, P.; Lu, X.; Xie, L.; Drewnowski, J. Development of Strategies for AOB and NOB Competition Supported by Mathematical Modeling in Terms of Successful Deammonification Implementation for Energy-Efficient WWTPs. Processes 2021, 9, 562. [Google Scholar] [CrossRef]
- Yuan, Y.; Xie, Y.; Xu, P.; Li, X. Verification of inhibition effects of anoxic/aerobic alternation on NOB in a nitrosation system under mainstream conditions. J. Water Process Eng. 2022, 45, 102479. [Google Scholar] [CrossRef]
- Strous, M.; Heijnen, J.; Kuenen, J.; Jetten, M.S.M. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl. Microbiol. Biotechnol. 1998, 50, 589–596. [Google Scholar] [CrossRef]
- Noophan, P.L.; Sripiboon, S.; Damrongsri, M.; Munakata-Marr, J. Anaerobic ammonium oxidation by Nitrosomonas spp. and anammox bacteria in a sequencing batch reactor. J. Environ. Manag. 2009, 90, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Clagnan, E.; Brusetti, L.; Pioli, S.; Visigalli, S.; Turolla, A.; Jia, M.; Bargna, M.; Ficara, E.; Bergna, G.; Canziani, R.; et al. Microbial community and performance of a partial nitritation/anammox sequencing batch reactor treating textile wastewater. Heliyon 2021, 7, e08445. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewski, M.; Cema, G.; Twardowski, T.; Ziembińska-Buczyńska, A. Performance of the anammox sequencing batch reactor treating synthetic and real landfill leachate. E3S Web Conf. 2018, 44, 00179. [Google Scholar] [CrossRef]
- Phan, T.N.; Van Truong, T.T.; Ha, N.B.; Nguyen, P.D.; Bui, X.T.; Dang, B.T.; Doan, V.T.; Park, J.; Guo, W.; Ngo, H.H. High rate nitrogen removal by ANAMMOX internal circulation reactor (IC) for old landfill leachate treatment. Bioresour. Technol. 2017, 234, 281–288. [Google Scholar] [CrossRef]
- Miodoński, S.; Muszyński-Huhajło, M.; Zięba, B.; Ratkiewicz, K.; Kołbuc, D.; Łagocka, M. Fast start-up of anammox process with hydrazine addition. SN Appl. Sci. 2019, 1, 523. [Google Scholar] [CrossRef]
- Vlaeminck, S.E.; Terada, A.; Smets, B.F.; Linden, D.V.D.; Boon, N.; Verstraete, W.; Carballa, M. Nitrogen Removal from Digested Black Water by One-Stage Partial Nitritation and Anammox. Environ. Sci. Technol. 2009, 43, 5035–5504. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Becker, A.C.F.; Naves, F.A.; de Angelis, D.D.F. Differential occurrence of heterotrophic bacteria to specific physicochemical characteristics of oil refinery wastewater and adjacent water bodies. J. Appl. Biotechnol. Bioeng. 2017, 2, 18–23. [Google Scholar] [CrossRef]
- Yamada, T.; Tsuji, H.; Daimon, H. Nitrate removal performance and diversity of active denitrifying bacteria in denitrification reactors using poly(L-lactic acid) with enhanced chemical hydrolyzability. Environ. Sci. Pollut. Res. Int. 2019, 36, 36236–36247. [Google Scholar] [CrossRef]
- Huno, S.K.M.; Rene, E.R.; van Hullebusch, E.D.; Annachhatre, A.P. Nitrate removal from groundwater: A review of natural and engineered processes. J. Water Supply Res. Technol. Aqua 2018, 67, 885–902. [Google Scholar] [CrossRef]
- Zhang, W.; Ruan, X.; Bai, Y.; Yin, L. The characteristics and performance of sustainable-releasing compound carbon source material applied on groundwater nitrate in-situ remediation. Chemosphere 2018, 205, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Salcedo Moyano, A.J.; Delforno, T.P.; Subtil, E.L. Simultaneous nitrification-denitrification (SND) using a thermoplastic gel as support: Pollutants removal and microbial community in a pilot-scale biofilm membrane bioreactor. Environ. Technol. 2021, 13, 4411–4425. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, C.M.; Camejo, P.; Oshlag, J.Z.; Noguera, D.R. Ammonia-oxidizing microbial communities in reactors with efficient nitrification at low-dissolved oxygen. Water Res. 2015, 70, 38–51. [Google Scholar] [CrossRef]
- Baskaran, V.; Patil, P.K.; Antony, M.L.; Avunje, S.; Nagaraju, V.T.; Ghate, S.D.; Nathamuni, S.; Dineshkumar, N.; Alavandi, S.V.; Vijayan, K.K. Microbial community profiling of ammonia and nitrite oxidizing bacterial enrichments from brackishwater ecosystems for mitigating nitrogen species. Sci. Rep. 2020, 10, 5201. [Google Scholar] [CrossRef]
- Nagpal, S.; Haque, M.M.; Singh, R.; Mande, S.S. iVikodak—A Platform and Standard Workflow for Inferring, Analyzing, Comparing, and Visualizing the Functional Potential of Microbial Communities. Front. Microbiol. 2019, 9, 3336. [Google Scholar] [CrossRef]




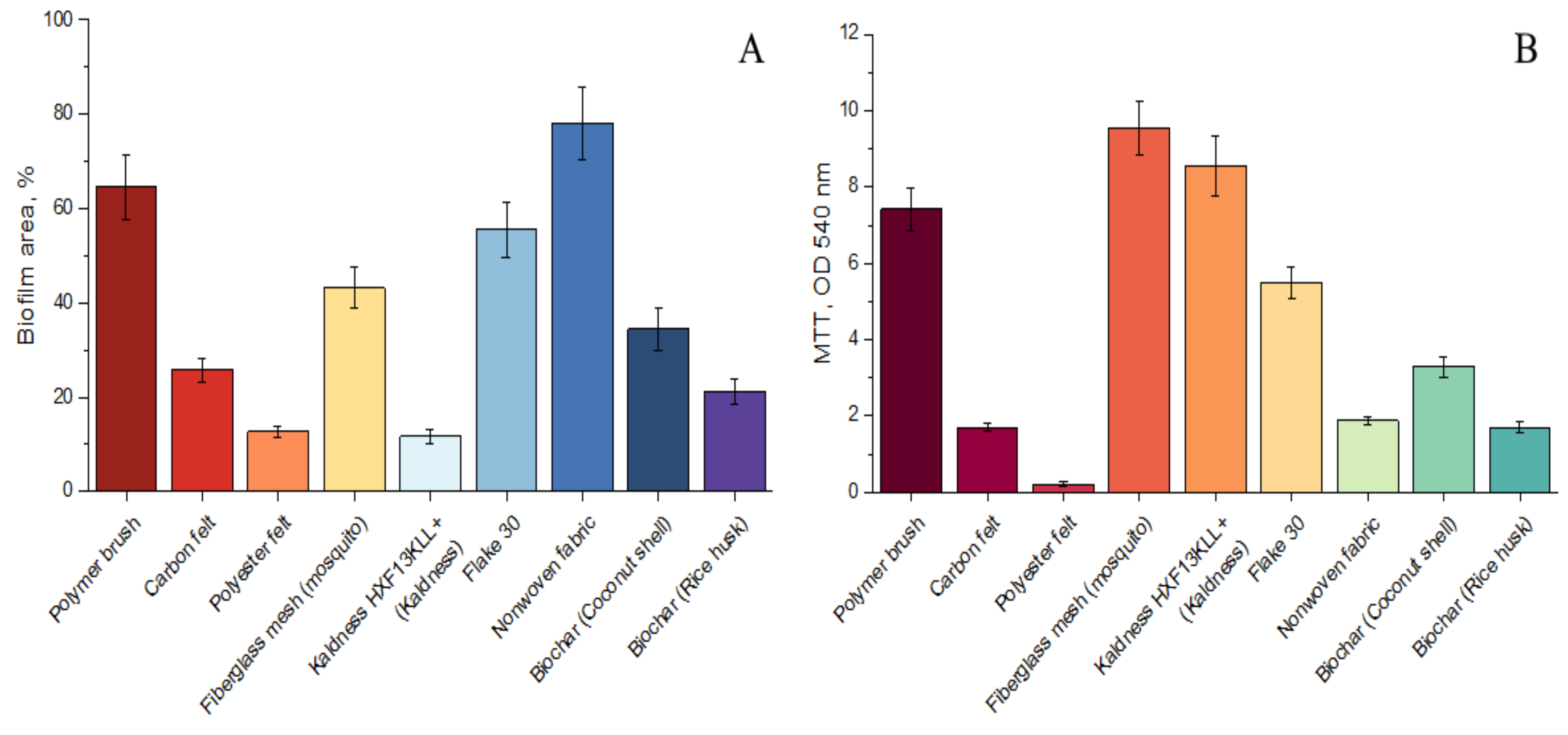





| Parameter | Value |
|---|---|
| NH4, mg/L | 58.4 |
| NO2, mg/L | 28.18 |
| NO3, mg/L | 7434 |
| HCO3, mg/L | 244 |
| K, mg/L | 447.6 |
| Na, mg/L | 879.7 |
| Mg, mg/L | 37.6 |
| Ca, mg/L | 1310 |
| Cl, mg/L | 1213 |
| SO4, mg/L | 1803 |
| Fetot, mg/L | 2.1 |
| Mn, mg/L | 15.7 |
| PO4, mg/L | 1.1 |
| U, µg/L | 256 |
| Corg, mg/L | 2.5 |
| pH | 7.1 |
| Eh, mV | 119 |
| T, °C | 7.5 |
| Carrier Material | BET Surface Area, m2/g |
|---|---|
| Polymer brush (Volgodonsk, Russia) | 1.5 |
| Carbon felt (ZLWMQMD 001 store, China) | 0.4259 |
| Polyester felt (JSC MONTEM, Russia) | 0.0536 |
| Fiberglass mesh (mosquito) (Phifer micro mesh, Tuscaloosa, AL, USA) | 0.003526 |
| HXF13KLL+ (Kaldness) (Hel-X, Marktrodach, Bavaria, Germany) | ~0.003 (955 m2/m3) |
| Flake 30 (Hel-X, Marktrodach, Bavaria, Germany) | ~0.01 (5000 m2/m3) |
| Nonwoven fabric (Geolia, Taiwan, China) | ~300 |
| Biochar (Coconut shell) (Noname, China) | 772 |
| Phases (Days) | N-NH4 in the Influent, mg/L | N-NO2 in the Influent, mg/L | рН | Daily Influent Flow Rate, mL/Day |
|---|---|---|---|---|
| 1 (1–80) | from 5 to 50 | from 5 to 40 | 7.5–8 | 122.4 |
| 2 (80–105) | 40–50 | 30–40 | 244.8 | |
| 3 (105–180) | 50–60 | 30–45 | 489.6 | |
| 4 (180–208) | 35–40 | 45–60 | 244.8 | |
| 4 (208–285) | 75–85 | 60–70 |
| Probe | Sequence, 5′-3′ | Target Group | Reference |
|---|---|---|---|
| Amx368 | CCT TTC GGG CAT TGC GAA | all anammox bacteria | [54] |
| Nsm156 | TAT TAG CAC ATC TTT CGA T | Nitrosomonas spp., Nitrosococcus spp. | [55] |
| Ntspa1151 | TTC TCC TGG GCA GTC TCT CC | Sublineage II of the genus Nitrospira sp. | [56] |
| Nit3 | CCT GTG CTC CAT GCT CCG (competitor: CCT GTG CTC CAG GCT CCG) | Nitrobacter spp. | [57] |
| Ntspa662 | GGA ATT CCG CGC TCC TCT (competitor: GGA ATT CCG CTC TCC TCT) | Nitrospira spp. | [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Botchkova, E.; Vishnyakova, A.; Popova, N.; Sukhacheva, M.; Kolganova, T.; Litti, Y.; Safonov, A. Characterization of Enrichment Cultures of Anammox, Nitrifying and Denitrifying Bacteria Obtained from a Cold, Heavily Nitrogen-Polluted Aquifer. Biology 2023, 12, 221. https://doi.org/10.3390/biology12020221
Botchkova E, Vishnyakova A, Popova N, Sukhacheva M, Kolganova T, Litti Y, Safonov A. Characterization of Enrichment Cultures of Anammox, Nitrifying and Denitrifying Bacteria Obtained from a Cold, Heavily Nitrogen-Polluted Aquifer. Biology. 2023; 12(2):221. https://doi.org/10.3390/biology12020221
Chicago/Turabian StyleBotchkova, Ekaterina, Anastasia Vishnyakova, Nadezhda Popova, Marina Sukhacheva, Tatyana Kolganova, Yuriy Litti, and Alexey Safonov. 2023. "Characterization of Enrichment Cultures of Anammox, Nitrifying and Denitrifying Bacteria Obtained from a Cold, Heavily Nitrogen-Polluted Aquifer" Biology 12, no. 2: 221. https://doi.org/10.3390/biology12020221
APA StyleBotchkova, E., Vishnyakova, A., Popova, N., Sukhacheva, M., Kolganova, T., Litti, Y., & Safonov, A. (2023). Characterization of Enrichment Cultures of Anammox, Nitrifying and Denitrifying Bacteria Obtained from a Cold, Heavily Nitrogen-Polluted Aquifer. Biology, 12(2), 221. https://doi.org/10.3390/biology12020221








