Simple Summary
Erwinia amylovora and E. pyrifoliae cause Erwinia blight, which damage pome fruits, and are highly contagious. We propose the use of bacteriophages to control these two pathogens simultaneously. Many drugs have been used in South Korea for the quick control of blight disease caused by both these species. This can result in antibiotic resistance; hence, phage cocktails have been suggested as an alternative. In this study, we observed that phage cocktails, including four isolated phages, exhibited extensive strain spectra and potential for rapid bacterial control. This study demonstrated the potential of a phage cocktail to replace antibiotics as biocontrol agents against Erwinia blight.
Abstract
The recent outbreak of blight in pome fruit plants has been a major concern as there are two indistinguishable Erwinia species, Erwinia amylovora and E. pyrifoliae, which cause blight in South Korea. Although there is a strict management protocol consisting of antibiotic-based prevention, the area and the number of cases of outbreaks have increased. In this study, we isolated four bacteriophages, pEp_SNUABM_03, 04, 11, and 12, that infect both E. amylovora and E. pyrifoliae and evaluated their potential as antimicrobial agents for administration against Erwinia-originated blight in South Korea. Morphological analysis revealed that all phages had podovirus-like capsids. The phage cocktail showed a broad spectrum of infectivity, infecting 98.91% of E. amylovora and 100% of E. pyrifoliae strains. The antibacterial effect was observed after long-term cocktail treatment against E. amylovora, whereas it was observed for both short- and long-term treatments against E. pyrifoliae. Genomic analysis verified that the phages did not encode harmful genes such as antibiotic resistance or virulence genes. All phages were stable under general orchard conditions. Collectively, we provided basic data on the potential of phages as biocontrol agents that target both E. amylovora and E. pyrifoliae.
1. Introduction
A major pathogenic bacterium of the pome fruit plant, Erwinia amylovora, has recently been introduced into South Korea [1,2,3,4]. E. amylovora has been reported to result in symptoms indistinguishable from those of E. pyrifoliae, an endemic pathogen in South Korea [5,6,7,8]. Both pathogens cause blight disease with the blackening of leaves, stems, and immature fruits, starting with flower infection [9,10,11,12,13]. As E. amylovora is regulated by law, the disease management protocol should be performed in a different way compared to E. pyrifoliae outbreaks [14]. Therefore, strict regulations are applied to E. amylovora outbreaks, with orchards being forcibly closed at 5% outbreak rates (or less) at the discretion of the government plant-disease control agent [15,16].
Periodic surveillance and prevention-based disease control programs must be performed to prevent the spread of these two pathogens [17]. The general protocol for fire blight prevention consists of three antibiotic administrations (once before flowering and twice during the flowering period). To prevent black shoot blight, antibiotics are administered twice after full bloom [15,18]. Despite the intensive disease control program and antibiotics for Erwinia-associated blight, the outbreak of fire blight has been on the rise, with an increased possibility of the evolution of antibiotic resistance among pathogenic strains [19,20]. Therefore, it is necessary to develop more effective agents other than antibiotics for the treatment of pathogenic Erwinia species [21,22,23].
Bacteriophages (phages) have been used as effective antimicrobial agents for the treatment of fire blight worldwide [24,25,26]. Phages are “smart biocontrol agents” as they replicate at the targeted infection site, enabling prolonged antimicrobial effects on-site [27,28]. The infection specificity of phages allows specific pathogens to be targeted while maintaining beneficial microbes in the environment [29,30]. To maximize the antimicrobial effects of phages, a combination of phages with different host spectra is used to exert antimicrobial effects over a wider range of pathogens; this pret-a-porter approach is one of the main paradigms for therapeutic phage preparation [31,32,33]. Furthermore, cocktail phage therapy, which is a combinatorial strategy, has been reported to have a synergistic effect [34,35,36,37,38].
This study investigated the biological control potential of the newly isolated Erwinia phages. The biological and genomic characteristics, including morphology, stability, and antimicrobial potential of four phages that showed infectivity toward both E. amylvoroa and E. pyrifoliae were examined in this study.
2. Materials and Methods
2.1. Phage Isolation
Water and soil samples were collected near the location where the blight outbreak occurred in South Korea to isolate phages that infect E. pyrifoliae. Phages were isolated as previously described [39]. Distilled water (10 mL) was added to the soil samples (1g). The samples were centrifuged at 10,000× g for 10 min to remove contaminants. A host strain suspension (1%, v/v) containing E. amylovora (TS3128) or E. pyrifoliae (KACC13945) was cultured overnight for approximately 18 h at 27 °C. The suspension was used to inoculate the samples and nutrient broth (NB; Difco) for phage enrichment and cultured for 24 h at 27 °C. After enrichment, serial dilutions of the culture broth were transferred onto bacterial lawns of the E. amylovora (TS3128) or E. pyrifoliae (KACC13945). Phage isolation was confirmed using a double-layer agar assay. The double-layer agar assay was used to verify bacteriolysis induced by the inhibition spots of phages. The samples showing plaque formation were centrifuged at 10,000× g and passed through 0.45 μm syringe filters. Pure phages were obtained by picking a single plaque and subjecting it to a double layer assay five times.
2.2. Phage Propagation and Purification
Phage propagation was conducted as previously described [40]. The overnight culture (1%) was inoculated with different multiplicity of infection (MOIs; 10, 5, 1 and 0.1) of phages to determine the optimum ratio for phage propagation and cultured for 24 h at 27 °C. Phage lysate was centrifuged at 12,000× g for 10 min and the supernatant was precipitated with 10% (w/v) polyethylene glycol/ 0.5 M NaCl. (final concentration). A cesium chloride (CsCl) gradient was used to purify the phage suspension [41]. The gradient layers were ultracentrifuged at 182,000× g for 3 h. Phage precipitation bands were collected and dialyzed using a dialysis bag (Slide-A-Lyzer™ Dialysis Cassettes, 10,000 MWCO).
2.3. Transmission Electron Microscopy (TEM)
Purified phage suspensions (10 μL) were mixed with the same volume of uranyl acetate (2%). The suspensions were incubated on a copper grid for 1 min. The excess sample was removed and washed with distilled water. Images of the phages were obtained using a Talos L120C (FEI, Hillsboro, OR, USA) at 120 kV. The dimensions of four independent phages were determined (n = 5).
2.4. Host Range
All the bacterial strains used in the host range assay were recent isolates from the blight tissues in South Korea. A total of 116 bacterial strains, including 92 E. amylovora and 24 E. pyrifoliae strains were spot assayed on nutrient agar (NA; Difco) plates with serial dilutions (10−1 to 10−8) of purified phage suspension; the plates were incubated for 24 h at 27 °C [40]. Plaque formation on the spot areas resulted in the bacterial strain being considered susceptible and is represented as “+” in Table S1. The experiments were performed in triplicates.
2.5. Stability Test
The thermal stability of the phages was evaluated as described by Kim et al. [42]. Phage suspensions (1 mL each, 2 × 108 PFU/mL) were incubated for 60 min at 4 (control), 20, 30, 40, and 50 °C. Approximately 100 μL aliquots of each suspension were used to determine the concentration of phages using a double-layer agar assay. The pH stability of the phages was evaluated by adjusting the pH of phage suspensions (2 × 108 PFU/mL) to 4.0, 5.0, 6.0, 7.0 (control), 8.0, and 9.0 with 0.1 M HCl and 0.1 M NaOH; each of the phage suspensions was then incubated for 60 min at 27 °C. They were then evaluated using a double-layer agar assay. All tests were performed in triplicate.
2.6. One-Step Growth Curve
The phage suspension (100 μL) was inoculated onto 10 mL of exponentially growing host strain culture (2 × 108 colony-forming units [CFU]/mL) at an MOI of 0.001 [43]. The phages were allowed to infect the bacterial cells for 10 min and the suspension was centrifuged at 12,000× g to remove unattached phages. The phage-infected bacterial pellets were then resuspended in preheated NB (10 mL) and incubated at 27 °C with shaking (150 rpm). Aliquots (100 μL) were collected at 5 min intervals for 50 min; the titers were then evaluated using a double-layer agar assay. The experiments were performed in triplicate.
2.7. Genome Analysis
Genomic DNA was extracted from phages as described previously [34,39]. Purified phage suspension (≥1010 PFU/mL) was digested with 10 IU of DNase I and RNase A to remove nucleotides originating from the hosts. The nucleases were heat-inactivated at 95 °C by the addition of EDTA. Proteinase K and SDS (10%) were added to the samples to degrade structural proteins. DNA was purified with phenol-chloroform-isopropanol and precipitated with absolute ethanol, followed by two washes with 70% ethanol. The phage genomic DNA was sequenced using an Illumina HiSeq platform at Macrogen (Seoul, South Korea). The short reads were assembled into contigs using de bruin graphs in CLC genomic workbench (v. 6.5.1). Open reading frames (ORFs) were identified using GenMarkS and Rapid Annotation using subsystem Technology (RAST) [44,45]. The presence of tRNA, and virulence and antibiotic genes was determined using tRNAscan-SE, VirulenceFinder, and ResFinder, respectively [46,47,48]. Comparative genome analysis was performed based on sequence similarity using tBLASTx [49]. Whole-genome phylogenetic analysis was performed using the Virus Classification and Tree Building Online Resource (VICTOR) with the recommended setting for complete nucleotide sequences [50].
2.8. Antibacterial Activity
The antibacterial effect of pEp_SNUABM_03, 04, 11, and 12 was evaluated over short (2 h) and long (8 h) periods of time. The assay was performed using two indicator strains, E. amylovora (TS3128) and E. pyrifoliae (KACC13945). The phage cocktail was prepared by combining the four phages at equal ratios (1:1:1:1) to obtain 2 × 108 PFU/mL. Exponentially growing indicator strains were inoculated into fresh NB to obtain 2 × 105 CFU/mL for 8 h and at 27 °C, and the phage suspension was inoculated into the broth at three concentrations (MOI 5, 1, and 0.1). The mixtures were cultured with shaking at 150 rpm, and CFUs were determined. The CFU values were determined by preparing serial dilutions in phosphate buffered saline and plating for the quantification of viable bacteria. All tests were performed in triplicate.
2.9. Statistical Analysis
Statistical differences were analyzed using Sigmaplot 12.5 (Systat Software Inc., Evanston, IL, USA) using analysis of variance with the Holm–Sidak test. Statistical significance was set at p < 0.05.
3. Results
3.1. TEM—Biological Analysis
Morphological observations using TEM revealed four distinct phages that belong to Podoviridae (Figure 1). Structural observations of pEp_SNUABM_03, 04, 11, and 12 revealed short tails with head diameters of 56 ± 2, 55 ± 3, 56 ± 3, and 63 ± 2 nm (n = 5), respectively (Table 1).
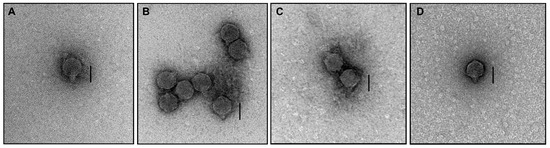
Figure 1.
Morphological observation by transmission electron micrographs of Erwinia pyrifoliae phages (A) pEp_SNUABM_03, (B) pEp_SNUABM_04, (C) pEp_SNUABM_11, and (D) pEp_SNUABM_12. Scale bar = 50 nm.

Table 1.
Morphological characteristics of Erwinia phages.
3.2. Stability Test
The test was conducted under normal-orchard environmental temperature and pH conditions (Figure 2). Thermal stability tests showed that pEp_SNUABM_03 and 11 were stable at 4 (control), 20, 30, 40, and 50 °C for 1 h, and virions of pEp_SNUABM_04 were vulnerable to high temperature (50 °C; P < 0.05). The phage pEp_SNUABM_12 was sensitive to temperature changes (P < 0.05). The pH stability test revealed that pEp_SNUABM_04, 11, and 12 were all stable, whereas the stability of pEp_SNUABM_03 decreased at pH 9 (P < 0.05).
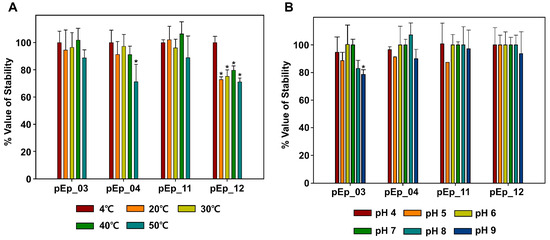
Figure 2.
Stability of phages pEp_SNUABM_03, pEp_SNUABM_04, pEp_SNUABM_11, and pEp_SNUABM_12 at thermal (A) and pH (B) stresses. Phages were incubated for 1 h under each condition and the phage titer was determined on the host strain. One-way ANOVA with Holm–Sidak tests were performed to determine significant differences (p < 0.05; n = 3).
3.3. One-Step Growth Curve
All four phages exhibited similar biological characteristics. Hence pEp_SNUABM_03 was used as a representative phage for one-step growth analysis (Figure 3). After the 10-min latent period, the first burst size of the phage growth was 76.83 PFU per bacterial cell for pEp_SNUABM_03.
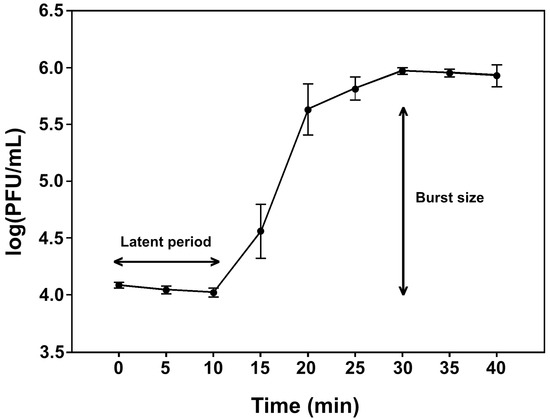
Figure 3.
One-step growth curve of the pEp_SNUABM_03 in E. pyrifoliae strain KACC13945. The values are presented as mean ± standard deviation.
3.4. Genome Analysis
The general characteristics of phages pEp_SNUABM_03, pEp_SNUABM_04, pEp_SNUABM_11, and pEp_SNUABM_12 are listed in Table 2. A total number of reads 3,864,800 (pEp_SNUABM_03), 3,730,842 (pEp_SNUABM_04), 3,426,138 (pEp_SNUABM_11), 3,818,762 (pEp_SNUABM_12) were obtained from the Illumina sequencer, which was assembled into the single contig. The circular genomes of phages pEp_SNUABM_03, pEp_SNUABM_04, pEp_SNUABM_11, and pEp_SNUABM_12 contained 39,879, 39,649, 39,626, and 39,980 bp with GC contents of 52.13%, 52.19%, 52.10%, and 51.19%, respectively (Table 2). A total of 52, 52, 49, and 50 ORFs were identified in the genomes of pEp_SNUABM_03, pEp_SNUABM_04, pEp_SNUABM_11, and pEp_SNUABM_12, respectively. The function of the predicted ORFs was categorized into five groups: structural and packaging proteins, nucleotide metabolism-related proteins, lysis proteins, additional function proteins, and hypothetical proteins (Figure 4).

Table 2.
General genomic features of Erwinia phages.
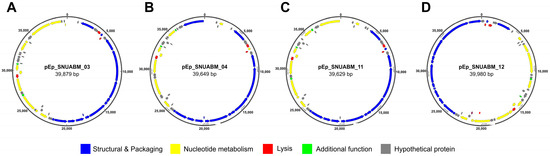
Figure 4.
Genome map of Erwinia phages (A) pEp_SNUABM_03, (B) pEp_SNUABM_04, (C) pEp_SNUABM_ 11, and (D) pEp_SNUABM_12 The color-coded ORFs are classified based on their function (Scale = base pair).
The phylogenetic positions of phages pEp_SNUABM_03, pEp_SNUABM_04, pEp_SNUABM_11, and pEp_SNUABM_12, which have the morphology of podovirus, were analyzed using the complete genome sequences of closely related phages infecting Enterobacterales (Erwinia, Dickeya, and Pectobacterium). All phages were classified under the subfamily Studiervirinae in the family Autographiviridae (Figure 5). Phage pEp_SNUABM_12 clustered with Ningirsuvirus and the dickey phage Ninurta, whereas the other three phages were unclassified. Phages pEp_SNUABM_03, 04, and 11 were clustered with Erwinia phage vB_EamP-L1 belonging to Elunavirus. This cluster was most closely related to FE 44, another Erwinia phage belonging to Berlinvirus. Two clusters of the newly isolated phages branched from a common ancestor.
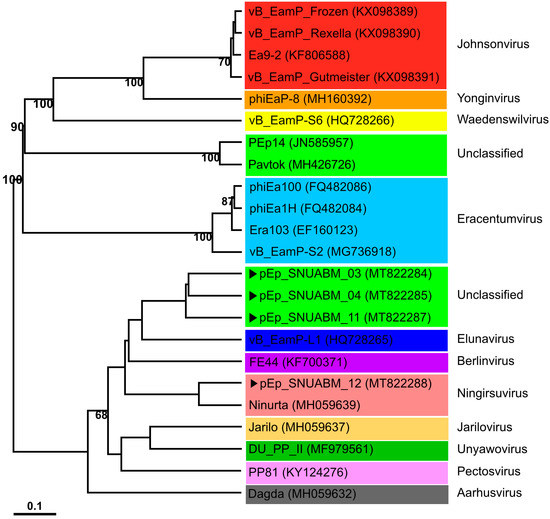
Figure 5.
Whole-genome phylogenetic analysis of newly isolated Erwinia phages. The four phages isolated in this study are indicated with arrows (▶). The different genera (Johnsonvirus, red box; Yonginvirus, orange box, Waedenswilvirus, yellow box; unclassified, light green; Eracentumvirus, sky-blue box; Elunavirus, deep blue box; Berlinvirus, purple box; Ningsuvirus, pink box; Jarilovirus, light orange box; Unyawovirus, green box; Pectosvirus, purple box and Aarhusvirus, gray box) are indicated using colors.
Comparative genome analysis supported the genomic distance between phages in the two clusters. The genomes of three unclassified phages, pEp_SNUABM_03, 04, and 11, showed highly conserved synteny revealing around 98% of nucleotide identity among them (thick blue), whereas the similarity level was low (nucleotide identity: around 70%; pale blue) with the closest neighbor, vB_EamP_L1 (Figure 6; Table S2). Phage pEp_SNUABM_12 showed high synteny with Ninurta (nucleotide identity: 94.66%), another member of Ningirsuvirus (Figure 6; Table S2) and genetic distance with pEp_SNUABM_03, 04, and 11. The three unclassified Autographiviridae phages shared more than 47 core genes, which accounted for more than 90% of their genes (Table S3). The shared genes among the four phages isolated in this study decreased to only 37 genes, as revealed by the comparative blast analysis (Tables S4–S7).
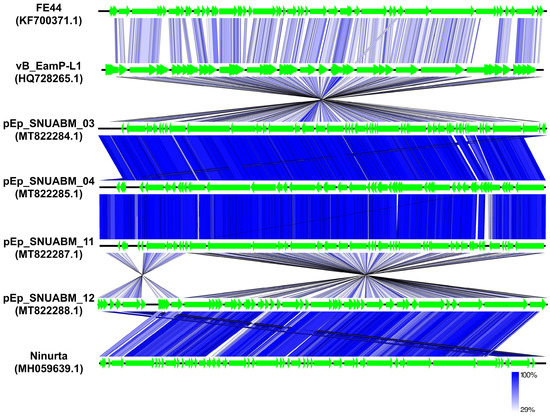
Figure 6.
Comparative whole-genome analysis of Erwinia phages pEp_SNUABM_03, pEp_SNUABM_04, pEp_SNUABM_11, and pEp_SNUABM_12 among phages infecting Enterobacterales species. The tBLASTx comparison analysis was constructed with tBLASTx algorithm using Easyfig.
3.5. Host Range
Host range analysis was performed against 116 Erwinia strains including 92 Erwinia amylovora and 24 Erwinia pyrifoliae (Table 3). pEp_SNUABM_03 and 04 showed broad-host-spectrum infectivity to both E. amylovora (98.91%, 91/92; 97.83%, 90/92) and E. pyrifoliae (91.67%, 22/24; 95.83%, 23/24) strains, respectively. Although pEp_SNUABM_11 had a relatively narrow host range compared to pEp_SNUABM_03 and 04, it was highly infective (E. amylovora: 76.09%, 70/92; E. pyrifoliae: 79.17%, 19/24). Phage pEp_SNUABM_12 showed specific infectivity in E. pyrifoliae (95.83%, 23/24). pEp_SNUABM_12 was able to infect only two E. amylovora strains (2.17%, 2/92). The phage cocktail infected almost all E. amylovora (98.91%, 91/92) and E. pyrifoliae (100%, 24/24) strains.

Table 3.
Host range analysis of individual and combined Erwinia phages, alone and as and the combined cocktail.
3.6. Antibacterial Activity of Phages on E. amylovora
The antibacterial efficacy of the newly isolated phages was evaluated at three concentrations (MOI 0.1, 1, and 5) over short (2 h) and long (8 h) time periods (Figure 7). Phages pEp_SNUABM_03, 04, 11 and 12 co-cultured with E. amylovora TS3128 at an MOI of 0.1 resulted in a slight inhibition of bacterial growth in the short term; pEp_SNUABM_04 showed significant inhibition after administration (p < 0.05). In the long term, the antibacterial effect was significant for all phages (p < 0.001), pEp_SNUABM_03 (−4.03 logCFU/mL), 04 (−3.70 logCFU/mL), 11 (−3.14 logCFU/mL), and 12 (−2.37 logCFU/mL). At an MOI of 1, all phages showed a significant inhibitory effect against TS3128 after short-term administration (p < 0.05). In the long term, all phages showed a significantly increased antibacterial effect, pEp_SNUABM_03 (−4.24 logCFU/mL), 04 (−3.78 logCFU/mL), 11 (−2.86 logCFU/mL), and 12 (−3.18 logCFU/mL) (p < 0.001). Phages pEp_SNUABM_03, 04, 11 and 12, were co-cultured with TS3128 at an MOI of 5 and exhibited significant inhibition of bacterial growth in the short term for all phages (p < 0.05). In the long term, there were notable reductions in bacterial counts for all phages; pEp_SNUABM_03 (−4.24 logCFU/mL), 04 (−3.97 logCFU/mL), 11 (−2.77 logCFU/mL), and 12 (−3.29 logCFU/mL) (p < 0.001).
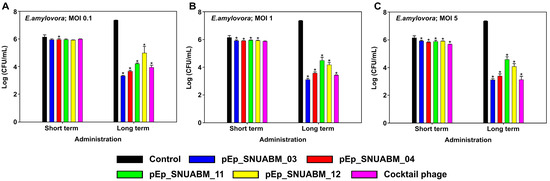
Figure 7.
Evaluation of antibacterial activity of phages on Erwinia amylovora. The assay was performed at an MOI of 0.1 (A), 1 (B), and 5 (C). Statistical significance was calculated using a one-way analysis of variance (ANOVA) with Holm-Sidak tests (p < 0.001).
The phage cocktail consisted of an equal ratio of the four phages, resulting in the same overall concentration as solely administered phages. Although one-fourth of each of the phages were combined, the antibacterial effect of the cocktail phage suspension administered over the long term, −3.42 logCFU/mL (MOI 0.1), −3.93 logCFU/mL(MOI 1), and −4.23 logCFU/mL (MOI 5), was higher than the average CFU reduction exhibited by individual phages, which is indicative of a synergistic effect.
3.7. Antibacterial Activity of Phages on E. pyrifoliae
The antibacterial effects of the four phages were evaluated at three concentrations (MOI 0.1, 1, and 5) over short (2 h) and long (8 h) periods of time (Figure 8). All phages showed rapid antibacterial effects against E. pyrifoliae. When E. pyrifoliae KACC13945 and phages pEp_SNUABM_03, 04, 11, and 12 were co-cultured at an MOI of 0.1, bacterial growth was inhibited in the short term, with pEp_SNUABM_11 showing significant inhibition (p < 0.05). In the long term, the antibacterial effect significantly decreased for all phages (p < 0.001), pEp_SNUABM_03 (−5.17 logCFU/mL), 04 (−5.27 logCFU/mL), 11 (−4.43 logCFU/mL), and 12 (−5.10 logCFU/mL). At an MOI of 1, all phages rapidly inhibited bacterial growth after short-term administration and showed a significant inhibitory effect against KACC13945 (p < 0.001). In the long term, the antibacterial effect was sustained in all phages; pEp_SNUABM_03 (−5.33 logCFU/mL), 04 (−5.20 logCFU/mL), 11 (−3.19 logCFU/mL), and 12 (−5.07 logCFU/mL) (p < 0.001). Phages pEp_SNUABM_03, 04, 11, and 12 co-cultured with KACC13945 at an MOI of 5 showed considerable reductions in bacterial counts in the short term for all phages (p < 0.001). In the long term, the antibacterial effect was maintained, and the bacterial counts were significantly reduced for all phages (p < 0.001); pEp_SNUABM_03 (−5.43 logCFU/mL), 04 (−5.17 logCFU/mL), 11 (−2.31 logCFU/mL), and 12 (−5.03 logCFU/mL).
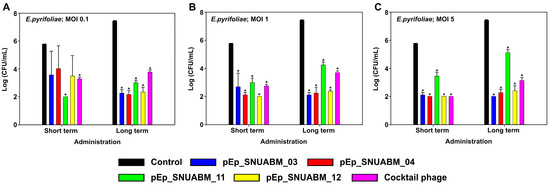
Figure 8.
Evaluation of antibacterial activity of phages on Erwinia pyrifoliae. The assay was performed at an MOI of 0.1 (A), 1 (B), and 5 (C). Statistical significance was determined using a one-way analysis of variance (ANOVA) with Holm-Sidak tests (p < 0.001).
The antibacterial efficacy of the phage cocktail suspension administered over a short term was −2.49 logCFU/mL (MOI 0.1), −3.03 logCFU/mL (MOI 1), and −3.77 logCFU/mL (MOI 5). Whereas the average CFU reduction in each phage, −2.50 logCFU/mL (MOI 0.1), −3.15 logCFU/mL (MOI 1), and −3.38 logCFU/mL (MOI 5), did not exhibit any synergy effect of the cocktail phage. However, there was a significant decrease in the bacterial count in the short-term phage cocktail treatment.
4. Discussion
Erwinia-associated blight disease in rosaceous fruit plants in South Korea is caused by E. pyrifoliae infection [6]. However, the recent outbreak of fire blight caused by E. amylovora has rendered the disease management protocol complicated, as a co-outbreak with E. pyrifoliae was identified [5,51]. In contrast to E. pyrifoliae, fire blight caused by E. amylovora is registered as a legal communicable disease in plants in South Korea, and there is a distinct disease management protocol [13,16,52]. To provide an effective control method against both pathogens, we isolated and characterized the potential of bacteriophages against Erwinia-originated blight disease in South Korea.
The rosaceous fruit plant industry has tried to use phages as biocontrol agents against E. amylovora outbreaks worldwide [53,54]. A number of phages have been isolated, and their potential as antimicrobial agents has been confirmed [34,55,56]. A cocktail phage suspension that combines phages with different infection mechanisms is preferred over individual phage isolates to minimize resistance and maximize the antibacterial effect for effective disease control [34,57,58]. As Erwinia bacteriophages have a broad host range, the major objective of their combined administration is to improve their antimicrobial potential [36,59]. The four phages used in this study also had a broad host range, except for pEp_SNUABM_12, which specifically infects E. pyrifoliae (Table 3). Phages use distinct infection strategies based on their tail structure, and the infectivity of the four phages is distinct from each other [60,61]. This suggests that they have different infection strategies that would prevent the prevalence of resistant bacterial strains [28,62].
Several studies have shown that phage resistance in bacterial strains is present in the form of a trade-off [63,64]; bacteria acquire phage resistance in return for fitness loss, including growth, virulence, and antibiotic susceptibility [65,66,67]. Attenuation or loss of virulence has been observed in several strains of Pectobacterium atrosepticum and Pseudomonas plecoglossicida resistant against phages PPpW-3 and/or PPpW-4, respectively [68,69]. Impaired growth characteristics have been reported in phage-resistant E. amylovora and P. syringae, which significantly affected their virulence [70,71]. Phage-resistant Escherichia coli, and E. amylovora strains become more susceptible to antibiotics [34,72]. Furthermore, E. amylovora bacteriophages showed transient resistance in infected bacterial strains, with phage infectivity being restored after the phage was eliminated.
Synergism is one of the major incentives for combining several phages in a cocktail suspension [36,37]. A synergistic effect refers to the antimicrobial potential of cocktail phages being greater than the sum of the individual phages; an additive effect occurs when a cocktail phage provides the sum of the effects of individual phages; an antagonistic effect refers to the antimicrobial potential of the cocktail phages being less than that of the sum of the individual phages [73]. The best selection for phage cocktail components results in synergy; as observed in our study (Figure 7), there should be no antagonistic effect between the cocktail phages. As phages can interrupt secondary infections by closely related phages, it is recommended that antagonistic phages be excluded at the first selection step.
The stability of phages under environmental stress should be verified before their application. The major stress factors expected are acidity, temperature, and UV radiation [74]. Although increased stability of the phages better facilitates their application as biocontrol agents, there are several ways to bypass environmental stresses (Figure 2). Control agents can be administered in the morning or encapsulated to minimize exposure to temperature and light, or acidity, respectively [75,76].
Although the efficacy and stability of phages are guaranteed, safety is a major concern. Generally, phages with an obligatory lytic life cycle are preferred as biocontrol agents against Erwinia-originated blight diseases (Figure 4). On the other hand, lysogenic phages have a greater potential for transducing harmful genes including those associated with antimicrobial resistance, virulence, and toxins [77]. However, if the transduction issue is eliminated, lysogenic phages may also be good candidates for controlling fire blight [78].
In the present study, the efficacy of the four phages and the phage cocktail against Erwinia strains indicates its possible use as a biocontrol agent under field conditions. The antibacterial effect can be further improved through modifications in the cocktail ratio as the phages exhibited synergy. To be applied in the actual environment, future studies should focus on the biocontrol efficacy of optimum phage cocktails in planta and carry out acute ecotoxic tests in fish to rule out possible environmental health hazards.
5. Conclusions
We isolated four phages, pEp_SNUABM_03, 04, 11, and 12, effective against both E. amylovora and E. pyrifoliae pathogens, and investigated their biological and genomic properties. Phages showed infectivity to both pathogens of Erwinia and were able to control these pathogens effectively over a long period of time. The cocktail treatment has the advantage of broadening the host spectrum as well as inducing synergistic effects. In addition, the stability and safety of phages for use as biocontrol agents were verified. Taken together, combining several phages that have distinct infection strategies and administering the cocktail phage suspension would be a remarkable way to control both Erwinia amylovora and E. pyrifoliae caused blight disease in South Korea. However, intensive verifications such as combined treatment with conventional agents, antibacterial efficacy in planta, and field tests, should be performed in further studies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology12020180/s1, Table S1: Host range of phage pEp_SNUABM_03, pEp_SNUABM_04, pEp_SNUABM_11, pEp_SNUABM_12, and Cocktail phage; Table S2: Nucleotide identity (%) among the closely related phages; Table S3: Coregene of Erwinia phages pEp_SNUABM_03, pEp_SNUABM_04, pEp_SNUABM_11, pEp_SNUABM_12; Table S4: Functional classification of ORFs in Erwinia phage pEp_SNUABM_03; Table S5: Functional classification of ORFs in Erwinia phage pEp_SNUABM_04; Table S6: Functional classification of ORFs in Erwinia phage pEp_SNUABM_11 Table S7: Functional classification of ORFs in Erwinia phage pEp_SNUABM_12.
Author Contributions
Conceptualization, S.J.J. and S.G.K.; methodology, S.G.K.; software, S.J.J.; validation, S.J.J., S.G.K. and S.S.G.; formal analysis, S.J.J. and J.W.K.; investigation, S.J.J., S.G.K. and S.B.L.; resources, W.J.J. and Y.M.L.; data curation, M.H.H., J.P., C.C. and E.R.; writing—original draft preparation, S.G.K. and S.J.J.; writing—review and editing, S.G.K. and S.C.P.; visualization, S.J.J.; supervision, S.C.P.; project administration, S.G.K.; funding acquisition, S.C.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by funded by the Rural Development Administration, Republic of Korea (PJ014965022022).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome sequences of the four Erwinia phages (pEp_SNUABM_03, pEp_SNUABM_04, pEp_SNUABM_11, and pEp_SNUABM_12) were deposited at GenBank under the accession numbers MT822284.1, MT822285.1, MT822287.1, and MT822288.1, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Piqué, N.; Miñana-Galbis, D.; Merino, S.; Tomás, J.M. Virulence factors of Erwinia amylovora: A review. Int. J. Mol. Sci. 2015, 16, 12836–12854. [Google Scholar] [CrossRef]
- Myung, I.-S.; Lee, J.-Y.; Yun, M.-J.; Lee, Y.-H.; Lee, Y.-K.; Park, D.-H.; Oh, C.-S. Fire blight of apple, caused by Erwinia amylovora, a new disease in Korea. Plant Dis. 2016, 100, 1774. [Google Scholar] [CrossRef]
- Llop, P.; Barbé, S.; López, M.M. Functions and origin of plasmids in Erwinia species that are pathogenic to or epiphytically associated with pome fruit trees. Trees 2012, 26, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Jik Lee, H.; Woo Lee, S.; Suh, S.; Hyun, I. Recent spread and potential pathways for fire blight in South Korea. EPPO Bull. 2022, 52, 135–140. [Google Scholar] [CrossRef]
- Park, D.H.; Yu, J.-G.; Oh, E.-J.; Han, K.-S.; Yea, M.C.; Lee, S.J.; Myung, I.-S.; Shim, H.S.; Oh, C.-S. First report of fire blight disease on Asian pear caused by Erwinia amylovora in Korea. Plant Dis. 2016, 100, 1946. [Google Scholar] [CrossRef]
- Rhim, S.; Völksch, B.; Gardan, L.; Paulin, J.; Langlotz, C.; Kim, W.; Geider, K. Erwinia pyrifoliae, an Erwinia species different from Erwinia amylovora, causes a necrotic disease of Asian pear trees. Plant Pathol. 1999, 48, 514–520. [Google Scholar] [CrossRef]
- Jock, S.; Geider, K. Molecular Differentiation of Erwinia amylovora Strains from North America and of two Asian pear pathogens by analyses of PFGE patterns and HrpN genes. Environ. Microbiol. 2004, 6, 480–490. [Google Scholar] [CrossRef]
- Waleron, M.; Waleron, K.; Geider, K.; Lojkowska, E. Application of RFLP analysis of RecA, GyrA and RpoS gene fragments for rapid differentiation of Erwinia amylovora from Erwinia strains isolated in Korea and Japan. Eur. J. Plant Pathol. 2008, 121, 161–172. [Google Scholar] [CrossRef]
- McGhee, G.C.; Schnabel, E.L.; Maxson-Stein, K.; Jones, B.; Stromberg, V.K.; Lacy, G.H.; Jones, A.L. Relatedness of chromosomal and plasmid DNAs of Erwinia pyrifoliae and Erwinia amylovora. Appl. Environ. Microbiol. 2002, 68, 6182–6192. [Google Scholar] [CrossRef]
- Park, J.; Lee, G.M.; Kim, D.; Park, D.H.; Oh, C.S. Characterization of the lytic bacteriophage PhiEaP-8 effective against both Erwinia amylovora and Erwinia pyrifoliae causing severe diseases in apple and pear. Plant Pathol. J. 2018, 34, 445–450. [Google Scholar] [CrossRef]
- Kim, W.S.; Jock, S.; Paulin, J.P.; Rhim, S.L.; Geider, K. Molecular detection and differentiation of Erwinia pyrifoliae and host range analysis of the Asian pear pathogen. Plant Dis. 2001, 85, 1183–1188. [Google Scholar] [CrossRef]
- Vrancken, K.; Holtappels, M.; Schoofs, H.; Deckers, T.; Valcke, R. Pathogenicity and infection strategies of the fire blight pathogen Erwinia amylovora in rosaceae: State of the art. Microbiology 2013, 159, 823–832. [Google Scholar] [CrossRef]
- Kim, W.S.; Gardan, L.; Rhim, S.L.; Geider, K. Erwinia pyrifoliae sp. nov., a novel pathogen that affects Asian pear trees (Pyrus pyrifolia Nakai). Int. J. Syst. Bacteriol. 1999, 49, 899–905. [Google Scholar] [CrossRef]
- Khan, M.A.; Zhao, Y.F.; Korban, S.S. Molecular mechanisms of pathogenesis and resistance to the bacterial pathogen Erwinia amylovora, causal agent of fire blight disease in Rosaceae. Plant Mol. Biol. Rep. 2012, 30, 247–260. [Google Scholar] [CrossRef]
- Park, D.; Lee, Y.-G.; Kim, J.-S.; Cha, J.-S.; Oh, C.-S. Current status of fire blight caused by Erwinia amylovora and action for its management in Korea. J. Plant Pathol. 2017, 99, 59–63. [Google Scholar]
- Ham, H.H.; Lee, Y.K.; Kong, H.G.; Hong, S.J.; Lee, K.J.; Oh, G.R.; Lee, M.H.; Lee, Y.H. Outbreak of fire blight of apple and Asian pear in 2015–2019 in Korea. Res. Plant Dis. 2020, 26, 222–228. [Google Scholar] [CrossRef]
- Palacio-Bielsa, A.; López-Quílez, A.; Llorente, I.; Ruz, L.; López, M.M.; Cambra, M.A. Criteria for efficient prevention of dissemination and successful eradication of Erwinia amylovora (the Cause of Fire Blight) in Aragón, Spain. Phytopathol. Mediterr. 2012, 51, 505–518. [Google Scholar]
- Ahn, M.I.; Yun, S.C. Application of the Maryblyt model for the infection of fire blight on apple trees at Chungju, Jecheon, and Eumsung during 2015–2020. Plant Pathol. J. 2021, 37, 543–554. [Google Scholar] [CrossRef]
- Norelli, J.L.; Jones, A.L.; Aldwinckle, H.S. Fire blight management in the twenty-first century: Using new technologies that enhance host resistance in apple. Plant Dis. 2003, 87, 756–765. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Duffy, B. Use of antibiotics in plant agriculture. Rev. Sci. Tech. 2012, 31, 199–210. [Google Scholar] [CrossRef]
- Sundin, G.W.; Castiblanco, L.F.; Yuan, X.; Zeng, Q.; Yang, C.H. Bacterial disease management: Challenges, experience, innovation and future prospects: Challenges in bacterial molecular plant pathology. Mol. Plant Pathol. 2016, 17, 1506–1518. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Roh, E.; Park, J.; Giri, S.S.; Kwon, J.; Kim, S.W.; Kang, J.W.; Lee, S.B.; Jung, W.J.; Lee, Y.M.; et al. The bacteriophage pEp_SNUABM_08 is a novel singleton siphovirus with high host specificity for Erwinia pyrifoliae. Viruses 2021, 13, 1231. [Google Scholar] [CrossRef] [PubMed]
- Sundin, G.W.; Wang, N. Antibiotic resistance in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2018, 56, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Sieiro, C.; Areal-Hermida, L.; Pichardo-Gallardo, Á.; Almuiña-González, R.; De Miguel, T.; Sánchez, S.; Sánchez-Pérez, Á.; Villa, T.G. A hundred years of bacteriophages: Can phages replace antibiotics in agriculture and aquaculture? Antibiotics 2020, 9, 493. [Google Scholar] [CrossRef]
- Svircev, A.; Roach, D.; Castle, A. Framing the future with bacteriophages in agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef]
- Jamal, M.; Bukhari, S.M.A.U.S.; Andleeb, S.; Ali, M.; Raza, S.; Nawaz, M.A.; Hussain, T.; Rahman, S.U.; Shah, S.S.A. Bacteriophages: An overview of the control strategies against multiple bacterial infections in different fields. J. Basic Microbiol. 2019, 59, 123–133. [Google Scholar] [CrossRef]
- Połaska, M.; Sokołowska, B. Bacteriophages—A new hope or a huge problem in the food industry. AIMS Microbiol. 2019, 5, 324–346. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef]
- Álvarez, B.; Biosca, E.G. Bacteriophage-based bacterial wilt biocontrol for an environmentally sustainable agriculture. Front. Plant Sci. 2017, 8, 1218. [Google Scholar] [CrossRef]
- Lehman, S.M. Development of a Bacteriophage-Based Biopesticide for Fire Blight; Doctorate Brock University: St. Catharines, ON, Canada, 2007. [Google Scholar]
- Pirnay, J.P.; De Vos, D.; Verbeken, G.; Merabishvili, M.; Chanishvili, N.; Vaneechoutte, M.; Zizi, M.; Laire, G.; Lavigne, R.; Huys, I.; et al. The phage therapy paradigm: Pret-a-porter or sur-mesure? Pharm. Res. 2011, 28, 934–937. [Google Scholar] [CrossRef]
- Russo, N.L.; Burr, T.J.; Breth, D.I.; Aldwinckle, H.S. Isolation of streptomycin-resistant isolates of Erwinia amylovora in New York. Plant Dis. 2008, 92, 714–718. [Google Scholar] [CrossRef]
- Kim, H.J.; Jun, J.W.; Giri, S.S.; Kim, S.G.; Kim, S.W.; Kwon, J.; Lee, S.B.; Chi, C.; Park, S.C. Bacteriophage cocktail for the prevention of multiple-antibiotic-resistant and mono-phage-resistant Vibrio coralliilyticus infection in pacific oyster (Crassostrea gigas) larvae. Pathogens 2020, 9, 831. [Google Scholar] [CrossRef]
- Kim, S.G.; Lee, S.B.; Jo, S.J.; Cho, K.; Park, J.K.; Kwon, J.; Giri, S.S.; Kim, S.W.; Kang, J.W.; Jung, W.J.; et al. Phage cocktail in combination with kasugamycin as a potential treatment for fire blight caused by Erwinia amylovora. Antibiotics 2022, 11, 1566. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J. Phage cocktail development for bacteriophage therapy: Toward improving spectrum of activity breadth and depth. Pharmaceuticals 2021, 14, 1019. [Google Scholar] [CrossRef]
- Nale, J.Y.; Vinner, G.K.; Lopez, V.C.; Thanki, A.M.; Phothaworn, P.; Thiennimitr, P.; Garcia, A.; AbuOun, M.; Anjum, M.F.; Korbsrisate, S.; et al. An optimized bacteriophage cocktail can effectively control Salmonella in vitro and in Galleria mellonella. Front. Microbiol. 2020, 11, 609955. [Google Scholar] [CrossRef]
- Ross, A.; Ward, S.; Hyman, P. More is better: Selecting for broad host range bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef]
- Kim, S.G.; Kwon, J.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.W.; Kang, J.W.; Lee, S.B.; Jung, W.J.; Park, S.C. Strategy for mass production of lytic Staphylococcus aureus bacteriophage pSa-3: Contribution of multiplicity of infection and response surface methodology. Microb. Cell Factories 2021, 20, 56. [Google Scholar] [CrossRef]
- Kim, S.G.; Lee, S.B.; Giri, S.S.; Kim, H.J.; Kim, S.W.; Kwon, J.; Park, J.; Roh, E.; Park, S.C. Characterization of novel Erwinia amylovora jumbo bacteriophages from Eneladusvirus genus. Viruses 2020, 12, 1373. [Google Scholar] [CrossRef]
- Kim, S.G.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.W.; Kang, J.W.; Han, S.J.; Kwon, J.; Jun, J.W.; Oh, W.T.; et al. Genomic characterization of bacteriophage pEt-SU, a novel PhiKZ-related virus infecting Edwardsiella tarda. Arch. Virol. 2020, 165, 219–222. [Google Scholar] [CrossRef]
- Kim, S.G.; Jun, J.W.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.W.; Kang, J.W.; Han, S.J.; Jeong, D.; Park, S.C. Isolation and characterisation of pVa-21, a giant bacteriophage with anti-biofilm potential against Vibrio alginolyticus. Sci. Rep. 2019, 9, 6284. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Giri, S.S.; Yun, S.K.; Kim, S.W.; Han, S.J.; Kwon, J.; Oh, W.T.; Lee, S.B.; Park, Y.H.; Park, S.C. Two novel bacteriophages control multidrug- and methicillin-resistant Staphylococcus pseudintermedius biofilm. Front. Med. 2021, 8, 524059. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Borodovsky, M. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005, 33, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 1–15. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. TRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Choi, J.H.; Kim, J.Y.; Park, D.H. Evidence of greater competitive fitness of Erwinia amylovora over E. pyrifoliae in Korean isolates. Plant Pathol. J. 2022, 38, 355–365. [Google Scholar] [CrossRef]
- Song, J.Y.; Yun, Y.H.; Kim, G.D.; Kim, S.H.; Lee, S.J.; Kim, J.F. Genome analysis of Erwinia amylovora strains responsible for a fire blight outbreak in Korea. Plant Dis. 2021, 105, 1143–1152. [Google Scholar] [CrossRef]
- Gill, J.J.; Svircev, A.M.; Smith, R.; Castle, A.J. Bacteriophages of Erwinia amylovora. Appl. Environ. Microbiol. 2003, 69, 2133–2138. [Google Scholar] [CrossRef]
- Erskine, J.M. Characteristics of Erwinia amylovora bacteriophage and its possible role in the epidemiology of fire blight. Can. J. Microbiol. 1973, 19, 837–845. [Google Scholar] [CrossRef]
- Boulé, J.; Sholberg, P.L.; Lehman, S.M.; O’Gorman, D.T.; Svircev, A.M. Isolation and characterization of eight bacteriophages infecting Erwinia amylovora and their potential as biological control agents in British Columbia, Canada. Can. J. Plant Pathol. 2011, 33, 308–317. [Google Scholar] [CrossRef]
- Thompson, D.W.; Casjens, S.R.; Sharma, R.; Grose, J.H. Genomic comparison of 60 completely sequenced bacteriophages that infect Erwinia and/or Pantoea bacteria. Virology 2019, 535, 59–73. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage Therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Born, Y.; Fieseler, L.; Marazzi, J.; Lurz, R.; Duffy, B.; Loessner, M.J. Novel virulent and broad-host-range Erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to Enterobacteriaceae phages. Appl. Environ. Microbiol. 2011, 77, 5945–5954. [Google Scholar] [CrossRef]
- Chaturongakul, S.; Ounjai, P. Phage–host interplay: Examples from tailed phages and gram-negative bacterial pathogens. Front. Microbiol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Casjens, S.R.; Molineux, I.J. Short noncontractile tail machines: Adsorption and DNA delivery by podoviruses. In Viral Molecular Machines; Springer: Berlin/Heidelberg, Germany, 2012; pp. 143–179. [Google Scholar] [CrossRef]
- Yehl, K.; Lemire, S.; Yang, A.C.; Ando, H.; Mimee, M.; Torres, M.T.; de la Fuente-Nunez, C.; Lu, T.K. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell 2019, 179, 459–469.e9. [Google Scholar] [CrossRef]
- Azam, A.H.; Tanji, Y. Bacteriophage-host arm race: An update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 2019, 103, 2121–2131. [Google Scholar] [CrossRef] [PubMed]
- Goldhill, D.H.; Turner, P.E. The evolution of life history trade-offs in viruses. Curr. Opin. Virol. 2014, 8, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Majkowska-Skrobek, G.; Markwitz, P.; Sosnowska, E.; Lood, C.; Lavigne, R.; Drulis-Kawa, Z. The evolutionary trade-offs in phage-resistant Klebsiella pneumoniae entail cross-phage sensitization and loss of multidrug resistance. Environ. Microbiol. 2021, 23, 7723–7740. [Google Scholar] [CrossRef] [PubMed]
- Segall, A.M.; Roach, D.R.; Strathdee, S.A. Stronger together? Perspectives on phage-antibiotic synergy in clinical applications of phage therapy. Curr. Opin. Microbiol. 2019, 51, 46–50. [Google Scholar] [CrossRef]
- Park, S.C.; Shimamura, I.; Fukunaga, M.; Mori, K.I.; Nakai, T. Isolation of bacteriophages specific to a fish pathogen, Pseudomonas plecoglossicida, as a candidate for disease control. Appl. Environ. Microbiol. 2000, 66, 1416–1422. [Google Scholar] [CrossRef]
- Park, S.C.; Nakai, T. Bacteriophage control of Pseudomonas plecoglossicida infection in ayu Plecoglossus altivelis. Dis. Aquat. Organ. 2003, 53, 33–39. [Google Scholar] [CrossRef]
- Meaden, S.; Paszkiewicz, K.; Koskella, B. The cost of phage resistance in a plant pathogenic bacterium is context-dependent. Evolution 2015, 69, 1321–1328. [Google Scholar] [CrossRef]
- Schnabel, E.L.; Jones, A.L. Isolation and characterization of five Erwinia amylovora bacteriophages and assessment of phage resistance in strains of Erwinia amylovora. Appl. Environ. Microbiol. 2001, 67, 59–64. [Google Scholar] [CrossRef]
- Zou, X.; Xiao, X.; Mo, Z.; Ge, Y.; Jiang, X.; Huang, R.; Li, M.; Deng, Z.; Chen, S.; Wang, L.; et al. Systematic strategies for developing phage resistant Escherichia coli strains. Nat. Commun. 2022, 13, 4491. [Google Scholar] [CrossRef]
- Schmerer, M.; Molineux, I.J.; Bull, J.J. Synergy as a Rationale for phage therapy using phage cocktails. PeerJ 2014, 2, e590. [Google Scholar] [CrossRef]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages—Review. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef]
- Kim, S.G.; Giri, S.S.; Jo, S.J.; Kang, J.W.; Lee, S.B.; Jung, W.J.; Lee, Y.M.; Kim, H.J.; Kim, J.H.; Park, S.C. Prolongation of fate of bacteriophages In Vivo by polylactic-co-glycolic-acid/alginate-composite encapsulation. Antibiotics 2022, 11, 1264. [Google Scholar] [CrossRef]
- Born, Y.; Bosshard, L.; Duffy, B.; Loessner, M.J.; Fieseler, L. Protection of Erwinia amylovora bacteriophage Y2 from UV-induced damage by natural compounds. Bacteriophage 2015, 5, e1074330. [Google Scholar] [CrossRef]
- Paul, J.H.; Jiang, S.C. Lysogeny and transduction. In Methods in Microbiology; Elsevier: Amsterdam, The Netherlands, 2001; Volume 30, pp. 105–125. ISBN 0580-9517. [Google Scholar] [CrossRef]
- Torres-Barceló, C. The disparate effects of bacteriophages on antibiotic-resistant bacteria. Emerg. Microbes Infect. 2018, 7, 168. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).