Selection and Evaluation of mRNA and miRNA Reference Genes for Expression Studies (qPCR) in Archived Formalin-Fixed and Paraffin-Embedded (FFPE) Colon Samples of DSS-Induced Colitis Mouse Model
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Protocol
2.2. Archived FFPE Colon Samples from 9 DSS Experiments and Selection
- FFPE samples from 9 DSS experiments;
- FFPE samples from females and males;
- FFPE samples from the distal part of the colon (last 1/3);
- FFPE samples have been histological examined and confirmed that:
- a.
- all layers of the colon wall and no other tissue is present on the FFPE samples;
- b.
- all samples from the DSS group have histologically confirmed colitis;
- c.
- all samples from the control group show a normal histological picture (no signs of lesion or inflammation).
- in the control group: 20 FFPE samples (from 20 untreated healthy mice: 10 males and 10 females),
- in the DSS group: 97 FFPE samples (from 97 DSS-treated mice: 50 males and 47 females).
2.3. RNA Isolation from Archived FFPE Samples
2.4. Selection of Reference Genes (mRNA/miRNA) for Archived FFPE Samples
2.5. Reverse Transcription and Quantitative Real-time Polymerase Chain Reaction (RT-qPCR)
2.5.1. mRNA
2.5.2. miRNA
2.6. Amplification Efficiency and RNA Integrity
| Control Group | DSS Group | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Assay ID | Length [bp] | Catalogue No. | Eff. [%] | R2 | Eff. [%] | R2 |
| EEF2 | Eucaryotic translation elongation factor 2 | Mm01171434_g1 | 74 | 4331182 | 95 | 0.998 | 102 | 0.997 |
| TBP | TATA box binding protein | Mm00446971_m1 | 93 | 4331182 | 125 | 0.974 | 107 | 0.971 |
| NONO | Non-POU domain containing, octamer binding protein | Mm07293722_g1 | 77 | 4351372 | 83 | 0.993 | 103 | 0.990 |
| PPIA | Peptidylprolyl isomerase A | Mm02342429_g1 | 112 | 4331182 | 90 | 0.998 | 85 | 0.998 |
| RPLP0 | Ribosomal protein large P0 | Mm01974474_gH | 89 | 4331182 | 89 | 0.998 | 91 | 0.997 |
| TNFR1 | Tumor necrosis factor receptor 1 | Mm00441875_m1 | 69 | 4331182 | 98 | 0.992 | 99 | 0.990 |
| TNFR2 | Tumor necrosis factor receptor 2 | Mm00441889_m1 | 64 | 4331182 | 98 | 0.994 | 92 | 0.994 |
| Control Group | DSS Group | |||||
|---|---|---|---|---|---|---|
| Mirna Symbol | Accession Number | Gene Globe ID/Cat. No. | Eff. [%] | R2 | Eff. [%] | R2 |
| U6 snRNA | / | YP02119464/339306 | 106 | 0.995 | 104 | 0.994 |
| miR-191-5p | MIMAT0000440 | YP00204306/339306 | 109 | 0.995 | 107 | 0.995 |
| miR-103a-3p | MIMAT0000101 | YP00204063/339306 | 106 | 0.995 | 103 | 0.996 |
| miR-16-5p | MIMAT0000069 | YP00205702/339306 | 100 | 0.997 | 96 | 0.998 |
| miR-181a-5p | MIMAT0000256 | YP00206081/339306 | 102 | 0.990 | 105 | 0.997 |
| miR-223-3p | MIMAT0000280 | YP00205986/339306 | 101 | 0.994 | 93 | 0.998 |
| miR-680 | MIMAT0003457 | YP00205101/339306 | 121 | 0.990 | 92 | 0.842 |
| miR-1224-5p | MIMAT0005460 | YP02115039/339306 | 151 | 0.885 | 145 | 0.882 |
| miR-5128 | MIMAT0020639 | YP02111071/339306 | 119 | 0.613 | 138 | 0.082 |
| miR-3968 | MIMAT0019352 | YP02104299/339306 | 112 | 0.933 | 82 | 0.911 |
| Let-7f-5p | MIMAT0000067 | YP00204359/339306 | 72 | 0.996 | 70 | 0.996 |
2.7. Reference Gene Stability Analysis
2.8. Statistical Analysis
2.9. Multivariate Analysis
3. Results
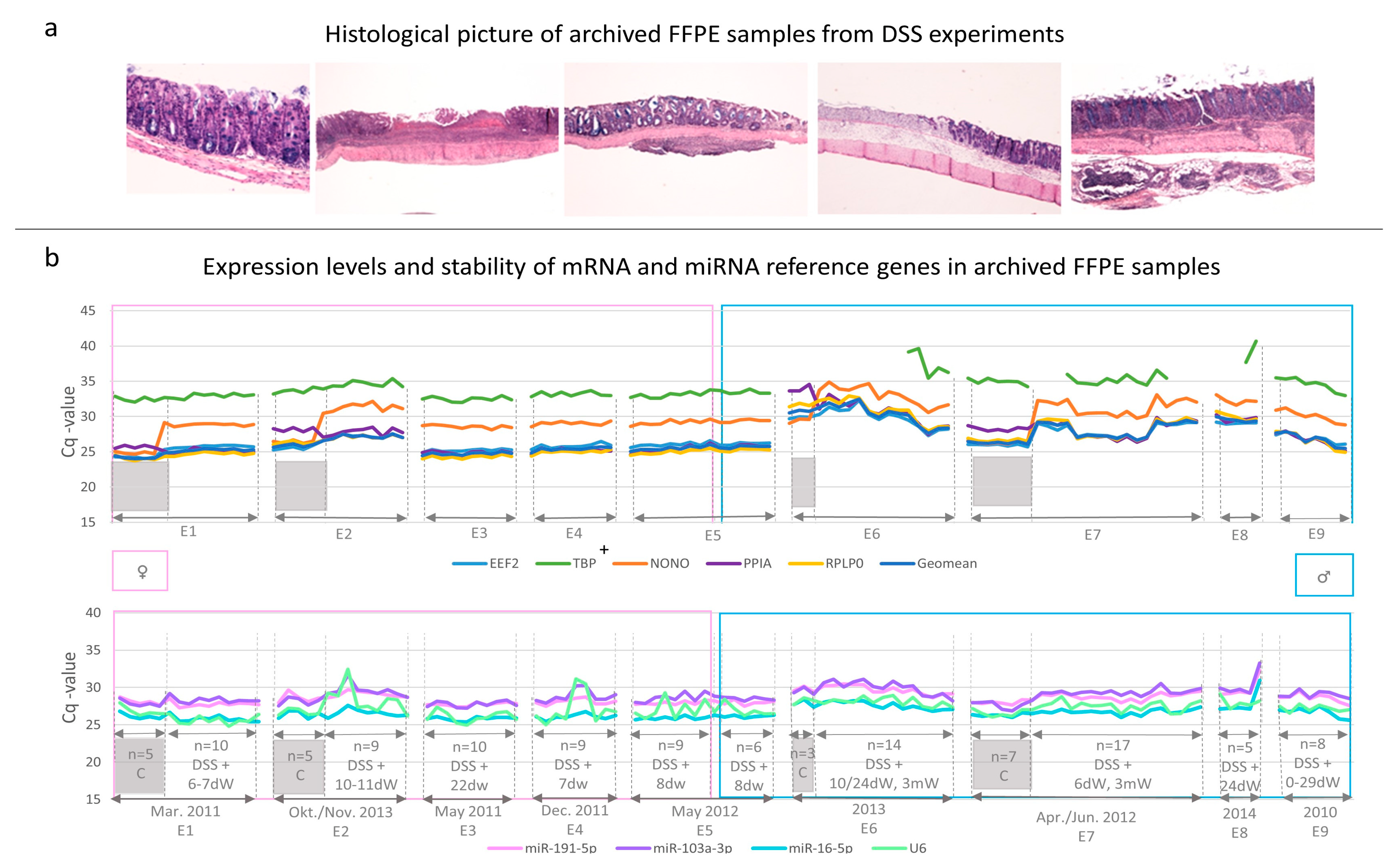
3.1. Histological Examination of Archived FFPE Samples
3.2. Efficiency and Specificity of mRNA/miRNA qPCR
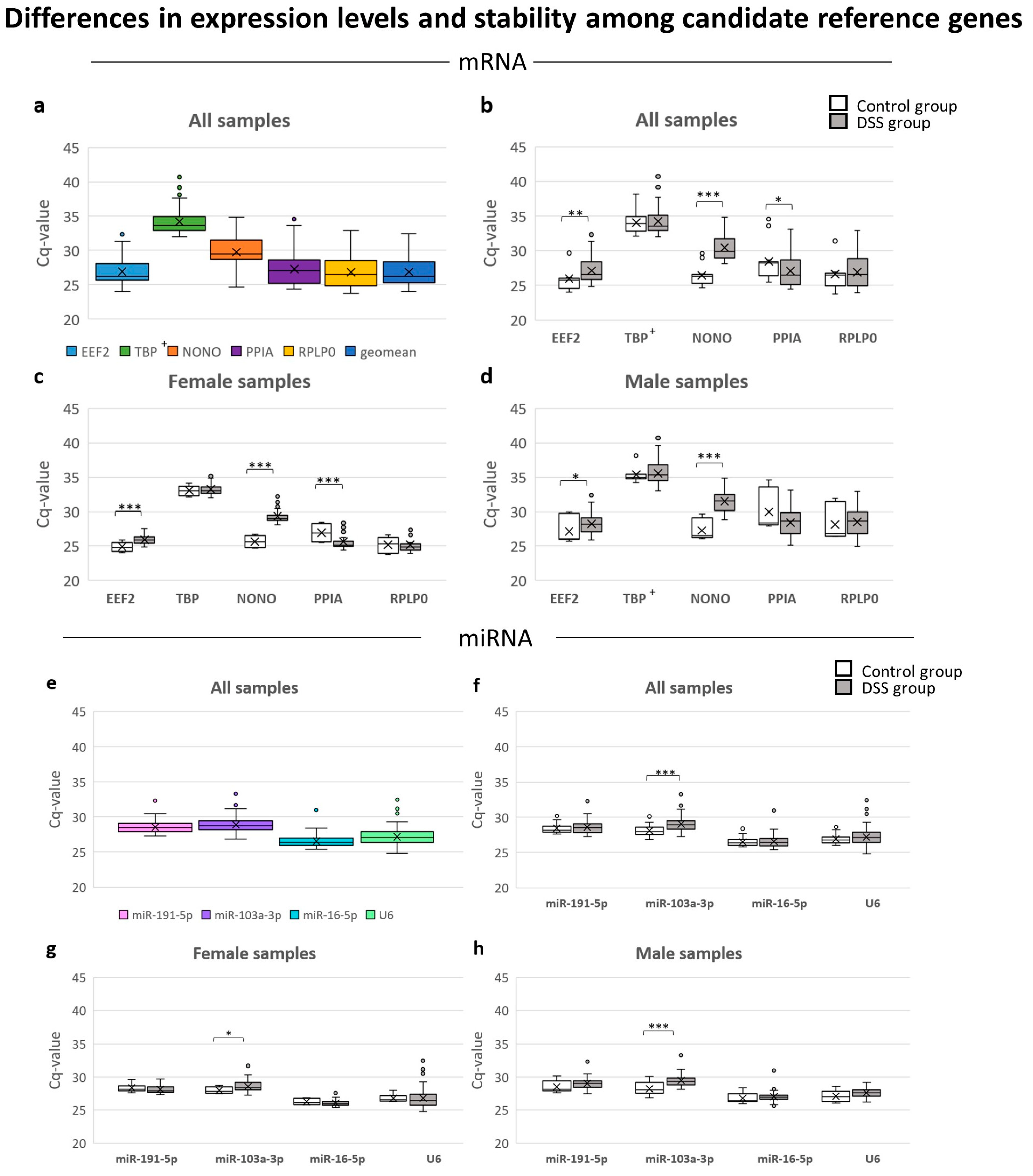
3.3. Candidate Reference Gene Expression Levels in Archived FFPE Samples
3.4. Experimental Factors Affecting Reference Gene Expression Patterns
3.4.1. Effects of DSS Treatment
3.4.2. Effects of Sex
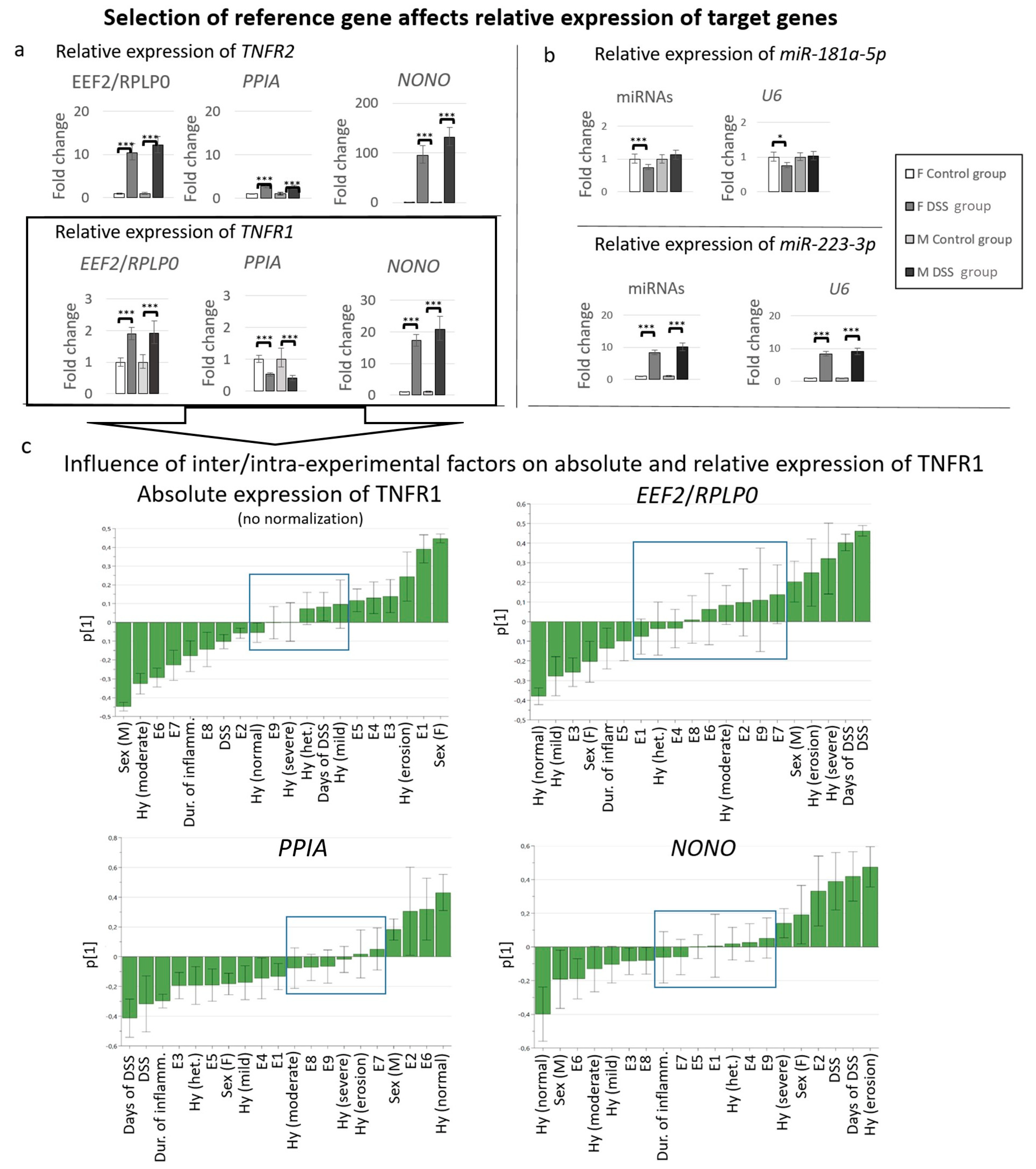
3.4.3. Effects of Inter/Intra-Experimental Factors
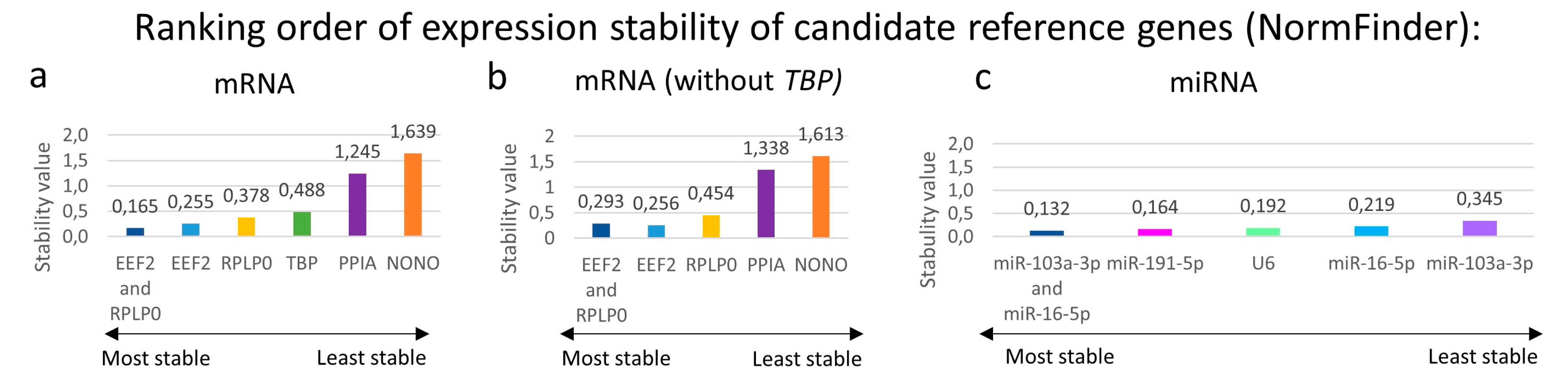
3.5. Stability of Reference Genes Was Evaluated by BestKeeper and NormFinder
3.5.1. BestKeeper
3.5.2. NormFinder:
3.6. Multivariate Analysis
3.7. Influence of the Use of Different Reference Genes on Expression Analysis of Selected Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Nolan, T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 2004, 15, 155–166. [Google Scholar] [PubMed]
- Coulson, D.T.; Brockbank, S.; Quinn, J.G.; Murphy, S.; Ravid, R.; Irvine, G.B.; Johnston, J.A. Identification of valid reference genes for the normalization of RT qPCR gene expression data in human brain tissue. BMC Mol. Biol. 2008, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Kosir, R.; Acimovic, J.; Golicnik, M.; Perse, M.; Majdic, G.; Fink, M.; Rozman, D. Determination of reference genes for circadian studies in different tissues and mouse strains. BMC Mol. Biol. 2010, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Axtner, J.; Sommer, S. Validation of internal reference genes for quantitative real-time PCR in a non-model organism, the yellow-necked mouse, Apodemus flavicollis. BMC Res. Notes 2009, 2, 264. [Google Scholar] [CrossRef] [PubMed]
- Das, R.K.; Banerjee, S.; Shapiro, B.H. Extensive sex- and/or hormone-dependent expression of rat housekeeping genes. Endocr. Res. 2013, 38, 105–111. [Google Scholar] [CrossRef]
- Eissa, N.; Hussein, H.; Wang, H.; Rabbi, M.F.; Bernstein, C.N.; Ghia, J.E. Stability of Reference Genes for Messenger RNA Quantification by Real-Time PCR in Mouse Dextran Sodium Sulfate Experimental Colitis. PLoS ONE 2016, 11, e0156289. [Google Scholar] [CrossRef]
- Eissa, N.; Kermarrec, L.; Hussein, H.; Bernstein, C.N.; Ghia, J.E. Appropriateness of reference genes for normalizing messenger RNA in mouse 2,4-dinitrobenzene sulfonic acid (DNBS)-induced colitis using quantitative real time PCR. Sci. Rep. 2017, 7, 42427. [Google Scholar] [CrossRef]
- Hildyard, J.C.W.; Finch, A.M.; Wells, D.J. Identification of qPCR reference genes suitable for normalizing gene expression in the mdx mouse model of Duchenne muscular dystrophy. PLoS ONE 2019, 14, e0211384. [Google Scholar] [CrossRef]
- Krzystek-Korpacka, M.; Diakowska, D.; Bania, J.; Gamian, A. Expression stability of common housekeeping genes is differently affected by bowel inflammation and cancer: Implications for finding suitable normalizers for inflammatory bowel disease studies. Inflamm. Bowel Dis. 2014, 20, 1147–1156. [Google Scholar] [CrossRef]
- Lu, X.; Liu, Y.; Zhang, D.; Liu, K.; Wang, Q.; Wang, H. Determination of the panel of reference genes for quantitative real-time PCR in fetal and adult rat intestines. Reprod. Toxicol. 2021, 104, 68–75. [Google Scholar] [CrossRef]
- Medrano, G.; Guan, P.; Barlow-Anacker, A.J.; Gosain, A. Comprehensive selection of reference genes for quantitative RT-PCR analysis of murine extramedullary hematopoiesis during development. PLoS ONE 2017, 12, e0181881. [Google Scholar] [CrossRef]
- Chapman, J.R.; Waldenstrom, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef]
- Drury, S.; Anderson, H.; Dowsett, M. Selection of REFERENCE genes for normalization of qRT-PCR data derived from FFPE breast tumors. Diagn. Mol. Pathol. 2009, 18, 103–107. [Google Scholar] [CrossRef]
- Kokkat, T.J.; Patel, M.S.; McGarvey, D.; LiVolsi, V.A.; Baloch, Z.W. Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv. Biobank. 2013, 11, 101–106. [Google Scholar] [CrossRef]
- Patel, P.G.; Selvarajah, S.; Guérard, K.P.; Bartlett, J.M.; Lapointe, J.; Berman, D.M.; Okello, J.B.; Park, P.C. Reliability and performance of commercial RNA and DNA extraction kits for FFPE tissue cores. PLoS ONE 2017, 12, e0179732. [Google Scholar] [CrossRef]
- Eichele, D.D.; Kharbanda, K.K. Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J. Gastroenterol. 2017, 23, 6016–6029. [Google Scholar] [CrossRef]
- Perse, M.; Cerar, A. Dextran sodium sulphate colitis mouse model: Traps and tricks. J. Biomed. Biotechnol. 2012, 2012, 718617. [Google Scholar] [CrossRef]
- Biassoni, R.; Raso, A. Quantitative Real-Time PCR: Methods and Protocols—Methods in Molecular Biology, 2nd ed.; Humana Press Inc.: New York, NY, USA, 2014. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: New York, NY, USA, 1989. [Google Scholar]
- Danese, E.; Minicozzi, A.M.; Benati, M.; Paviati, E.; Lima-Oliveira, G.; Gusella, M.; Pasini, F.; Salvagno, G.L.; Montagnana, M.; Lippi, G. Reference miRNAs for colorectal cancer: Analysis and verification of current data. Sci. Rep. 2017, 7, 8413. [Google Scholar] [CrossRef]
- Buonpane, C.; Ares, G.; Benyamen, B.; Yuan, C.; Hunter, C.J. Identification of suitable reference microRNA for qPCR analysis in pediatric inflammatory bowel disease. Physiol. Genom. 2019, 51, 169–175. [Google Scholar] [CrossRef]
- Lee, J.; Park, E.J.; Yuki, Y.; Ahmad, S.; Mizuguchi, K.; Ishii, K.J.; Shimaoka, M.; Kiyono, H. Profiles of microRNA networks in intestinal epithelial cells in a mouse model of colitis. Sci. Rep. 2015, 5, 18174. [Google Scholar] [CrossRef] [PubMed]
- Lou, C.; Li, Y. Functional role of microRNA-135a in colitis. J. Inflamm. 2018, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Shen, L.; Luo, H. Identification and Validation of Key miRNAs and a microRNA-mRNA Regulatory Network Associated with Ulcerative Colitis. DNA Cell Biol. 2021, 40, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; He, Y.; Feng, X.; Ye, S.; Wang, H.; Tan, W.; Yu, C.; Hu, J.; Zheng, R.; Zhou, Y. MicroRNA-206 is involved in the pathogenesis of ulcerative colitis via regulation of adenosine A3 receptor. Oncotarget 2017, 8, 705–721. [Google Scholar] [CrossRef]
- Yao, J.; Gao, R.Y.; Luo, M.H.; Wei, C.; Wu, B.H.; Guo, L.L.; Wang, L.S.; Wang, J.Y.; Li, D.F. Possible role of microRNA miRNA-IL-25 interaction in mice with ulcerative colitis. Bioengineered 2020, 11, 862–871. [Google Scholar] [CrossRef]
- Friedrich, M.; Pohin, M.; Powrie, F. Cytokine Networks in the Pathophysiology of Inflammatory Bowel Disease. Immunity 2019, 50, 992–1006. [Google Scholar] [CrossRef]
- Hadziselimovic, F.; Emmons, L.R.; Gallati, H. Soluble tumour necrosis factor receptors p55 and p75 in the urine monitor disease activity and the efficacy of treatment of inflammatory bowel disease. Gut 1995, 37, 260–263. [Google Scholar] [CrossRef]
- Gardiner, K.R.; Halliday, M.I.; Barclay, G.R.; Milne, L.; Brown, D.; Stephens, S.; Maxwell, R.J.; Rowlands, B.J. Significance of systemic endotoxaemia in inflammatory bowel disease. Gut 1995, 36, 897–901. [Google Scholar] [CrossRef]
- Noguchi, M.; Hiwatashi, N.; Liu, Z.; Toyota, T. Secretion imbalance between tumour necrosis factor and its inhibitor in inflammatory bowel disease. Gut 1998, 43, 203–209. [Google Scholar] [CrossRef]
- Wang, Y.; Han, G.; Chen, Y.; Wang, K.; Liu, G.; Wang, R.; Xiao, H.; Li, X.; Hou, C.; Shen, B.; et al. Protective role of tumor necrosis factor (TNF) receptors in chronic intestinal inflammation: TNFR1 ablation boosts systemic inflammatory response. Lab. Investig. 2013, 93, 1024–1035. [Google Scholar] [CrossRef]
- Liu, C.Y.; Tam, S.S.; Huang, Y.; Dubé, P.E.; Alhosh, R.; Girish, N.; Punit, S.; Nataneli, S.; Li, F.; Bender, J.M.; et al. TNF Receptor 1 Promotes Early-Life Immunity and Protects against Colitis in Mice. Cell Rep. 2020, 33, 108275. [Google Scholar] [CrossRef]
- Stillie, R.; Stadnyk, A.W. Role of TNF receptors, TNFR1 and TNFR2, in dextran sodium sulfate-induced colitis. Inflamm. Bowel Dis. 2009, 15, 1515–1525. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, F.L.; Wang, H.B.; Dong, N.; Zhu, X.M.; Wu, Y.; Wang, Y.T.; Yao, Y.M. TNF-alpha mRNA is negatively regulated by microRNA-181a-5p in maturation of dendritic cells induced by high mobility group box-1 protein. Sci. Rep. 2017, 7, 12239. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Latham, G.J. Normalization of microRNA quantitative RT-PCR data in reduced scale experimental designs. Methods Mol. Biol. 2010, 667, 19–31. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Kramer, R. Chemometric Tehniques for Quantitative Analysis, 1st ed.; CRC Press: Boca Raton, FL, USA, 1998. [Google Scholar]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Stanta, G. Guidelines for Molecular Analysis in Archive Tissues; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control selection for RNA quantitation. Biotechniques 2000, 29, 332–337. [Google Scholar] [CrossRef]
- Srinivasan, M.; Sedmak, D.; Jewell, S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 2002, 161, 1961–1971. [Google Scholar] [CrossRef]
- Ben-Ezra, J.; Johnson, D.A.; Rossi, J.; Cook, N.; Wu, A. Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by the polymerase chain reaction. J. Histochem. Cytochem. 1991, 39, 351–354. [Google Scholar] [CrossRef]
- Aggerholm-Pedersen, N.; Maretty-Nielsen, K.; Keller, J.; Baerentzen, S.; Schrøder, H.; Jørgensen, P.H.; Hansen, B.H.; Nielsen, O.S.; Safwat, A. The importance of standardized treatment in high-grade osteosarcoma: 30 years of experience from a hospital-based database. Acta Oncol. 2015, 54, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Navarro, I.; Gámez-Pozo, A.; González-Barón, M.; Pinto-Marín, Á.; Hardisson, D.; López, R.; Madero, R.; Cejas, P.; Mendiola, M.; Espinosa, E.; et al. Comparison of gene expression profiling by reverse transcription quantitative PCR between fresh frozen and formalin-fixed, paraffin-embedded breast cancer tissues. Biotechniques 2010, 48, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Kouadjo, K.E.; Nishida, Y.; Cadrin-Girard, J.F.; Yoshioka, M.; St-Amand, J. Housekeeping and tissue-specific genes in mouse tissues. BMC Genom. 2007, 8, 127. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, J.; Liu, D.; Su, Y. Normalizing genes for real-time polymerase chain reaction in epithelial and nonepithelial cells of mouse small intestine. Anal. Biochem. 2010, 399, 211–217. [Google Scholar] [CrossRef]
- Bamias, G.; Goukos, D.; Laoudi, E.; Balla, I.G.; Siakavellas, S.I.; Daikos, G.L.; Ladas, S.D. Comparative study of candidate housekeeping genes for quantification of target gene messenger RNA expression by real-time PCR in patients with inflammatory bowel disease. Inflamm. Bowel Dis. 2013, 19, 2840–2847. [Google Scholar] [CrossRef]
- Silver, N.; Cotroneo, E.; Proctor, G.; Osailan, S.; Paterson, K.L.; Carpenter, G.H. Selection of housekeeping genes for gene expression studies in the adult rat submandibular gland under normal, inflamed, atrophic and regenerative states. BMC Mol. Biol. 2008, 9, 64. [Google Scholar] [CrossRef]
- Della Beffa, C.; Klawonn, F.; Menetski, J.P.; Schumacher, H.R., Jr.; Pessler, F. Evaluation of glyceraldehyde-3-phosphate, prolylpeptidyl isomerase A, and a set of stably expressed genes as reference mRNAs in urate crystal inflammation. BMC Res. Notes 2011, 4, 443. [Google Scholar] [CrossRef]
- Glare, E.M.; Divjak, M.; Bailey, M.J.; Walters, E.H. beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax 2002, 57, 765–770. [Google Scholar] [CrossRef]
- Qureshi, M.A.A.; Li, J.; Stark, A.; Eriksson, P.; Ahmed, M. Validation of Reference Genes for mRNA Quantification in Adjuvant Arthritis. Open J. Rheumatol. Autoimmune Dis. 2012, 2, 64–72. [Google Scholar] [CrossRef]
- Congiu, M.; Slavin, J.L.; Desmond, P.V. Expression of common housekeeping genes is affected by disease in human hepatitis C virus-infected liver. Liver Int. 2011, 31, 386–390. [Google Scholar] [CrossRef]
- Yin, R.; Tian, F.; Frankenberger, B.; de Angelis, M.H.; Stoeger, T. Selection and evaluation of stable housekeeping genes for gene expression normalization in carbon nanoparticle-induced acute pulmonary inflammation in mice. Biochem. Biophys. Res. Commun. 2010, 399, 531–536. [Google Scholar] [CrossRef]
- DuBois, B.; Pearson, J.; Hastings, B.; Mahmood, T.; Chan, T.; Alnakhli, A.; Cherala, G. Maternal low-protein diet alters the expression of real-time quantitative polymerase chain reaction reference genes in an age-, sex-, and organ-dependent manner in rat offspring. Nutr. Res. 2013, 33, 235–241. [Google Scholar] [CrossRef]
- Verma, A.S.; Shapiro, B.H. Sex-dependent expression of seven housekeeping genes in rat liver. J. Gastroenterol. Hepatol. 2006, 21, 1004–1008. [Google Scholar] [CrossRef]
- Derks, N.M.; Müller, M.; Gaszner, B.; Tilburg-Ouwens, D.T.W.M.; Roubos, E.W.; Kozicz, L.T. Housekeeping genes revisited: Different expressions depending on gender, brain area and stressor. Neuroscience 2008, 156, 305–309. [Google Scholar] [CrossRef]
- Hall, J.S.; Taylor, J.; Valentine, H.R.; Irlam, J.J.; Eustace, A.; Hoskin, P.J.; Miller, C.J.; West, C.M. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. Br. J. Cancer 2012, 107, 684–694. [Google Scholar] [CrossRef]
- Li, J.; Smyth, P.; Flavin, R.; Cahill, S.; Denning, K.; Aherne, S.; Guenther, S.M.; O’Leary, J.J.; Sheils, O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007, 7, 36. [Google Scholar] [CrossRef]
- Lou, G.; Ma, N.; Xu, Y.; Jiang, L.; Yang, J.; Wang, C.; Jiao, Y.; Gao, X. Differential distribution of U6 (RNU6-1) expression in human carcinoma tissues demonstrates the requirement for caution in the internal control gene selection for microRNA quantification. Int. J. Mol. Med. 2015, 36, 1400–1408. [Google Scholar] [CrossRef]
- Benz, F.; Roderburg, C.; Cardenas, D.V.; Vucur, M.; Gautheron, J.; Koch, A.; Zimmermann, H.; Janssen, J.; Nieuwenhuijsen, L.; Luedde, M.; et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp. Mol. Med. 2013, 45, e42. [Google Scholar] [CrossRef]
- Kalpachidou, T.; Kummer, K.K.; Mitric, M.; Kress, M. Tissue Specific Reference Genes for MicroRNA Expression Analysis in a Mouse Model of Peripheral Nerve Injury. Front. Mol. Neurosci. 2019, 12, 283. [Google Scholar] [CrossRef]
- Zárybnický, T.; Matoušková, P.; Ambrož, M.; Šubrt, Z.; Skálová, L.; Boušová, I. The Selection and Validation of Reference Genes for mRNA and microRNA Expression Studies in Human Liver Slices Using RT-qPCR. Genes 2019, 10, 763. [Google Scholar] [CrossRef]
- Bass, B.P.; Engel, K.B.; Greytak, S.R.; Moore, H.M. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: How well do you know your FFPE specimen? Arch. Pathol. Lab. Med. 2014, 138, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.K.; Im, J.; Kwak, Y.; Han, N.; Nam, K.H.; Seo, A.N.; Lee, H.S. Effects of Fixation and Storager of Human Tissue Samples on Nucleic Acid Preservation. Korean J. Pathol. 2014, 48, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Webster, A.F.; Zumbo, P.; Fostel, J.; Gandara, J.; Hester, S.D.; Recio, L.; Williams, A.; Wood, C.E.; Yauk, C.L.; Mason, C.E. Mining the Archives: A Cross-Platform Analysis of Gene Expression Profiles in Archival Formalin-Fixed Paraffin-Embedded Tissues. Toxicol. Sci. 2015, 148, 460–472. [Google Scholar] [CrossRef] [PubMed]
- Boisen, M.K.; Dehlendorff, C.; Linnemann, D.; Schultz, N.A.; Jensen, B.V.; Høgdall, E.V.S.; Johansen, J.S. MicroRNA Expression in Formalin-fixed Paraffin-embedded Cancer Tissue: Identifying Reference MicroRNAs and Variability. BMC Cancer 2015, 15, 1024. [Google Scholar] [CrossRef]
- Bovell, L.; Shanmugam, C.; Katkoori, V.R.; Zhang, B.; Vogtmann, E.; Grizzle, W.E.; Manne, U. miRNAs are stable in colorectal cancer archival tissue blocks. Front. Biosci. 2012, 4, 1937–1940. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Giulian, A.; Hashimoto, M.; Erenpreisa, J.; Yoshikawa, K. Emergent self-organized criticality in gene expression dynamics: Temporal development of global phase transition revealed in a cancer cell line. PLoS ONE 2015, 10, e0128565. [Google Scholar] [CrossRef]
- Zimatore, G.; Tsuchiya, M.; Hashimoto, M.; Kasperski, A.; Giuliani, A. Self-organization of whole-gene expression through coordinated chromatin structural transition. Biophys. Rev. 2021, 2, 031303. [Google Scholar] [CrossRef]
| mRNA/miRNA | Minimum | Maximum | Median | Mean | SD |
|---|---|---|---|---|---|
| EEF2 | 24.02 | 32.34 | 26.28 | 26.94 | 1.748 |
| TBP+ | 31.99 | 40.71 | 33.67 | 34.20 | 1.722 |
| NONO | 24.67 | 34.87 | 29.45 | 29.75 | 2.244 |
| PPIA | 24.43 | 34.58 | 27.07 | 27.31 | 2.324 |
| RPLP0 | 23.76 | 32.92 | 26.48 | 26.85 | 2.409 |
| miR-191-5p | 27.31 | 32.29 | 28.46 | 28.56 | 0.849 |
| miR-103a-3p | 26.86 | 33.27 | 28.78 | 28.90 | 1.017 |
| miR-16-5p | 25.39 | 30.95 | 26.37 | 26.54 | 0.813 |
| U6 | 24.79 | 32.43 | 27.08 | 27.18 | 1.238 |
| BestKeeper | EEF2 | TBP+ | NONO | PPIA | RPLP0 | miR-191-5p | miR-103a-3p | miR-16-5p | U6 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Control group (n = 20) | Geo Mean [CP] | 26.93 | 34.04 | 30.29 | 30.59 | 28.90 | 28.44 | 28.11 | 26.52 | 26.92 |
| SD [± CP] | 1.22 | 1.16 | 1.09 | 1.81 | 1.69 | 0.57 | 0.59 | 0.50 | 0.58 | |
| DSS group (n = 97) | Geo Mean [CP] | 27.08 | 34.19 | 30.39 | 30.41 | 28.70 | 28.58 | 29.05 | 26.53 | 28.01 |
| SD [± CP] | 1.37 | 1.38 | 1.45 | 2.09 | 2.17 | 0.70 | 0.75 | 0.63 | 1.04 | |
| F control group (n = 10) | Geo Mean [CP] | 25.81 | 33.03 | 29.38 | 29.05 | 27.36 | 28.35 | 28.00 | 26.27 | 26.77 |
| SD [± CP] | 0.69 | 0.59 | 0.91 | 1.34 | 1.20 | 0.45 | 0.42 | 0.41 | 0.43 | |
| F DSS group (n = 47) | Geo Mean [CP] | 25.94 | 33.22 | 29.34 | 28.83 | 26.94 | 28.11 | 28.60 | 26.04 | 27.58 |
| SD [± CP] | 0.52 | 0.60 | 0.76 | 1.00 | 0.82 | 0.46 | 0.66 | 0.35 | 1.25 | |
| M control group (n = 10) | Geo Mean [CP] | 28.10 | 35.34 | 31.22 | 32.22 | 30.53 | 28.52 | 28.23 | 26.76 | 27.08 |
| SD [± CP] | 1.70 | 0.73 | 1.48 | 2.56 | 2.29 | 0.71 | 0.76 | 0.63 | 0.69 | |
| M DSS group (n = 50) | Geo Mean [CP] | 28.20 | 35.50 | 31.41 | 31.97 | 30.45 | 29.02 | 29.48 | 27.00 | 28.43 |
| SD [± CP] | 1.33 | 1.42 | 1.35 | 1.98 | 2.01 | 0.63 | 0.65 | 0.57 | 0.59 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Unkovič, A.; Boštjančič, E.; Belič, A.; Perše, M. Selection and Evaluation of mRNA and miRNA Reference Genes for Expression Studies (qPCR) in Archived Formalin-Fixed and Paraffin-Embedded (FFPE) Colon Samples of DSS-Induced Colitis Mouse Model. Biology 2023, 12, 190. https://doi.org/10.3390/biology12020190
Unkovič A, Boštjančič E, Belič A, Perše M. Selection and Evaluation of mRNA and miRNA Reference Genes for Expression Studies (qPCR) in Archived Formalin-Fixed and Paraffin-Embedded (FFPE) Colon Samples of DSS-Induced Colitis Mouse Model. Biology. 2023; 12(2):190. https://doi.org/10.3390/biology12020190
Chicago/Turabian StyleUnkovič, Ana, Emanuela Boštjančič, Aleš Belič, and Martina Perše. 2023. "Selection and Evaluation of mRNA and miRNA Reference Genes for Expression Studies (qPCR) in Archived Formalin-Fixed and Paraffin-Embedded (FFPE) Colon Samples of DSS-Induced Colitis Mouse Model" Biology 12, no. 2: 190. https://doi.org/10.3390/biology12020190
APA StyleUnkovič, A., Boštjančič, E., Belič, A., & Perše, M. (2023). Selection and Evaluation of mRNA and miRNA Reference Genes for Expression Studies (qPCR) in Archived Formalin-Fixed and Paraffin-Embedded (FFPE) Colon Samples of DSS-Induced Colitis Mouse Model. Biology, 12(2), 190. https://doi.org/10.3390/biology12020190






