Spatially and Temporally Confined Response of Gastrointestinal Antibiotic Resistance Gene Levels to Sulfadiazine and Extracellular Antibiotic Resistance Gene Exposure in Mice
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids and Strains
2.2. Conjugation Assay
2.3. Exposure of Gastrointestinal Microbiomes to Sulfadiazine, pR55, and Sulfadiazine + pR55
2.4. Quantitative Methods
2.5. Statistics
3. Results and Discussion
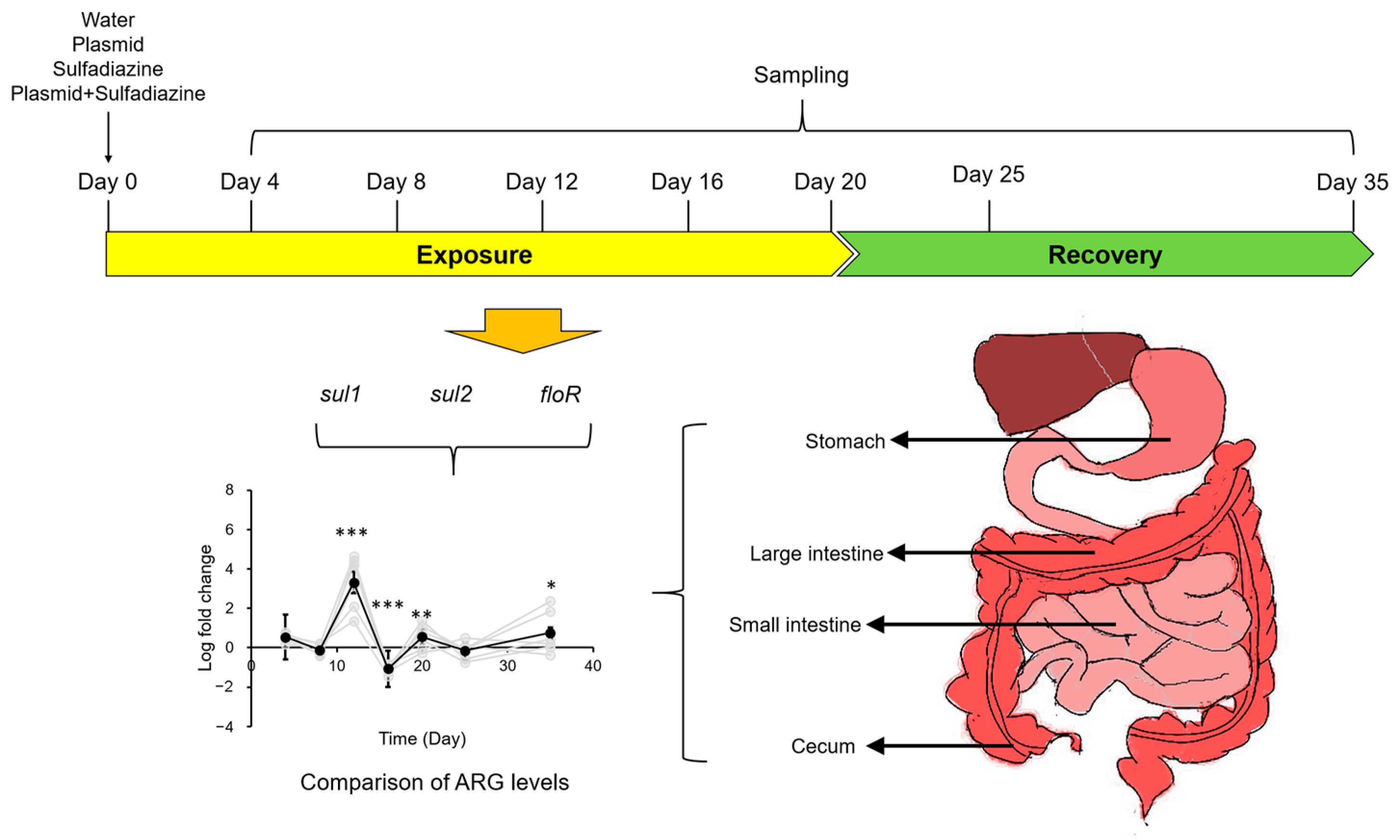
3.1. Experimental Setup
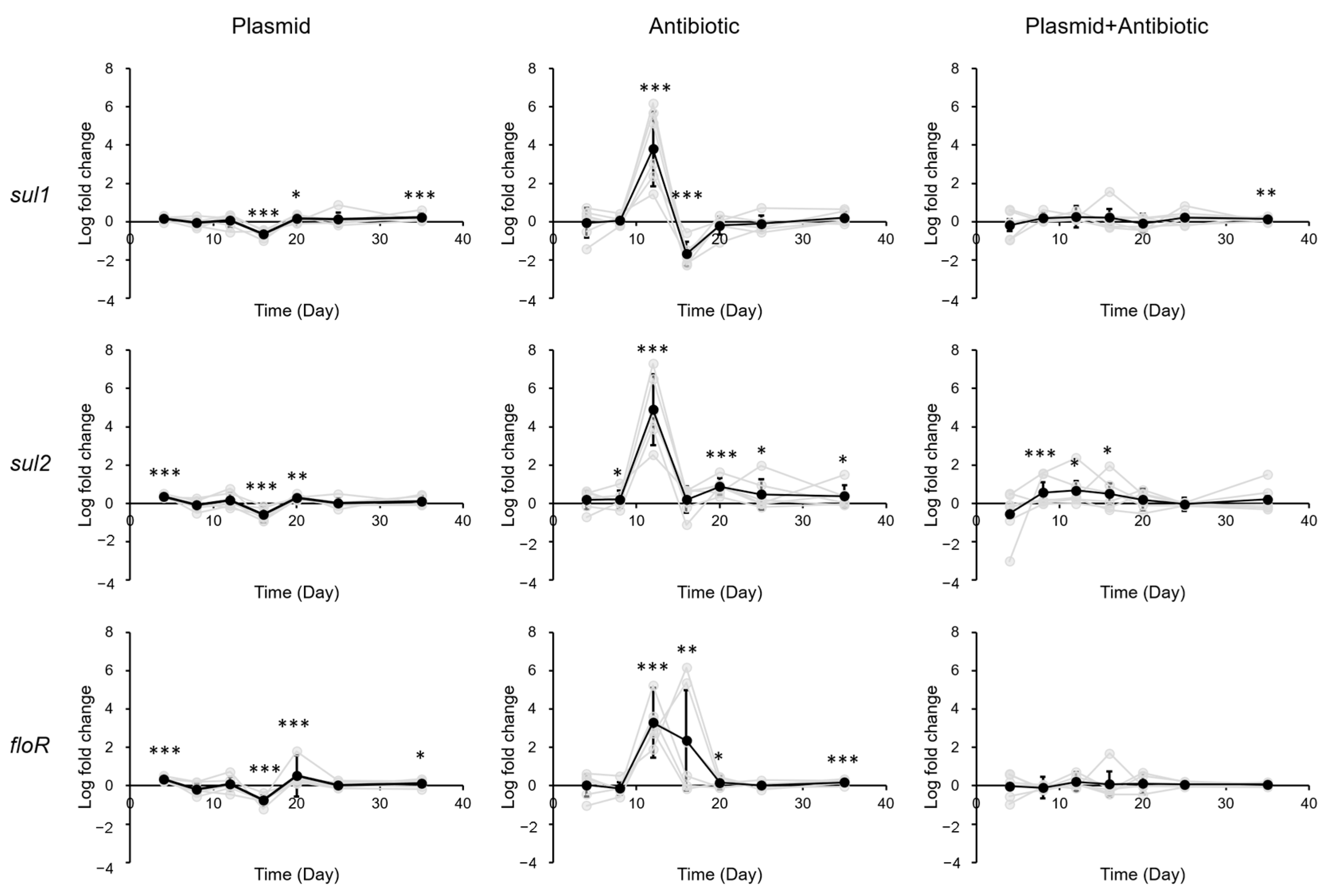
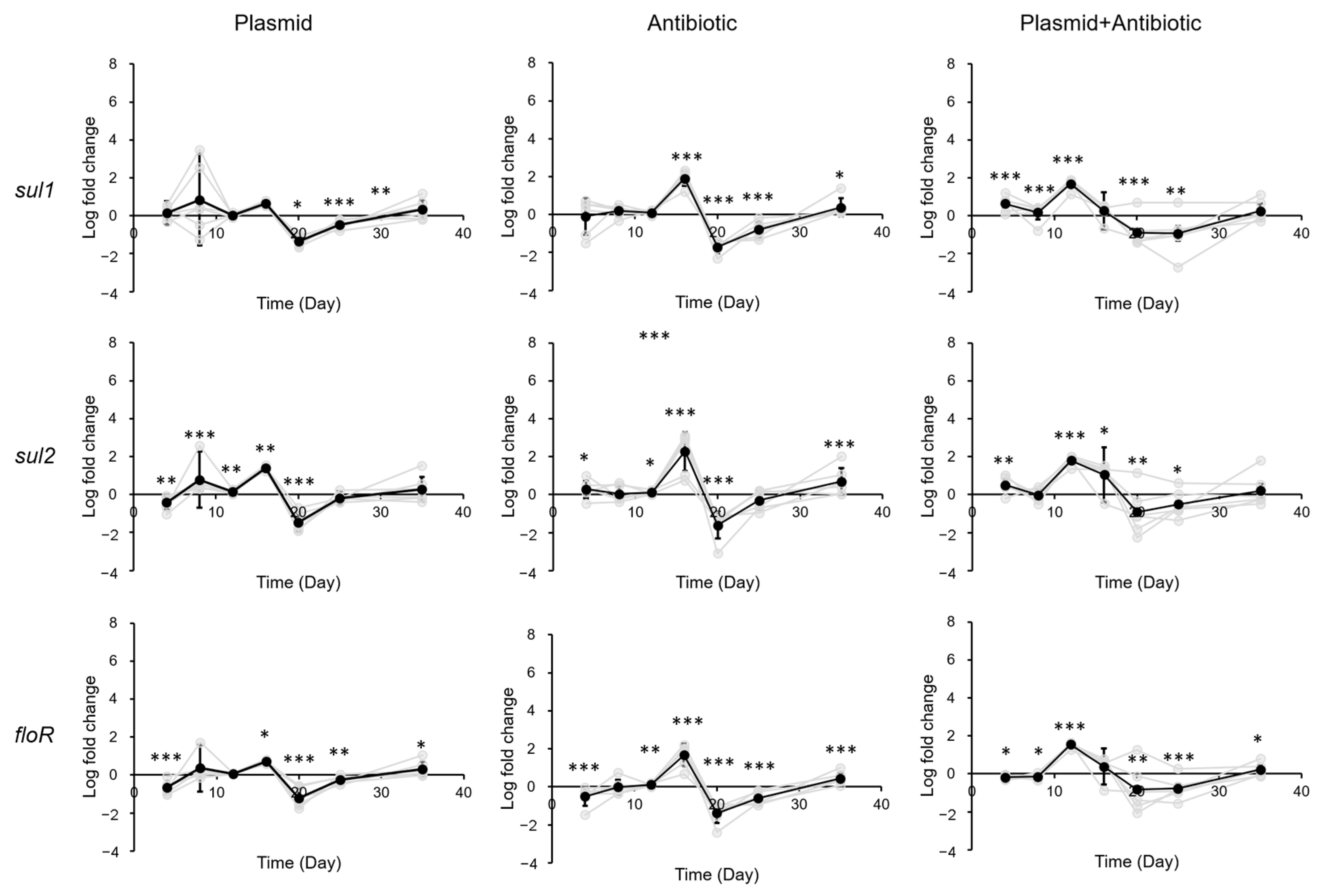
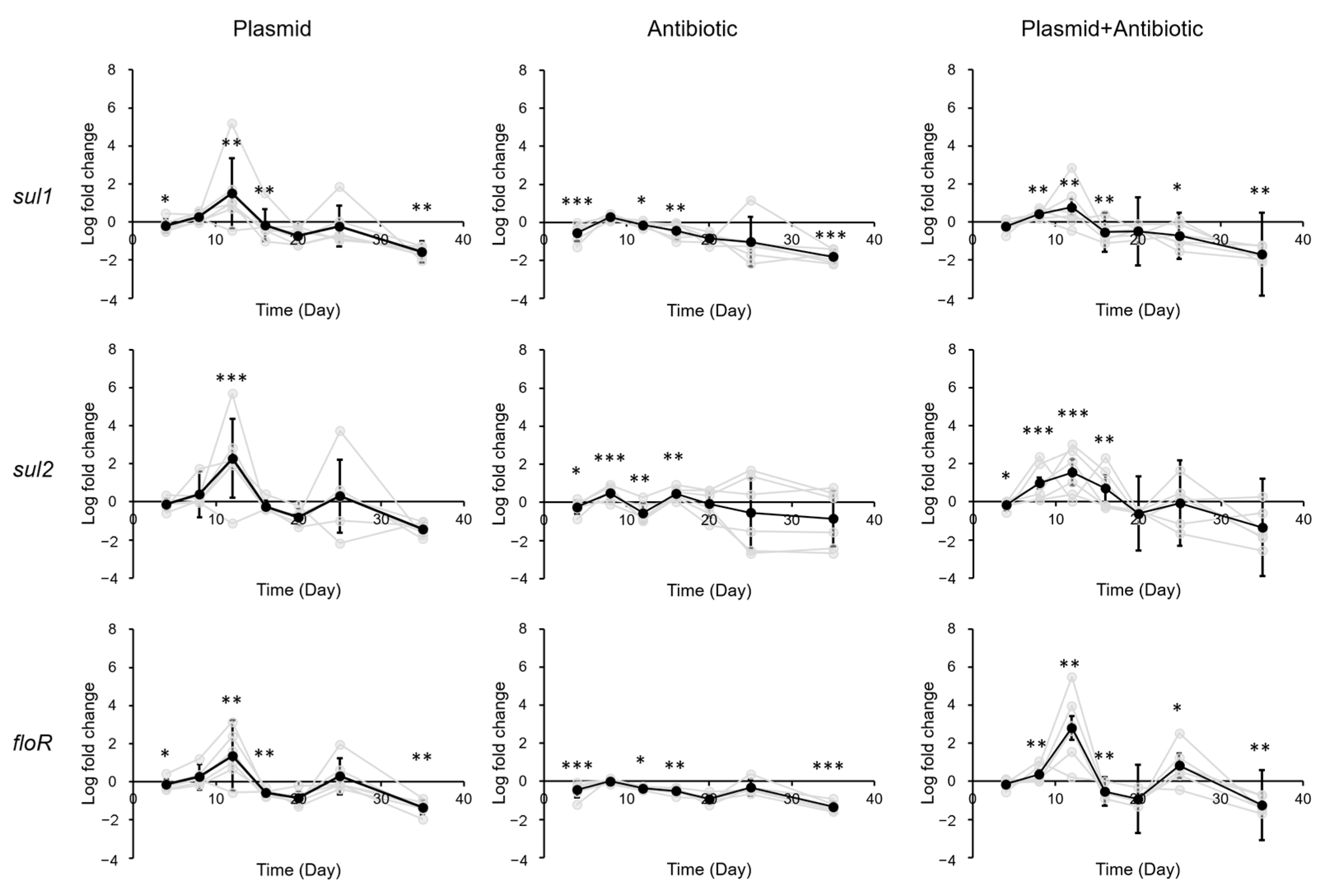
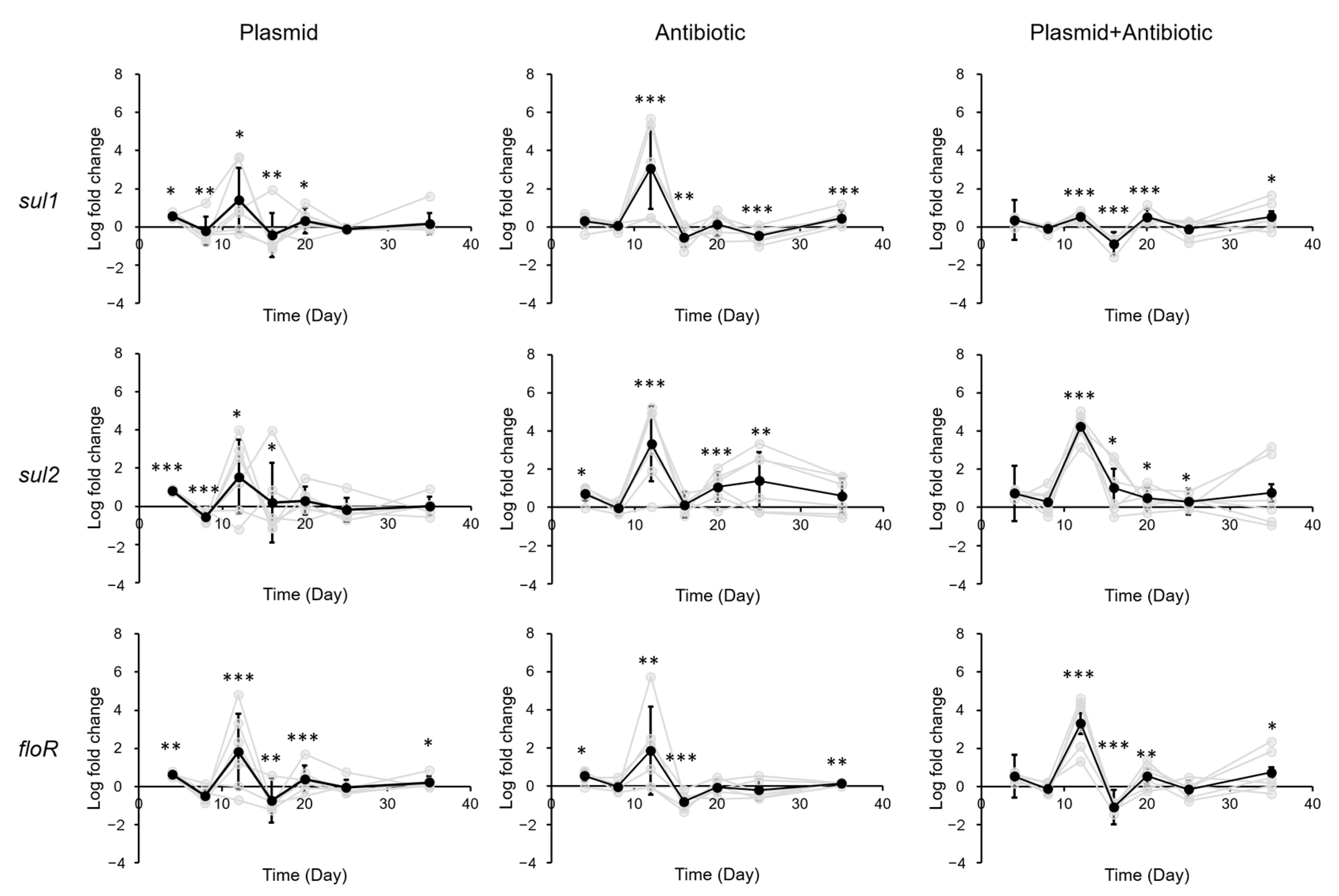
3.2. Impact of Sulfadiazine and eARG Exposure on ARG Levels in Gastrointestinal Tract of Mice
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cambray, G.; Guerout, A.-M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Liao, X.-P.; D’Souza, A.W.; Boolchandani, M.; Li, S.-H.; Cheng, K.; Luis Martínez, J.; Li, L.; Feng, Y.-J.; Fang, L.-X.; et al. Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat. Commun. 2020, 11, 1427. [Google Scholar] [CrossRef] [PubMed]
- Schubbert, R.; Renz, D.; Schmitz, B.; Doerfler, W. Foreign (M13) DNA ingested by mice reaches peripheral leukocytes, spleen, and liver via the intestinal wall mucosa and can be covalently linked to mouse DNA. Proc. Natl. Acad. Sci. USA 1997, 94, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-N.; Liu, L.; Qiu, Z.-G.; Shen, Z.-Q.; Guo, X.; Yang, D.; Li, J.; Liu, W.; Jin, M.; Li, J.-W. A new adsorption-elution technique for the concentration of aquatic extracellular antibiotic resistance genes from large volumes of water. Water Res. 2016, 92, 188–198. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Z.; Hu, J.; Yan, L.; He, Y.; Li, X.; Wang, M.; Sun, X.; Xu, H. The biological and chemical contents of atmospheric particulate matter and implication of its role in the transmission of bacterial pathogenesis. Environ. Microbiol. 2021, 23, 5481–5486. [Google Scholar] [CrossRef]
- Yan, L.; Liu, D.; Wang, X.-H.; Wang, Y.; Zhang, B.; Wang, M.; Xu, H. Bacterial plasmid-mediated quinolone resistance genes in aquatic environments in China. Sci. Rep. 2017, 7, 40610. [Google Scholar] [CrossRef]
- Ma, L.; Li, B.; Jiang, X.-T.; Wang, Y.-L.; Xia, Y.; Li, A.-D.; Zhang, T. Catalogue of antibiotic resistome and host-tracking in drinking water deciphered by a large scale survey. Microbiome 2017, 5, 154. [Google Scholar] [CrossRef]
- Cerqueira, F. Antibiotic resistance gene distribution in agricultural fields and crops. A soil-to-food analysis. Environ. Res. 2019, 177, 108608. [Google Scholar] [CrossRef]
- Meletiadis, J.; Turlej-Rogacka, A.; Lerner, A.; Adler, A.; Tacconelli, E.; Mouton, J.W.; the SATURN Diagnostic Study Group. Amplification of antimicrobial resistance in gut flora of patients treated with ceftriaxone. Antimicrob. Agents Chemother. 2017, 61, e00473-17. [Google Scholar] [CrossRef]
- Card, R.M.; Cawthraw, S.A.; Nunez-Garcia, J.; Ellis, R.J.; Kay, G.; Pallen, M.J.; Woodward, M.J.; Anjum, M.F. An in vitro chicken gut model demonstrates transfer of a multidrug resistance plasmid from Salmonella to commensal Escherichia coli. mBio 2017, 8, e00777-17. [Google Scholar] [CrossRef]
- Pehrsson, E.C.; Tsukayama, P.; Patel, S.; Mejía-Bautista, M.; Sosa-Soto, G.; Navarrete, K.M.; Calderon, M.; Cabrera, L.; Hoyos-Arango, W.; Bertoli, M.T.; et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature 2016, 533, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Yang, D.; Jin, M.; Liu, W.; Zhao, X.; Li, C.; Zhao, T.; Wang, J.; Gao, Z.; Shen, Z.; et al. Aquatic animals promote antibiotic resistance gene dissemination in water via conjugation: Role of different regions within the zebra fish intestinal tract, and impact on fish intestinal microbiota. Mol. Ecol. 2017, 26, 5318–5333. [Google Scholar] [CrossRef] [PubMed]
- Lakticová, V.; Hutton-Thomas, R.; Meyer, M.; Gurkan, E.; Rice, L.B. Antibiotic-induced Enterococcal expansion in the mouse intestine occurs throughout the small bowel and correlates poorly with suppression of competing flora. Antimicrob. Agents Chemother. 2006, 50, 3117–3123. [Google Scholar] [CrossRef]
- Mh, M.; Gw, B.; Rw, G.; Lj, H. Effects of heat stress on the antimicrobial drug resistance of Escherichia coli of the intestinal flora of swine. J. Appl. Microbiol. 2000, 88, 836–844. [Google Scholar] [CrossRef]
- Doublet, B.; Boyd, D.; Douard, G.; Praud, K.; Cloeckaert, A.; Mulvey, M.R. Complete nucleotide sequence of the multidrug resistance IncA/C plasmid pR55 from Klebsiella pneumoniae isolated in 1969. J. Antimicrob. Chemother. 2012, 67, 2354–2360. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shi, L.; Wang, B.; Wei, X.; Zhang, J.; Guo, T.; Kong, J.; Wang, M.; Xu, H. In vitro assessment of antimicrobial resistance dissemination dynamics during multidrug-resistant-bacterium invasion events by using a continuous-culture device. Appl. Environ. Microbiol. 2021, 87, e02659-20. [Google Scholar] [CrossRef] [PubMed]
- Borgia, S.; Lastovetska, O.; Richardson, D.; Eshaghi, A.; Xiong, J.; Chung, C.; Baqi, M.; McGeer, A.; Ricci, G.; Sawicki, R.; et al. Outbreak of carbapenem-resistant Enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clin. Infect. Dis. 2012, 55, e109–e117. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-N.; Wu, Y.-Q.; Qu, Y.; Wei, J.-D.; Zhong, G.-S. Comparison of four methods for extracting microbial DNA from mouse intestine. Chin. J. Ecol. 2015, 34, 1183–1188. [Google Scholar]
- Wang, Y.; Ren, M.; Wang, Y.; Wang, L.; Liu, H.; Shi, M.; Zhong, Y. Constitutive overexpression of cellobiohydrolase 2 in Trichoderma reesei reveals its ability to initiate cellulose degradation. Eng. Microbiol. 2023, 3, 100059. [Google Scholar] [CrossRef]
- Wang, M.; Shen, W.; Yan, L.; Wang, X.-H.; Xu, H. Stepwise Impact of urban wastewater treatment on the bacterial community structure, antibiotic contents, and prevalence of antimicrobial resistance. Environ. Pollut. 2017, 231, 1578–1585. [Google Scholar] [CrossRef]
- Jiang, L.; Hu, X.; Xu, T.; Zhang, H.; Sheng, D.; Yin, D. Prevalence of antibiotic resistance genes and their relationshiip with antibiotics in the Huangpu River and the drinking water sources, Shanghai, China. Sci. Total Environ. 2013, 458–460, 267–272. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, J.-L. Multipurpose assessment for the quantification of Vibrio spp. and total bacteria in fish and seawater using multiplex real-time polymerase chain reaction. J. Sci. Food Agric. 2014, 94, 2807–2817. [Google Scholar] [CrossRef] [PubMed]
- Shen, W. Elimination of Antibiotics and Antibiotics Resistance Genes in Sewage Treatment Plants and Preliminary Study on the Degradation of Antibiotics by Microorganisms. Master’s Thesis, Shandong University, Jinan, China, 2018. [Google Scholar]
- Huddleston, J.R. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug Resist. 2014, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L.; Rojo, F. Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 768–789. [Google Scholar] [CrossRef]
- San Millan, A.; MacLean, R.C. Fitness costs of plasmids: A limit to plasmid transmission. Microbiol. Spectr. 2017, 5, MTBP-0016-2017. [Google Scholar] [CrossRef] [PubMed]
- Hunt, R.H.; Camilleri, M.; Crowe, S.E.; El-Omar, E.M.; Fox, J.G.; Kuipers, E.J.; Malfertheiner, P.; McColl, K.E.L.; Pritchard, D.M.; Rugge, M.; et al. The stomach in health and disease. Gut 2015, 64, 1650–1668. [Google Scholar] [CrossRef]
- Chaucheyras-Durand, F.; Sacy, A.; Karges, K.; Apper, E. Gastro-intestinal microbiota in equines and its role in health and disease: The black box opens. Microorganisms 2022, 10, 2517. [Google Scholar] [CrossRef]
- Bowcutt, R.; Forman, R.; Glymenaki, M.; Carding, S.R.; Else, K.J.; Cruickshank, S.M. Heterogeneity across the murine small and large Intestine. World J. Gastroenterol. 2014, 20, 15216–15232. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Zhang, J.; Wang, B.; Wang, W.; Sun, Y.; Li, L.; Xu, H.; Wang, M. Spatially and Temporally Confined Response of Gastrointestinal Antibiotic Resistance Gene Levels to Sulfadiazine and Extracellular Antibiotic Resistance Gene Exposure in Mice. Biology 2023, 12, 210. https://doi.org/10.3390/biology12020210
Wei X, Zhang J, Wang B, Wang W, Sun Y, Li L, Xu H, Wang M. Spatially and Temporally Confined Response of Gastrointestinal Antibiotic Resistance Gene Levels to Sulfadiazine and Extracellular Antibiotic Resistance Gene Exposure in Mice. Biology. 2023; 12(2):210. https://doi.org/10.3390/biology12020210
Chicago/Turabian StyleWei, Xin, Jian Zhang, Bianfang Wang, Wenjia Wang, Yuqing Sun, Ling Li, Hai Xu, and Mingyu Wang. 2023. "Spatially and Temporally Confined Response of Gastrointestinal Antibiotic Resistance Gene Levels to Sulfadiazine and Extracellular Antibiotic Resistance Gene Exposure in Mice" Biology 12, no. 2: 210. https://doi.org/10.3390/biology12020210
APA StyleWei, X., Zhang, J., Wang, B., Wang, W., Sun, Y., Li, L., Xu, H., & Wang, M. (2023). Spatially and Temporally Confined Response of Gastrointestinal Antibiotic Resistance Gene Levels to Sulfadiazine and Extracellular Antibiotic Resistance Gene Exposure in Mice. Biology, 12(2), 210. https://doi.org/10.3390/biology12020210






