Simple Summary
Salt stress produced ion toxicity on plant cells and limited the of culture of cultivated Rosa rugosa. GT genes in salt stresses responses have been emerging. From the GT gene family of the salt-tolerant wild Rosa rugosa, four NaCl stress responsive genes (RrGT-1, RrSIP1, RrSIP2, RrGTγ-4) were identified. RrSIP1 and RrGTγ-4, RrGT-1 and RrSIP2 located on chloroplasts and cell nucleus, respectively. RrSIP1, RrSIP2 and RrGTγ-4 could play roles in regulation of sodion and potassium transport. And RrGT-1 expressed higher specifically in wild Rosa rugosa than in the salt-sensitive cultivated Rosa rugosa. These four genes would be candidates for further study of regulation mechanism of salt-tolerance of wild Rosa rugosa and would supply gene resources for tolerance improvement of cultivated Rosa rugosa.
Abstract
Rosa rugosa was a famous aromatic plant while poor salt tolerance of commercial cultivars has hindered its culture in saline-alkali soil. In many plants, the roles of GT (or trihelix) genes in salt stresses responses have been emerging. In the wild R. rugosa, a total of 37 GTs (RrGTs) were grouped into GT-1, GT-2, GTγ, SH4, and SIP1 lineages. SIP1 lineage expanded by transposition. The motifs involved in the binding of GT cis-elements were conserved. Four RrGTs (RrGT11/14/16/18) significantly differentially expressed in roots or leaves under salt stress. The responsive patterns within 8 h NaCl treatment indicated that RrGTγ-4 (RrGT18) and RrGT-1 (RrGT16) were significantly induced by salt in roots of R. rugosa. Subcellular localizations of RrSIP1 (RrGT11) and RrGTγ-4 were on chloroplasts while RrGT-1 and RrSIP2 (RrGT14) located on cell nucleus. Regulation of ion transport could be the most important role of RrSIPs and RrGTγ-4. And RrGT-1 could be a halophytic gene with higher transcription abundance than glycophytic GT-1. These results provide key clue for further investigations of roles of RrGTs in salt stress response and would be helpful in the understanding the salt tolerance regulation mechanism of R. rugosa.
1. Introduction
GT (or trihelix) family is a plant-specific transcription factor (TF) family with a DNA-binding domain rich in proline and glutamine residues [1,2]. The conserved DNA-binding domain containing three tandem helix structures (helix-loop-helix-loop-helix), namely trihelix domain [1,2]. The highly similar structure of trihelix domains and Myb/SANT-LIKE DNA-binding domains indicated that GT factors evolved from Myb/SANT-LIKE proteins [3]. GT elements are photosensitive cis-elements with A/T-rich motifs like ‘(T/A) -A- (T/A)’ [4]. Interactions between GT factors and GT elements are implicated in the light dependent expression of many plant genes. e.g., the first identified GT gene GT-1 binds two light-responsive elements of Rubisco small subunit 3A (rbcS-3A) of Pisum sativum L. [5]. A total of 30 GTs in Arabidopsis thaliana (L.) Heynh (AtGTs) were identified and classified into GT-1, GT-2, GTγ, SH4, and SIP1 lineages [6]. Most AtGTs played complex transcriptional regulation roles in light-responsive gene regulation [7], immunity (ARABIDOPSIS SH4-RELATED3, ASR3) [8], perianth architecture regulation and fertility (PETAL LOSS, PTL) [9] and seed maturation (Arabidopsis 6b-interacting protein 1-like 1, ASIL1) [6]. Recently, roles of abiotic stress responses were identified in GT family, like AtGTs involved in salt-induced gene expression (Arabidopsis GT-1-like transcription factor, GT-3A) [10], hypoxia-responsive response (AT3G10040) [11] and salt and/or other abiotic stress tolerances (Arabidopsis SIP1 clade Trihelix1, AST1; GT-4, GT2L) [12,13,14]. Salt stress produced osmotic stress and ion toxicity on plant cells, resulting in the destruction of organelles and excess accumulation of activated oxygen [15,16]. Further, the photosynthesis decreasing and poor growth under salt stress contributed to the losses of quality and yield of economic crops [17]. The expression profile studies predicted some GTs which responded to salt stress in many crops, like cotton [18], wheat [19], Brassica napus L. [20], quinoa [21], Sorghum bicolor L. [22], soybean [23] and rice [24]. Function studies have proved salt tolerance regulation roles of several crop GTs. e.g., OsGTγ-1 (Os02g33770) and OsGTγ-2 (Os11g06410) of rice positively regulate salt tolerance [24]. GmGT-2A, GmGT-2B of soybean [25] and GhGT26 of cotton [26] enhance salinity tolerance in overexpressed Arabidopsis.
Salinity stress is one of the most severe abiotic stresses and poses a continuing threat to economic crops [27], and for this reason the research are starting to consider and valorize wild species suitable for saline environments [28,29]. Rosa rugosa cultivars are widely used for spicery of food industry or essential oil of cosmetics industry [30,31]. These R. rugosa cultivars lost salt tolerance along with the breeding processes of floral traits, resulting in their limiting planting areas although there were vast saline-alkali soils in China [32,33,34]. e.g., the commercial cultivar R. rugosa ‘Zizhi’ (Zizhi) which planted in the narrow hilly lands of Shandong Province (China) was a typical glycophyte. While the wild R. rugosa which distributed naturally in the coastal area of northeast China belong to halophytes. The wild R. rugosa kept strong salt tolerance to adapt to the high salinity beach, as observed in other plant species in the coastal areas in the world [35]. In genetic engineering of salt-tolerance for glycophytic crop, homologous genes from halophytes should be more efficient since halophytes are more salt-tolerant than glycophytes [28,36]. The mining of salt responsive TF of wild R. rugosa would supply a base for salt tolerance improvement of R. rugosa cultivars [34].
With the genome of wild R. rugosa, this study aimed to screen GTs of R. rugosa (RrGTs) involved in the salt response. The phylogeny, synteny and sequence analyses would give a systematic understanding of lineage, gene duplication events, conserved motifs and gene structures of RrGT family. Expression profiles of salt treated R. rugosa were built to detect significantly induced/reduced RrGTs. These candidates were pointcuts of further study of salt stress responses regulation. Our study preliminary studied the roles of RrGTs under slat stress and would be helpful in understanding the regulatory mechanism of salt tolerance of R. rugosa.
2. Materials and Methods
2.1. Identification of RrGT Family
The R. rugosa and Rosa chinensis Jacq. genomes were obtained from GDR (Genome Database for Rosaceae, https://www.rosaceae.org/, accessed on 1 May 2022). Based on the hidden Markov model Myb/SANT-like DNA-binding domain (PF13837, http://Pfam.sanger.ac.uk/, accessed on 1 May 2022), candidate proteins were screened from the genome using HMMER 3.3.2 [37] with a cutoff threshold value of E-5. The ambiguous or incomplete candidates were rejected by TF verification of PlantTFDB (http://planttfdb.cbi.pku.edu.cn/, accessed on 1 May 2022).
2.2. Phylogenetic Analyses of RrGT Proteins
GT genes of Oryza sativa (OsGTs) and Arabidopsis thaliana were obtained from PlantTFDB (http://planttfdb.cbi.pku.edu.cn/, accessed on 1 May 2022). Based on the alignment of GT domains using MAFFT 7.55 (https://mafft.cbrc.jp/alignment/server/, accessed on 1 May 2022), a neighbor-join (NJ) phylogenetic tree was constructed by MEGA 7 with 1000 bootstrapping replications [38]. Besides, the p-distance model of the substitution type, pairwise deletion of Gaps/Missing data and uniform rates among sites were selected for the phylogenetic analysis.
2.3. Synteny Analysis of RrGTs
The homologous gene pairs (E < 10−5, top five matches) within the R. rugosa genome or among R. rugosa, R. chinensis and Fragaria vesca L. genomes (obtained from GDR) were identified by BLASTP (BLAST+ 2.13.0) search. Based on the location of homologous pairs, MCScanX (mcscan2) [39] identified the syntenic regions and predicted the gene duplication events. RrGTs and corresponding homologous GTs on syntenic regions were highlighted by Synteny plot tool of TBtools [40].
2.4. Gene Structure, Motif Analysis and Cis-Acting Elements of RrGT Family
The top 10 conserved motifs were predicted by MEME web tools (https://meme-suite.org/meme/, accessed on 2 May 2022) under default parameters. Cis-elements on the 2000 bp sequence upstream to the initiation codon were predicted by PLANTCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 2 May 2022). The gene structures and motifs were illustrated by the Gene structure view tool of TBtools [40].
2.5. Expression Analysis under Salt Stress
Previous transcriptome data [32,41] of wild R. rugosa and R. rugosa ‘Zizhi’ provided the per kilobase of exon model per million mapped fragments (FPKM) of RrGTs and the fold-changes of differentially expressed RrGTs.
One-month-old wild R. rugosa seedlings were treated with 340 mM NaCl solution for 0.5 h, 1 h, 2 h, 4 h and 8 h and roots of these samples were collected with three biological repetitions. Total RNAs were extracted and reverse-transcribed as cDNA templets by RNAprep Pure plant kit (Tiagen, Beijing, China) and HiScript® III RT SuperMix (Vazyme, Nanjing, China). Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted using ChamQ SYBR Color qPCR Master Mix (Vazyme, Nanjing, China) on the CFX96 platform (Bio-Rad, China). All the steps of RNA extraction, reverse-transcription and qRT-PCR were conducted following the manufacturers’ recommended instructions. Table S5 listed the primers of RrGTs and reference genes (phD and 5.8s).
2.6. Subcellular Localization Analysis
The subcellular localization of RrGTs were predicted by webtools Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/, accessed on 2 May 2022) and WoLF PSORT (https://wolfpsort.hgc.jp/, accessed on 2 May 2022). Coding sequences of 4 candidate RrGTs were cloned into 35S: green fluorescent protein (GFP) vector pNC-AMP-GFP-C and transformed to Arabidopsis protoplasts for overexpression of RrGT-GFP fusion proteins. The fluorescence of chloroplasts, GFP and 4′,6-diamidino-2-phenylindole (DAPI, Nuclear marker) were observed by the laser confocal microscopy.
3. Results
3.1. Lineages and Synteny of RrGT Family
A total of 37 RrGTs scattered across the seven chromosomes of R. rugosa genome (Figure 1B, Table S2). The NJ-tree of RrGTs, OsGTs and AtGTs divided 18, 7, 5, 4, 3 RrGTs to SIP1, GT-2, GT-1, SH4 and GTγ lineages, respectively (Figure 1A). The gene number of RrGT of each linage is similar to that of OsGTs or AtGTs of corresponding lineage except SIP1 (Table S1).

Figure 1.
The phylogeny (A) and chromosome location (B) of GT family of Rosa rugosa (RrGT). The dendrogram of GTs of R. rugosa, Rosa chinensis (XP), Oryza sativa (LOC) and Arabidopsis thaliana (AT) was generated using the neighbor-joining method with 1000 bootstrap replicates (numbers in branches). Nodes were colored according to the five lineages of GT family and RrGTs were highlighted by red dots. The gene ID was listed in Table S2.
WGD (whole genome duplication) or segmental duplication produced three paralogous RrGT pairs, namely RrTH2-RrTH32, RrTH3-RrTH30, RrTH26-RrTH28 on intra-species synteny regions of Chr1-Chr7, Chr1-Chr6 and Chr6-Chr6, respectively (Figure 2A, Tables S2 and S3) and 22 orthologous gene pairs of RrGTs-RcGTs (GTs of R. chinensis)-FvGTs (GTs of F. vesca) on inter-species synteny regions among R. rugosa, R. chinensis and F. vesca (Figure 2B, Table S2). Other RrGTs were predicted as ‘dispersed type’ which might arise from transposition and no tandem duplication events were identified. RrTH12, RrTH30, RrTH32 produced five redundant homologous pairs on the synteny regions of non-homologous chromosomes (Figure 2B, red and green lines). After removing the small synteny regions which including less than 20 gene pairs (Figure S1), only one redundant pair was credible (Figure 2B, red line). The consistent collinearity indicated that the lineage evolution of GT families among R. rugosa, R. chinensis and F. vesca was conserved (Figure 2B).

Figure 2.
The intra-species (A) and inter-species (B) synteny of RrGT family. (A) The paralogous RrGTs on synteny regions (dark lines) of chromosome 1/6/7. Three RrGT pairs are highlighted (Table S2). (B) Synteny regions (dark lines) of genomes of R. rugosa (red), Fragaria vesca (green), R.chinensis (brown) and orthologous GTs (color lines). The orthologous GT pairs are linked by black lines. Green lines indicate GT pairs on overlapped synteny regions.
It was worth noting that 7 RrTHs (RrTH1/4/12/17/21/31/37) of SIP1 lineage and RrTH10 of GT-2 lineage lacked homologous GTs in synteny regions (Table S2). All these RrTHs in SIP1 lineage were dispersed type and it indicated transposition should contributed to the SIP1 lineage expansion of R. rugosa.
3.2. Gene Structures and Conseved Motifs of RrGT Family
The length of RrGT proteins varied from 199 to 896 amino acids (aa), with a molecular weight range of 21.977 (RrTH37) to 99.117 kDa (RrTH33). RrGT proteins of GT-2 lineage were longer due to their two trihelix domains. And the N-terminal trihelix domain (corresponding to the ‘7-5-10’ motif group) was spaced from the C-terminal trihelix domain by motif 8. In other linages, ‘2-5-1-4’ motif group or ‘7-5-1-4’ motif group constituted the trihelix domains (Figure 3, Table S4). Besides, motif 6, motif 9, motif 3 and motif 8 were specific to SIP1 lineage and GT-1/GT-2 lineage, respectively.

Figure 3.
The conserved motifs and exon–intron structures of RrGT family. Five lineages are indicated by colored branches of NJ-dendrogram. The top 10 conserved motifs (boxes with numbers) were located by amino acid scale plate. The exons (green boxes) and introns (lines) were located by the nucleotide scale plate.
Over half RrGTs (20 RrGTs, 54.05%) included 2 exons and 10 RrGTs (27.03%) were intron-free. One RrGT contained 3, 7, 8 and 9 exons and two RrGT genes contained 5 exons, respectively. Exons of RrGT33 were up to 17 but most (exon 1–exon 12) coded the long nonconservative N-terminal with no conserved motifs.
3.3. Expression Analysis of RrGTs
In the RNA-seq expression profiles of wild R. rugosa under salt stress, 15 and 10 RrGTs were clustered as high- and middle-abundance genes, respectively (Figure 4, hierarchical clustering). In the 12 low abundant genes, except RrGT5/6/25/32 (10 > FPKM > 1) belonging to GT-2 and SH4 lineages, other 8 extremely low abundant RrGTs (FPKM < 1) were corresponding to the SIP1 lineage RrGTs without corresponding collinear RcGTs and FvGTs (Table S2).

Figure 4.
Expression profiles of RrGTs under salt stress in roots and leaves of R. rugosa. Four columns indicated roots or leaves of R. rugosa seedings treated by NaCl solution (R1h or L1h) and water (RCK or LCK) for 1 h. Normalized FPKM (per kilobase of exon model per million mapped fragments) (Table S2) are indicated by the boxes with gradient colors. Log2(fold-change) (Table S2) using RCK and LCK as controls were indicated by size and gradient color of circles. Rows were clustered by hierarchical clustering of Euclidean distance. Five lineages were represented by colored nodes of clustergram. Differentially expressed RrGTs were labeled in red.
Four differentially expressed genes were predicted as salt responsive RrGTs (Figure 4, red labeled RrGTs). Under salt stress, RrTH18 and RrH16 upregulated 16.19 fold in roots and 3.47 fold in leaves, respectively. RrTH11 and RrTH14 downregulated 0.35 fold, 0.36 fold in leaves, respectively (Table S2). The four candidate genes RrTH18, RrTH11, RrTH14 and RrH16 were named as RrGTγ-4, RrSIP1, RrSIP2 and RrGT-1, respectively.
The expression patterns of the four candidate genes were checked in wild R. rugosa seedings treated by water (CK) and 340 mM NaCl solution (Figure 5). The salt responsive expression of RrSIP1 was down-regulated in 0.5 h, kept low abundance in 1 h and 2 h, then significantly up-regulated in 4 h and 8 h. Interestingly, water stress (CK) induced significant upregulation in 0.5 h and downregulation after 0.5 h of RrSIP1 which was opposite to its salt stress response. The expression of RrSIP2 only significantly downregulated in 2 h salt stress. The expression of RrGT-1 was stable before 2h then significantly upregulated to 19.08 fold in 8 h. RrGTγ-4 responded to salt stress tempestuously and rapidly. Its expression significantly upregulated to 14.5 fold from 1 h, reached 669 fold in 2 h then downregulated to 8.2 fold in 8 h. Water stress also induced the upregulations of RrGTγ-4 but the fold changes were far from inducement of salt stress.
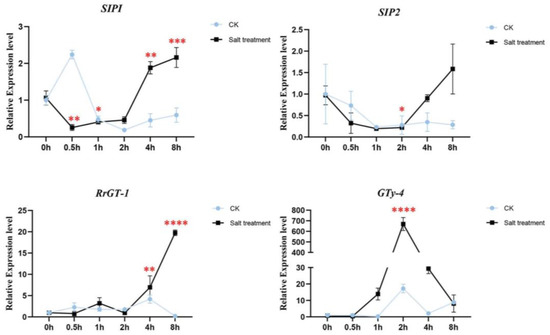
Figure 5.
The salt responsive expression levels of RrGTγ-4, RrSIP1, RrSIP2 and RrGT-1 in roots of R. rugosa. The wild R. rugosa seedings were treated by water (CK) and 340 mM NaCl solution (Salt treatment) from 0.5 h to 8 h. Untreated seedings were the reference samples (0 h). *, **, *** and **** indicated the threshold value of significance of t-test 0.05, 0.01, 0.001 and 0.0001, respectively.
3.4. Subcellular Localization of RrGT Candidates
The predicted subcellular localizations (Table S2) of RrGT family were diverse (Nuclei, chloroplasts, mitochondria, cytoplasm and so on). And different prediction tools were inconsistent in prediction of several RrGTs. e.g., RrSIP1 and RrSIP2 were predicted as proteins located on nucleus by Plant-mPLoc but on chloroplasts by WoLF PSORT. We detected the subcellular localizations of above 4 candidates (RrGTγ-4, RrSIP1, RrSIP2 and RrGT-1) to exclude the inconsistent prediction.
The fusion proteins of RrSIP1-GFP or RrGTγ-4-GFP (green) overlapped with chloroplasts (red) indicated that RrSIP1 and RrGTγ-4 were located on chloroplasts. The colocalizations (cyan) of fusion proteins (green) and nuclear marker (blue) indicated that RrGT-1 and RrSIP2 were located on nucleus (Figure 6).
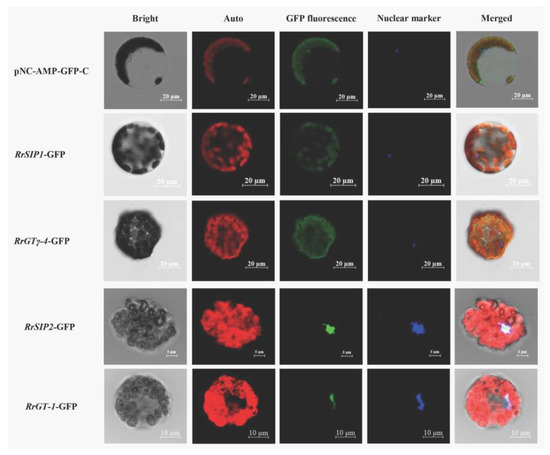
Figure 6.
The subcellular localizations of RrGTγ-4, RrSIP1, RrSIP2 and RrGT-1. Arabidopsis protoplasts transformed by GFP empty vector (pNC-AMP-GFP-C) or 35S: GTs-GFP vectors were observed using the laser confocal microscopy. The merged figures (Merged) were based on bright fields (Bright) and fluorescence of chloroplasts (Auto), green fluorescent protein (GFP), 4’,6-diamidino-2-phenylindole (DAPI, Nuclear marker) on dark fields.
4. Discussion
4.1. The Species Specific Expansion of SIP1 Lineage
Expansion of SIP1 lineage has been observed in the Brassica plants. The majority of SIP1 genes of Brassica rapa L. retained two or three copies of corresponding AtGTs and it was more than other four lineages (one copy) [42]. The Brassica specific whole genome triplication contributed to the SIP1 lineage expansion since its divergence from A. thaliana [42,43]. But the conserved number of SIP1 genes of R. rugosa relative plants (8 GTs of F. vesca and 10 GTs of R. chinensis) indicated genome triplication or WGD did not contribute to R. rugosa SIP1 lineage expansion, and the specific expansion happened after the divergence from R. chinensis (Table S1). The 7 RrGTs (RrTH1/12/17/21/31/37) without corresponding collinear RcGTs and FvGTs were predicted as dispersed duplication types, which indicated proximal duplication and tandem duplication did not contribute to the SIP1 lineage expansion. And 5 of above genes (RrTH1/12/21/31/37) were clustered into the Rosa-specific lineages of the dendrogram with RrTH34/35 and transcript abundances of the 5 genes were extremely low in roots and leaves (Figure 4). These observations indicated part of RrGTs in SIP1 lineage could arise from transposition, which contributed to SIP1 lineage expansion of R. rugosa.
4.2. Salt Responsive Candidates of RrGTs
In GT-1 lineage, GT-3A (AT5G01380) binds to GT elements of salt-induced SCaM-4 gene (Ca2+-binding protein) of Arabidopsis and soybean [10]. The interaction of GT-4 (AT3G25990) and TEM2 (a B3 and AP2/ERF domain-containing protein) improved Arabidopsis salt tolerance by activating Cor15A (AT2G42540) [14]. RrGT-1 clustered with GT-3A and AT2G38250 (no reports on abiotic stress) in same cluster of GT-1 lineage (Figure 1). RrGT-1 expressed much higher in roots (FPKM = 60.15) than in leaves (FPKM = 2.5). RrGT-1 upregulated within 1 h salt stress only in leaves but strongly upregulated in roots under 8 h salt stress. The inconsistent inducing timing indicated RrGT-1 responded to salt more promptly in leaves than in roots.
In GTγ lineage, HRA1 (HYPOXIA RESPONSE ATTENUATOR 1, AT3G10040) attenuates the anaerobic response induced by ERF-VIIs by protein interaction with RAP2.12 [11]. In rice, three OsGTγ genes were induced by salt, abscisic acid (ABA) or other abiotic stresses [24,44]. OsGTγ-1 was significantly induced by salt and its mutant increased salt stress sensitivity while OsGTγ-1 overexpression enhanced salt tolerance [24]. Similar, knockout and overexpression of OsGTγ-2 increased salt stress sensitivity and tolerance, respectively. In our study, RrGTγ-4 clustered with OsGTγ-1 and HRA1 in same cluster of GTγ lineage. RrGTγ-4 only highly expressed in roots under salt stress (FPKM = 271.54) and its abundance was much higher than other RrGTs (FPKM < 83). qRT-PCR proved the rapid and dramatic salt-inducing of RrGTγ-4 in roots within 1 h and peaked at 2 h, which was similar to OsGTγ-1. RrGTγ-4 should be a key candidate involved in salt tolerance regulation of R. rugosa roots.
In SIP1 lineage, NtSIP1 was firstly identified from Agrobacterium 6b-interacting proteins of Nicotiana tabacum L. [45]. AST1 (At3g24860) binds to a Novel AGAG-Box of stress tolerance genes to regulate Arabidopsis salt tolerance positively [12]. BnSIP1-1 overexpression improved both osmotic and salt stresses tolerance during seed germination, but only improved osmotic stress tolerance of transgenic B. napus plants [20]. In our study, RrSIP1, RrSIP2 were highly expressed in roots and leaves (FPKM > 20) and downregulated under salt stress only in leaves. Though most studies focus on stress induced GT genes, the two salt-stress-reduced RrGTs could be candidates involved in the negative regulation of salt tolerance.
Though no candidate RrGTs of GT-2 lineage significantly induced or reduced by salt, some studies have indicated GT-2 lineage also played roles in stress responses. e.g., Arabidopsis gtl1 (GTL1, AT1G33240) mutant improved water use efficiency by reducing leaf transpiration [46], and GT2L (AT5G28300) induced significantly by salt, drought, cold and ABA and it responded to cold and salt stresses by interaction with calcium/calmodulin [13].
4.3. Potential Target Genes and Regulation Roles of RrGTs
Previous study indicated that R. rugosa responds to ion stress by gene expression regulation but rather gene dosage since its ion transporter gene number was conserved [34]. In rice, OsGTγ-2 directly interacted with the GT-1 element ‘GAAAAA’ of three ion transporter genes (OsHKT2; 1, OsNHX1 and OsHKT1; 3) [44]. In the ion transporter genes of the R. rugosa, GT-1 elements were found in the promoters (1000 bp upstream) of RrHKT (evm.model.Chr5.2560, only one HKT in the genome) and 6 of all 8 RrNHXs (evm.model.Chr1.2289, evm.model.Chr1.4465 which is the only one RrSOS1 [34], evm.model.Chr2.1941, evm.model.Chr2.1942, evm.model.Chr4.416 and evm.model.Chr5.7017). Most transporter genes contained 1-4 GT-1 elements (-300 to -100 mostly) while only evm.model.Chr5.7017 (AtNHX5/6 homolog) identified 7 GT-1 elements all across the promoter (−700 to −100). It indicated that RrGTγ-4 could be regulator of these ion transporter genes and coordinated ion transport under salt stress.
SIPs mostly take part in ABA signaling to resist growth inhibiting effect of salt stress. In apple, MdSIP1-2 promoted lateral root development promotion which was associated with ABA sensitivity, drought and salt stress tolerance [47]. AST1 and BnSIP1-1 reduced water loss rat in the overexpressed seedings [12,20]. The osmotic stress marker genes were highly expressed both in the BnSIP1-1 transgenic plants and seedings whereas ion transporter genes (BnSOS1, BnNHX1, and BnHKT) were only significantly higher expressed in transgenic seedings (not in plants). SIP genes seem to exert different regulatory mechanisms along with different development process. Interestingly, the AGAG-Box ‘GGTAAA’ was found in two RrNHXs (evm.model.Chr5.2640, evm.model.Chr4.534) which lacked GT-1 elements. It indicated that excess ion response and plant developing regulation were crossed in R. rugosa by RrSIPs.
The genes of halophytes might be more superior for plant growth in saline-rich areas than their orthologs of glycophytes [28,29]. The 4 RrGT candidates were homologous to corresponding glycophytic GTs of Zizhi (over 90% protein identity). In roots, only RrGT-1 (FPKM = 60.15, this study) expressed significantly higher than GT-1 of Zizhi (FPKM = 2.34 [41]). RrGT-1 could be a halophytic gene with higher transcription levels in wild R. rugosa. While the post translational regulation not transcription contributed mostly of salt-tolerance difference between halophytes and glycophytes. Whether the GTs were involved in the post translational regulation was not studied yet and it could be an important way to study the roles of other 3 RrGTs specific to halophytes [28,29].
5. Conclusions
In this study, we identified the RrGT family with 37 members from the R. × rugosa genome. RrGTs belonged to 5 lineages and SIP1 lineage expanded significantly. The conserved motif groups corresponding to trihelix domains were most conserved. RrGTγ-4 (RrGT18), RrGT-1 (RrGT16) located on chloroplasts and RrSIP1 (RrGT11), RrSIP2 (RrGT14) located on nucleus. The four genes significantly differentially expressed under salt stress in roots or leaves. Regulation of ion transport could be the most important role of RrSIP genes and RrGTγ-4 in response to salt stress of wild R. rugosa. And RrGT-1 could be a halophytic gene with higher transcription abundance than glycophytic GT-1. Together, ion transport regulation roles of GT needed to be illuminated and the regulation role of GTs specific to halophytes would be a breakthrough point for further researches.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology12020176/s1. Figure S1: The synteny analysis of RrGTs, RcGTs and FvGTs; Table S1: Numer of GT genes of different plants; Table S2: Gene information, synteny analysis by MCscanX, RNA-seq and prediction of subcellular localizations of RrGTs; Table S3: The intra-species synteny regions; Table S4: Top 10 conserved motifs of RrGT family; Table S5: Primers for qRT-PCR.
Author Contributions
Conceptualization, J.W. and L.F.; Data curation, Y.C. and X.S.; Writing—Original draft, J.W.; Writing—Review & editing, J.W. and L.F. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by the National Natural Science Foundation of China (grant numbers 32002076, 31972454, 32171861), the National Key R&D Program of China (grant number 2018YFD1000400), the Jiangsu Provincial Natural Science Foundation (grant number SBK2020043530), the Jiangsu Provincial Natural Science Research Project of Universities (grant number 20KJB210004), and the Jiangsu Provincial Agricultural Science and Technology Independent Innovation Fund (grant numbers CX(20)3023, CX(20)3026).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Villain, P.; Mache, R.; Zhou, D.X. The mechanism of GT element-mediated cell type-specific transcriptional control. J. Biol. Chem. 1996, 271, 32593. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.X. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 1999, 4, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Nagano, Y. Several features of the GT-factor trihelix domain resemble those of the Myb DNA-binding domain. Plant Physiol. 2000, 124, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Green, P.J.; Kay, S.A.; Chua, N.H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 1987, 6, 2543–2549. [Google Scholar] [CrossRef]
- Green, P.J.; Yong, M.H.; Cuozzo, M.; Kano-Murakami, Y.; Silverstein, P.; Chua, N.H. Binding site requirements for pea nuclear protein factor GT-1 correlate with sequences required for light-dependent transcriptional activation of the rbcS-3A gene. EMBO J. 1988, 7, 4035–4044. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.J.; Lydiate, D.J.; Li, X.; Lui, H.; Gjetvaj, B.; Hegedus, D.D.; Rozwadowski, K. Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 2009, 21, 54–71. [Google Scholar] [CrossRef]
- Murata, J.; Takase, H.; Hiratsuka, K. Characterization of a Novel GT-box Binding Protein from Arabidopsis. Plant Biotechnol. 2002, 19, 103–112. [Google Scholar] [CrossRef]
- Li, B.; Jiang, S.; Yu, X.; Cheng, C.; Chen, S.; Cheng, Y.; Yuan, J.S.; Jiang, D.; He, P.; Shan, L. Phosphorylation of trihelix transcriptional repressor ASR3 by MAP KINASE4 negatively regulates Arabidopsis immunity. Plant Cell 2015, 27, 839–856. [Google Scholar] [CrossRef]
- Brewer, P.B.; Howles, P.A.; Dorian, K.; Griffith, M.E.; Ishida, T.; Kaplan-Levy, R.N.; Kilinc, A.; Smyth, D.R. PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 2004, 131, 4035–4045. [Google Scholar] [CrossRef]
- Park, H.C.; Kim, M.L.; Kang, Y.H.; Jeon, J.M.; Yoo, J.H.; Kim, M.C.; Park, C.Y.; Jeong, J.C.; Moon, B.C.; Lee, J.H.; et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004, 135, 2150–2161. [Google Scholar] [CrossRef] [PubMed]
- Giuntoli, B.; Lee, S.C.; Licausi, F.; Kosmacz, M.; Oosumi, T.; van Dongen, J.T.; Bailey-Serres, J.; Perata, P. A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol. 2014, 12, e1001950. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Shi, X.; He, L.; Guo, Y.; Zang, D.; Li, H.; Zhang, W.; Wang, Y. Arabidopsis thaliana Trihelix Transcription Factor AST1 Mediates Salt and Osmotic Stress Tolerance by Binding to a Novel AGAG-Box and Some GT Motifs. Plant Cell Physiol. 2018, 59, 946–965. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Qiu, Y.; Du, L.; Poovaiah, B.W. Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci. 2012, 185–186, 274–280. [Google Scholar] [CrossRef]
- Wang, X.H.; Li, Q.T.; Chen, H.W.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Trihelix transcription factor GT-4 mediates salt tolerance via interaction with TEM2 in Arabidopsis. BMC Plant Biol. 2014, 14, 339. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.L.; Shi, H. Physiological and molecular mechanisms of plant salt tolerance. Photosynth Res. 2013, 115, 1–22. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Blumwald, E. Developing salt-tolerant crop plants: Challenges and opportunities. Trends Plant Sci. 2005, 10, 615–620. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Kirungu, J.N.; Lu, P.; Yang, X.; Dong, Q.; Cai, X.; Xu, Y.; Wang, X.; Zhou, Z.; Hou, Y.; et al. Genome wide identification of the trihelix transcription factors and overexpression of Gh_A05G2067 (GT-2), a novel gene contributing to increased drought and salt stresses tolerance in cotton. Physiol. Plant 2019, 167, 447–464. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Pan, Q.; Chen, S.; Feng, C.; Hai, J.; Li, H. Comparison of Trihelix transcription factors between wheat and Brachypodium distachyon at genome-wide. BMC Genom. 2019, 20, 142. [Google Scholar] [CrossRef]
- Luo, J.; Tang, S.; Mei, F.; Peng, X.; Li, J.; Li, X.; Yan, X.; Zeng, X.; Liu, F.; Wu, Y.; et al. BnSIP1-1, a Trihelix Family Gene, Mediates Abiotic Stress Tolerance and ABA Signaling in Brassica napus. Front. Plant Sci. 2017, 8, 44. [Google Scholar] [CrossRef]
- Li, K.; Fan, Y.; Zhou, G.; Liu, X.; Chen, S.; Chang, X.; Wu, W.; Duan, L.; Yao, M.; Wang, R.; et al. Genome-wide identification, phylogenetic analysis, and expression profiles of trihelix transcription factor family genes in quinoa (Chenopodium quinoa Willd.) under abiotic stress conditions. BMC Genom. 2022, 23, 499. [Google Scholar] [CrossRef]
- Li, K.; Duan, L.; Zhang, Y.; Shi, M.; Chen, S.; Yang, M.; Ding, Y.; Peng, Y.; Dong, Y.; Yang, H.; et al. Genome-wide identification and expression profile analysis of trihelix transcription factor family genes in response to abiotic stress in sorghum [Sorghum bicolor (L.) Moench]. BMC Genom. 2021, 22, 738. [Google Scholar] [CrossRef]
- Osorio, M.B.; Bücker-Neto, L.; Castilhos, G.; Turchetto-Zolet, A.C.; Wiebke-Strohm, B.; Bodanese-Zanettini, M.H.; Margis-Pinheiro, M. Identification and in silico characterization of soybean trihelix-GT and bHLH transcription factors involved in stress responses. Genet. Mol. Biol. 2012, 35, 233–246. [Google Scholar] [CrossRef]
- Fang, Y.; Xie, K.; Hou, X.; Hu, H.; Xiong, L. Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol. Genet. Genom. 2010, 283, 157–169. [Google Scholar] [CrossRef]
- Xie, Z.M.; Zou, H.F.; Lei, G.; Wei, W.; Zhou, Q.Y.; Niu, C.F.; Liao, Y.; Tian, A.G.; Ma, B.; Zhang, W.K.; et al. Soybean Trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PLoS ONE 2009, 4, e6898. [Google Scholar] [CrossRef]
- Li, Y.; Hu, Z.; Dong, Y.; Xie, Z. Trihelix Transcriptional Factor GhGT26 of Cotton Enhances Salinity Tolerance in Arabidopsis. Plants 2022, 11, 2694. [Google Scholar] [CrossRef]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Himabindu, Y.; Chakradhar, T.; Reddy, M.C.; Kanygin, A.; Redding, K.E.; Chandrasekhar, T. Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 2016, 124, 39–63. [Google Scholar] [CrossRef]
- Mishra, A.; Tanna, B. Halophytes: Potential Resources for Salt Stress Tolerance Genes and Promoters. Front. Plant Sci. 2017, 8, 829. [Google Scholar] [CrossRef]
- Li, C.; Luo, Y.; Zhang, W.; Cai, Q.; Wu, X.; Tan, Z.; Chen, R.; Chen, Z.; Wang, S.; Zhang, L. A comparative study on chemical compositions and biological activities of four essential oils: Cymbopogon citratus (DC.) Stapf, Cinnamomum cassia (L.) Presl, Salvia japonica Thunb. and Rosa rugosa Thunb. J. Ethnopharmacol. 2021, 280, 114472. [Google Scholar] [CrossRef]
- Cui, W.H.; Du, X.Y.; Zhong, M.C.; Fang, W.; Suo, Z.Q.; Wang, D.; Dong, X.; Jiang, X.D.; Hu, J.Y. Complex and reticulate origin of edible roses (Rosa, Rosaceae) in China. Hortic Res. 2022, 9, uhab051. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, W.; Cheng, Y.; Feng, L. Genome-Wide Identification of LATERAL ORGAN BOUNDARIES DOMAIN (LBD) Transcription Factors and Screening of Salt Stress Candidates of Rosa rugosa Thunb. Biology 2021, 10, 992. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Z.; Zhang, Q.; Ci, H.; Wang, P.; Yu, L.; Jia, G. Genome-wide transcriptome analysis of the salt stress tolerance mechanism in Rosa chinensis. PLoS ONE 2018, 13, e0200938. [Google Scholar] [CrossRef]
- Zang, F.; Ma, Y.; Tu, X.; Huang, P.; Wu, Q.; Li, Z.; Liu, T.; Lin, F.; Pei, S.; Zang, D.; et al. A high-quality chromosome-level genome of wild Rosa rugosa. DNA Res. 2021, 28, dsab017. [Google Scholar] [CrossRef]
- Perrino, E.V.; Signorile, G.; Marvulli, M. A first checklist of the vascular flora of the Polignano a Mare coast (Apulia, southern Italy). Nat. Croat. 2013, 22, 295–318. [Google Scholar]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot 2013, 112, 1209–1221. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, J.; Wang, P.; Xu, M.; Chen, Y.; Feng, L. Systematic Identification and Analysis of OSC Gene Family of Rosa rugosa Thunb. Int. J. Mol. Sci. 2022, 23, 13884. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, P.; Liu, T.; Ren, H.; Li, Y.; Hou, X. Genome-wide Analysis and Expression Divergence of the Trihelix family in Brassica Rapa: Insight into the Evolutionary Patterns in Plants. Sci. Rep. 2017, 7, 6463. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.P.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef]
- Liu, X.; Wu, D.; Shan, T.; Xu, S.; Qin, R.; Li, H.; Negm, M.; Wu, D.; Li, J. The trihelix transcription factor OsGTγ-2 is involved adaption to salt stress in rice. Plant Mol. Biol. 2020, 103, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Kitakura, S.; Terakura, S.; Yoshioka, Y.; Machida, C.; Machida, Y. Interaction between Agrobacterium tumefaciens oncoprotein 6b and a tobacco nucleolar protein that is homologous to TNP1 encoded by a transposable element of Antirrhinum majus. J. Plant Res. 2008, 121, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.Y.; Mano, N.; Finkler, A.; Weng, H.; Day, I.S.; Reddy, A.S.N.; Poovaiah, B.W.; Fromm, H.; Hasegawa, P.M.; Mickelbart, M.V. A Ca(2+)/CaM-regulated transcriptional switch modulates stomatal development in response to water deficit. Sci. Rep. 2019, 9, 12282. [Google Scholar] [CrossRef]
- Liu, H.F.; Zhang, T.T.; Liu, Y.Q.; Kang, H.; Rui, L.; Wang, D.R.; You, C.X.; Xue, X.M.; Wang, X.F. Genome-wide analysis of the 6B-INTERACTING PROTEIN1 gene family with functional characterization of MdSIP1-2 in Malus domestica. Plant Physiol. Biochem. 2023, 195, 89–100. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).