Viroids: Non-Coding Circular RNAs Able to Autonomously Replicate and Infect Higher Plants
Abstract
Simple Summary
Abstract
1. Overview of Viroids
2. Discovery and Possible Origin of Viroids
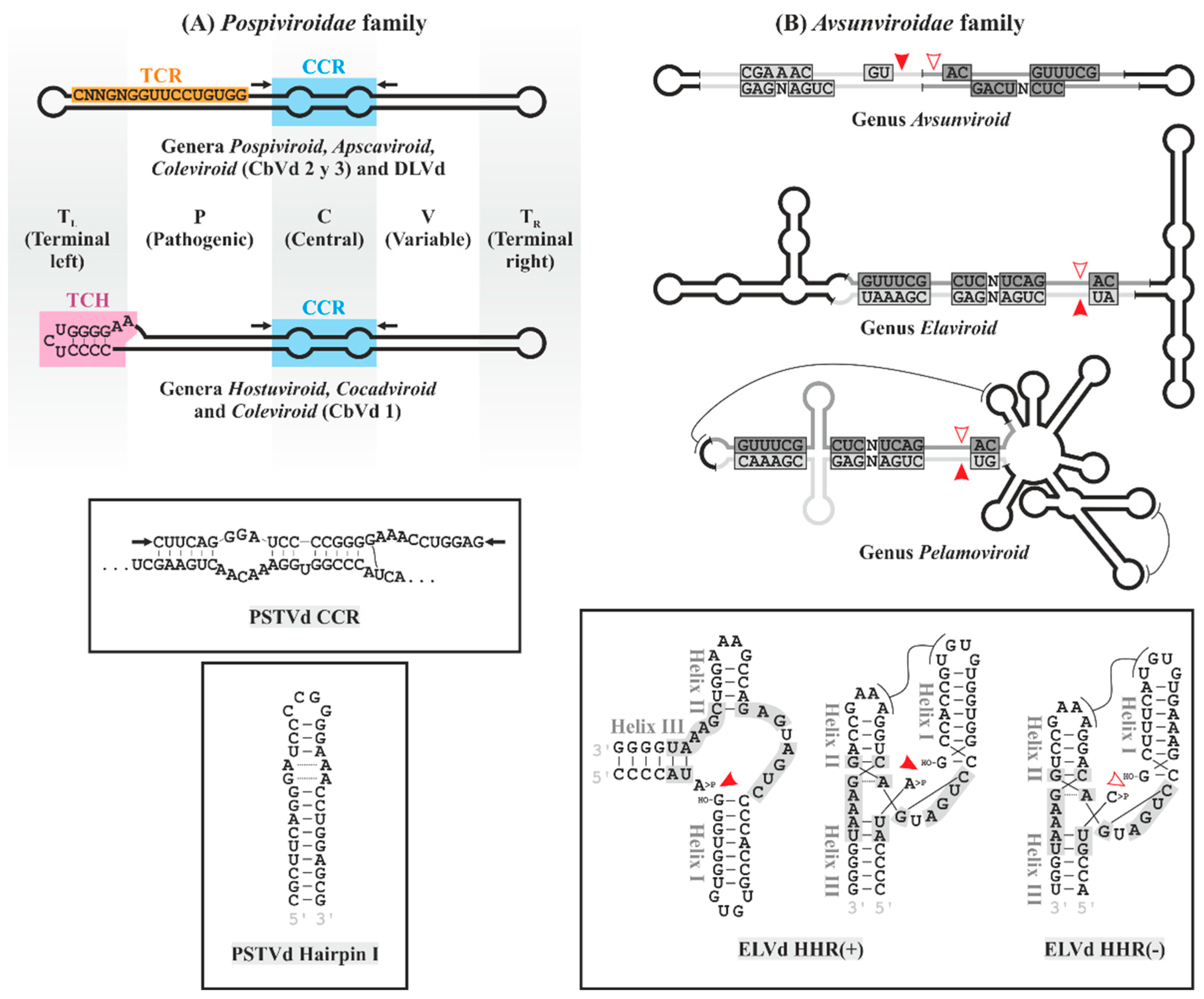
3. General Structure and Phylogenetic Classification of Viroids
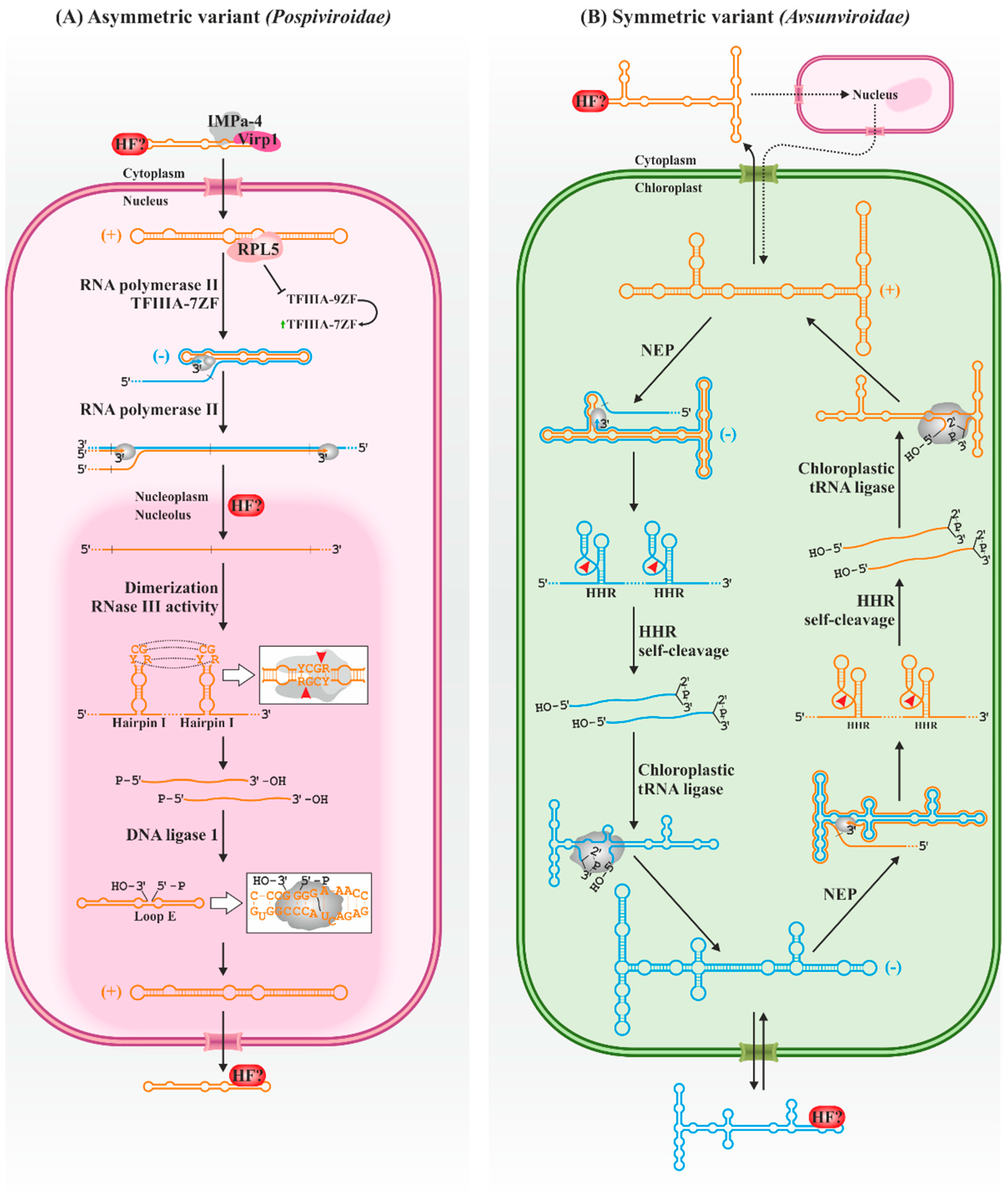
4. Viroid Replication
5. Movement of Viroids within the Plant
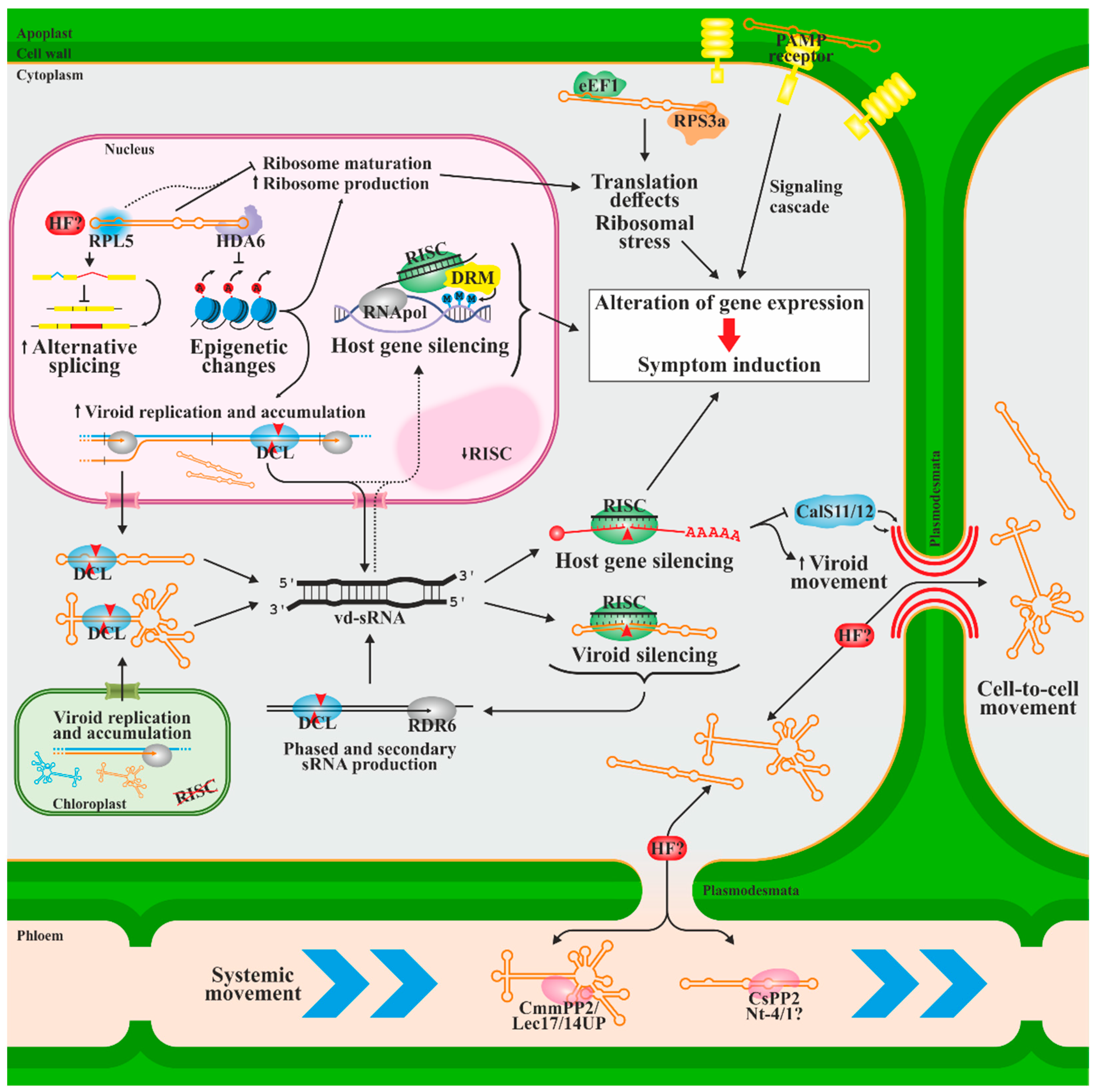
6. Host Defense and Pathogenesis
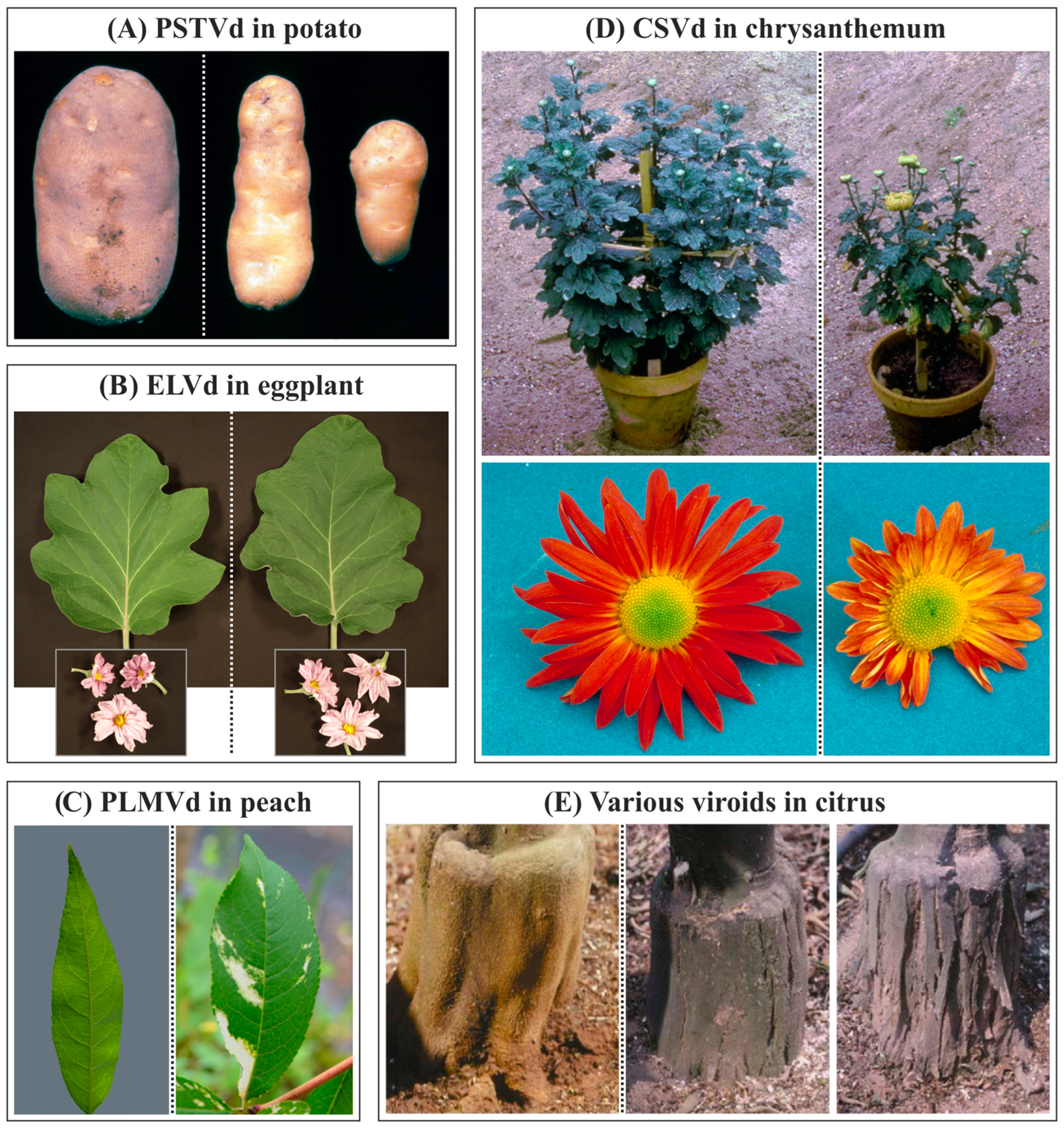
7. Host Range and Symptoms
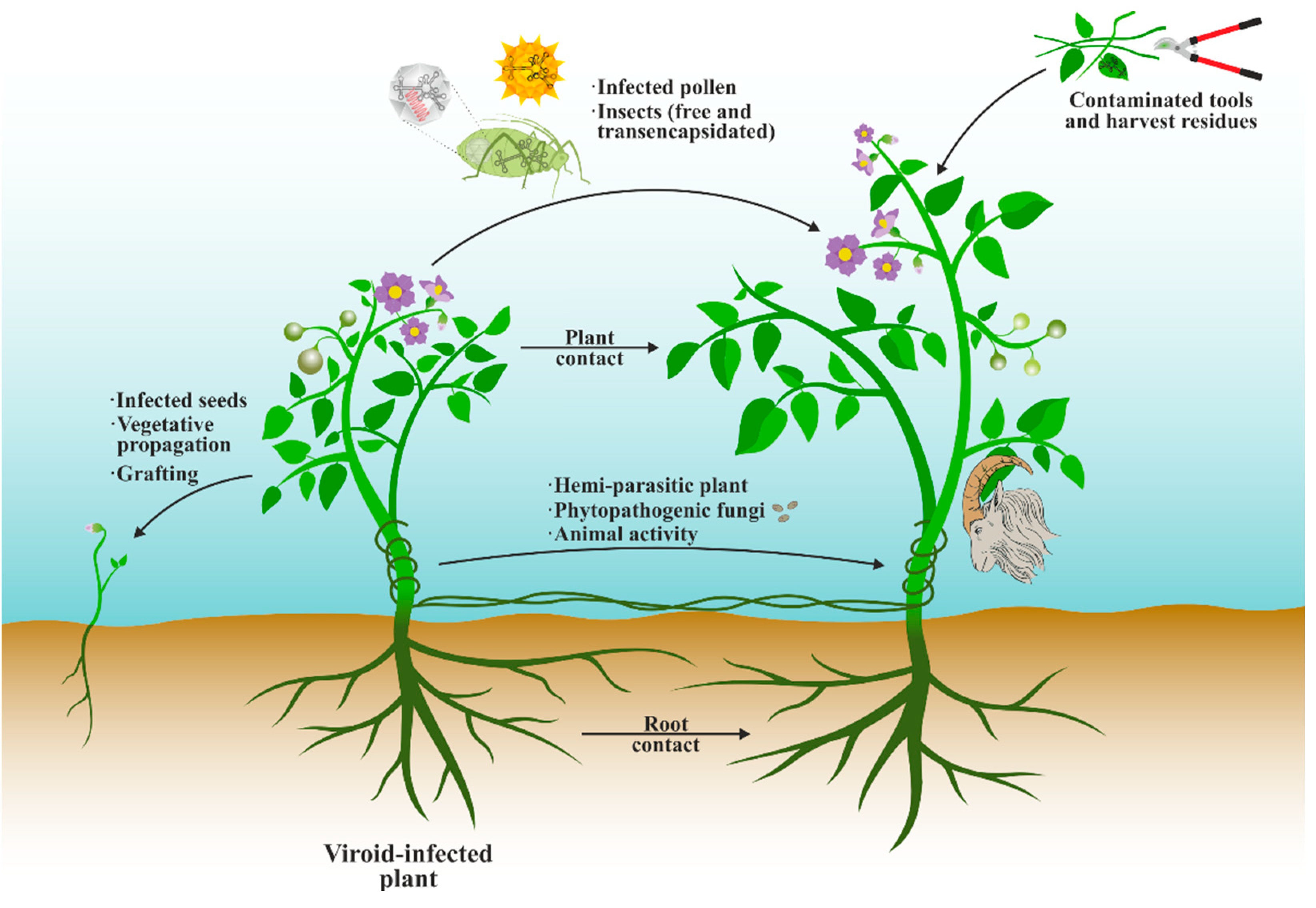
8. Transmission between Plants and Control Strategies
9. Biotechnological Aspects of Viroids
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Martin, W.H. “Spindle Tuber”, a New Potato Trouble. In Hints: To Potato Growers; New Jersey State Potato Association: East Windsor, NJ, USA, 1922; Volume 3, p. 8. [Google Scholar]
- Schultz, E.S.; Folsom, D. Transmission, Variation, and Control of Certain Degeneration Diseases of Irish Potatoes. J. Agric. Res. 1923, 25, 43–118. [Google Scholar]
- Diener, T.O.; Raymer, W.B. Potato Spindle Tuber Virus: A Plant Virus with Properties of a Free Nucleic Acid. Science 1967, 158, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Bagnall, R.H. Infectious Nucleic Acid from Host Tissues Infected with the Potato Spindle Tuber Virus. Phytopathology 1968, 58, 696–699. [Google Scholar]
- Diener, T. Potato Spindle Tuber “Virus”: IV. A Replicating, Low Molecular Weight RNA. Virology 1971, 45, 411–428. [Google Scholar] [CrossRef]
- Semancik, J.; Weathers, L. Exocortis virus of citrus: Association of infectivity with nucleic acid preparations. Virology 1968, 36, 326–328. [Google Scholar] [CrossRef]
- Diener, T.; Lawson, R. Chrysanthemum stunt: A viroid disease. Virology 1973, 51, 94–101. [Google Scholar] [CrossRef]
- Diener, T.O. Are viroids escaped introns? Proc. Natl. Acad. Sci. USA 1981, 78, 5014–5015. [Google Scholar] [CrossRef]
- Kiefer, M.C.; Owens, R.A.; Diener, T.O. Structural similarities between viroids and transposable genetic elements. Proc. Natl. Acad. Sci. USA 1983, 80, 6234–6238. [Google Scholar] [CrossRef]
- Catalán, P.; Elena, S.F.; Cuesta, J.A.; Manrubia, S. Parsimonious Scenario for the Emergence of Viroid-Like Replicons De Novo. Viruses 2019, 11, 425. [Google Scholar] [CrossRef]
- Jain, N.; Blauch, L.R.; Szymanski, M.R.; Das, R.; Tang, S.K.Y.; Yin, Y.W.; Fire, A.Z. Transcription polymerase–catalyzed emergence of novel RNA replicons. Science 2020, 368, eaay0688. [Google Scholar] [CrossRef]
- Pérez-Vargas, J.; de Oliveira, R.P.; Jacquet, S.; Pontier, D.; Cosset, F.-L.; Freitas, N. HDV-Like Viruses. Viruses 2021, 13, 1207. [Google Scholar] [CrossRef] [PubMed]
- Forgia, M.; Navarro, B.; Daghino, S.; Cervera, A.; Gisel, A.; Perotto, S.; Aghayeva, D.N.; Akinyuwa, M.F.; Gobbi, E.; Zheludev, I.N.; et al. Extant Hybrids of RNA Viruses and Viroid-like Elements. bioRxiv 2022. [Google Scholar] [CrossRef]
- Diener, T.O. Circular RNAs: Relics of precellular evolution? Proc. Natl. Acad. Sci. USA 1989, 86, 9370–9374. [Google Scholar] [CrossRef] [PubMed]
- Moelling, K.; Broecker, F. Viroids and the Origin of Life. Int. J. Mol. Sci. 2021, 22, 3476. [Google Scholar] [CrossRef]
- Elena, S.F.; Dopazo, J.; Flores, R.; Diener, T.O.; Moya, A. Phylogeny of viroids, viroidlike satellite RNAs, and the viroidlike domain of hepatitis delta virus RNA. Proc. Natl. Acad. Sci. USA 1991, 88, 5631–5634. [Google Scholar] [CrossRef]
- Elena, S.F.; Dopazo, J.; de la Peña, M.; Flores, R.; Diener, T.O.; Moya, A. Phylogenetic Analysis of Viroid and Viroid-Like Satellite RNAs from Plants: A Reassessment. J. Mol. Evol. 2014, 53, 155–159. [Google Scholar] [CrossRef]
- Gross, H.J.; Domdey, H.; Lossow, C.; Jank, P.; Raba, M.; Alberty, H.; Sänger, H.L. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 1978, 273, 203–208. [Google Scholar] [CrossRef]
- Keese, P.; Symons, R.H. Domains in viroids: Evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA 1985, 82, 4582–4586. [Google Scholar] [CrossRef]
- Sano, T.; Candresse, T.; Hammond, R.W.; Diener, T.O.; Owens, R.A. Identification of multiple structural domains regulating viroid pathogenicity. Proc. Natl. Acad. Sci. USA 1992, 89, 10104–10108. [Google Scholar] [CrossRef]
- Zhong, X.; Archual, A.J.; Amin, A.A.; Ding, B. A Genomic Map of Viroid RNA Motifs Critical for Replication and Systemic Trafficking. Plant Cell 2008, 20, 35–47. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, C.; Li, J.; Li, C.; Tao, X.; Leontis, N.B.; Zirbel, C.L.; Bisaro, D.M.; Ding, B. Functional analysis reveals G/U pairs critical for replication and trafficking of an infectious non-coding viroid RNA. Nucleic Acids Res. 2020, 48, 3134–3155. [Google Scholar] [CrossRef] [PubMed]
- Diener, T. Potato Spindle Tuber Virus: A Plant Virus with Properties of a Free Nucleic Acid: III. Subcellular Location of PSTV-RNA and the Question of Whether Virions Exist in Extracts or in Situ. Virology 1971, 43, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Sänger, H.L. An Infectious and Replicating RNA of Low Molecular Weight: The Agent of the Exocortis Disease of Citrus. In Workshop on Mechanisms and Prospects of Genetic Exchange, Berlin, December 11 to 13, 1971; Pergamon Press: Oxford, UK, 1972; pp. 103–116. [Google Scholar] [CrossRef]
- Takahashi, T.; Yaguchi, S.; Oikawa, S.; Kamita, N. Subcellular Location of Hop Stunt Viroid1. J. Phytopathol. 1982, 103, 285–293. [Google Scholar] [CrossRef]
- Bonfiglioli, R.G.; McFadden, G.I.; Symons, R.H. In situ hybridization localizes avocado sunblotch viroid on chloroplast thylakoid membranes and coconut cadang cadang viroid in the nucleus. Plant J. 1994, 6, 99–103. [Google Scholar] [CrossRef]
- Qi, Y.; Ding, B. Differential Subnuclear Localization of RNA Strands of Opposite Polarity Derived from an Autonomously Replicating Viroid. Plant Cell 2003, 15, 2566–2577. [Google Scholar] [CrossRef] [PubMed]
- Symons, R.H. Avocado sunblotch viroid: Primary sequence and proposed secondary structure. Nucleic Acids Res. 1981, 9, 6527–6537. [Google Scholar] [CrossRef]
- Lima, M.I.; Fonseca, M.E.N.; Flores, R.; Kitajima, E.W. Detection of avocado sunblotch viroid in chloroplasts of avocado leaves by in situ hybridization. Arch. Virol. 1994, 138, 385–390. [Google Scholar] [CrossRef]
- Bussière, F.; Lehoux, J.; Thompson, D.A.; Skrzeczkowski, L.J.; Perreault, J.-P. Subcellular Localization and Rolling Circle Replication of Peach Latent Mosaic Viroid: Hallmarks of Group A Viroids. J. Virol. 1999, 73, 6353–6360. [Google Scholar] [CrossRef]
- Navarro, J.-A.; Daròs, J.-A.; Flores, R. Complexes Containing Both Polarity Strands of Avocado Sunblotch Viroid: Identification in Chloroplasts and Characterization. Virology 1999, 253, 77–85. [Google Scholar] [CrossRef]
- Hernández, C.; Flores, R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. USA 1992, 89, 3711–3715. [Google Scholar] [CrossRef]
- Navarro, B.; Flores, R. Chrysanthemum chlorotic mottle viroid: Unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. USA 1997, 94, 11262–11267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qi, S.; Tang, N.; Zhang, X.; Chen, S.; Zhu, P.; Ma, L.; Cheng, J.; Xu, Y.; Lu, M.; et al. Discovery of Replicating Circular RNAs by RNA-Seq and Computational Algorithms. PLOS Pathog. 2014, 10, e1004553. [Google Scholar] [CrossRef] [PubMed]
- Fadda, Z.; Daròs, J.A.; Fagoaga, C.; Flores, R.; Duran-Vila, N. Eggplant Latent Viroid, the Candidate Type Species for a New Genus within the Family Avsunviroidae (Hammerhead Viroids). J. Virol. 2003, 77, 6528–6532. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Randles, J.; Bar-Joseph, M.; Diener, T. A proposed scheme for viroid classification and nomenclature. Arch. Virol. 2014, 143, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Branch, A.D.; Robertson, H.D.; Dickson, E. Longer-than-unit-length viroid minus strands are present in RNA from infected plants. Proc. Natl. Acad. Sci. USA 1981, 78, 6381–6385. [Google Scholar] [CrossRef]
- Branch, A.D.; Robertson, H.D. A Replication Cycle for Viroids and Other Small Infectious RNA’s. Science 1984, 223, 450–455. [Google Scholar] [CrossRef]
- Zaitlin, M.; Niblett, C.; Dickson, E.; Goldberg, R. Tomato DNA contains no detectable regions complementary to potato spindle tuber viroid as assayed by solution and filter hybridization. Virology 1980, 104, 1–9. [Google Scholar] [CrossRef]
- Branch, A.D.; Dickson, E. Tomato DNA contains no detectable regions complementary to potato spindle tuber viroid as assayed by Southern hybridization. Virology 1980, 104, 10–26. [Google Scholar] [CrossRef]
- Rohde, W.; Sänger, H.L. Detection of complementary RNA intermediates of viroid replication by Northern blot hybridization. Biosci. Rep. 1981, 1, 327–336. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Owens, R.A.; Erbe, E.; Hadidi, A.; Steere, R.L.; Diener, T.O. Separation and infectivity of circular and linear forms of potato spindle tuber viroid. Proc. Natl. Acad. Sci. USA 1977, 74, 3859–3863. [Google Scholar] [CrossRef] [PubMed]
- Grill, L.K.; Semancik, J.S. RNA sequences complementary to citrus exocortis viroid in nucleic acid preparations from infected Gynura aurantiaca. Proc. Natl. Acad. Sci. USA 1978, 75, 896–900. [Google Scholar] [CrossRef] [PubMed]
- Bruening, G.; Gould, A.R.; Murphy, P.J.; Symons, R.H. Oligomers of avocado sunblotch viroid are found in infected avocado leaves. FEBS Lett. 1982, 148, 71–78. [Google Scholar] [CrossRef]
- Owens, R.A.; Diener, T.O. RNA intermediates in potato spindle tuber viroid replication. Proc. Natl. Acad. Sci. USA 1982, 79, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Hadidi, A.; Hashimoto, J.; Diener, T. Potato spindle tuber viroid-specific double-stranded RNA in extracts from infected tomato leaves. Ann. de l’Institut Pasteur/Virol. 1982, 133, 15–31. [Google Scholar] [CrossRef]
- Branch, A.D.; Benenfeld, B.J.; Robertson, H.D. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl. Acad. Sci. USA 1988, 85, 9128–9132. [Google Scholar] [CrossRef]
- Feldstein, P.A.; Hu, Y.; Owens, R.A. Precisely full length, circularizable, complementary RNA: An infectious form of potato spindle tuber viroid. Proc. Natl. Acad. Sci. USA 1998, 95, 6560–6565. [Google Scholar] [CrossRef]
- Seo, H.; Wang, Y.; Park, W.J. Time-Resolved Observation of the Destination of Microinjected Potato Spindle Tuber Viroid (PSTVd) in the Abaxial Leaf Epidermal Cells of Nicotiana benthamiana. Microorganisms 2020, 8, 2044. [Google Scholar] [CrossRef]
- Seo, H.; Kim, K.; Park, W. Effect of VIRP1 Protein on Nuclear Import of Citrus Exocortis Viroid (CEVd). Biomolecules 2021, 11, 95. [Google Scholar] [CrossRef]
- Maniataki, E.; Tabler, M.; Tsagris, M. Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA 2003, 9, 346–354. [Google Scholar] [CrossRef]
- de Alba, A.E.M.; Sägesser, R.; Tabler, M.; Tsagris, M. A Bromodomain-Containing Protein from Tomato Specifically Binds Potato Spindle Tuber Viroid RNA In Vitro and In Vivo. J. Virol. 2003, 77, 9685–9694. [Google Scholar] [CrossRef] [PubMed]
- Gozmanova, M.; Denti, M.A.; Minkov, I.N.; Tsagris, M.; Tabler, M. Characterization of the RNA motif responsible for the specific interaction of potato spindle tuber viroid RNA (PSTVd) and the tomato protein Virp1. Nucleic Acids Res. 2003, 31, 5534–5543. [Google Scholar] [CrossRef] [PubMed]
- Kalantidis, K.; Denti, M.A.; Tzortzakaki, S.; Marinou, E.; Tabler, M.; Tsagris, M. Virp1 Is a Host Protein with a Major Role in Potato Spindle Tuber Viroid Infection in Nicotiana Plants. J. Virol. 2007, 81, 12872–12880. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Mudiyanselage, S.D.D.; Park, W.J.; Wang, M.; Takeda, R.; Bin Liu, B.; Wang, Y. A nuclear import pathway exploited by pathogenic noncoding RNAs. Plant Cell 2022, 34, 3543–3556. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, S.; Kalantidis, K.; Rao, A.L.N. A Bromodomain-Containing Host Protein Mediates the Nuclear Importation of a Satellite RNA of Cucumber Mosaic Virus. J. Virol. 2014, 88, 1890–1896. [Google Scholar] [CrossRef]
- Abraitiene, A.; Zhao, Y.; Hammond, R. Nuclear targeting by fragmentation of the Potato spindle tuber viroid genome. Biochem. Biophys. Res. Commun. 2008, 368, 470–475. [Google Scholar] [CrossRef]
- Marquez-Molins, J.; Navarro, J.A.; Seco, L.C.; Pallas, V.; Gomez, G. Might exogenous circular RNAs act as protein-coding transcripts in plants? RNA Biol. 2021, 18, 98–107. [Google Scholar] [CrossRef]
- Mühlbach, H.-P.; Sänger, H.L. Viroid replication is inhibited by α-amanitin. Nature 1979, 278, 185–188. [Google Scholar] [CrossRef]
- Flores, R.; Semancik, J.S. Properties of a cell-free system for synthesis of citrus exocortis viroid. Proc. Natl. Acad. Sci. USA 1982, 79, 6285–6288. [Google Scholar] [CrossRef]
- Yoshikawa, N.; Takahashi, T. Inhibition of Hop Stunt Viroid Replication by α-Amanitin. J. Plant Dis. Prot. 1986, 93, 62–71. [Google Scholar]
- Warrilow, D.; Symons, R.H. Citrus exocortis viroid RNA is associated with the largest subunit of RNA polymerase II in tomato in vivo. Arch. Virol. 1999, 144, 2367–2375. [Google Scholar] [CrossRef]
- Mudiyanselage, S.D.D.; Ma, J.; Pechan, T.; Pechanova, O.; Liu, B.; Wang, Y. A remodeled RNA polymerase II complex catalyzing viroid RNA-templated transcription. PLOS Pathog. 2022, 18, e1010850. [Google Scholar] [CrossRef]
- Wang, Y.; Qu, J.; Ji, S.; Wallace, A.J.; Wu, J.; Li, Y.; Gopalan, V.; Ding, B. A Land Plant-Specific Transcription Factor Directly Enhances Transcription of a Pathogenic Noncoding RNA Template by DNA-Dependent RNA Polymerase II. Plant Cell 2015, 28, 1094–1107. [Google Scholar] [CrossRef] [PubMed]
- Kolonko, N.; Bannach, O.; Aschermann, K.; Hu, K.-H.; Moors, M.; Schmitz, M.; Steger, G.; Riesner, D. Transcription of potato spindle tuber viroid by RNA polymerase II starts in the left terminal loop. Virology 2006, 347, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Bojić, T.; Beeharry, Y.; Zhang, D.J.; Pelchat, M. Tomato RNA polymerase II interacts with the rod-like conformation of the left terminal domain of the potato spindle tuber viroid positive RNA genome. J. Gen. Virol. 2012, 93, 1591–1600. [Google Scholar] [CrossRef]
- Eiras, M.; Nohales, M.A.; Kitajima, E.W.; Flores, R.; Daròs, J.A. Ribosomal protein L5 and transcription factor IIIA from Arabidopsis thaliana bind in vitro specifically Potato spindle tuber viroid RNA. Arch. Virol. 2011, 156, 529–533. [Google Scholar] [CrossRef]
- Jiang, J.; Smith, H.N.; Ren, D.; Mudiyanselage, S.D.D.; Dawe, A.L.; Wang, L.; Wang, Y. Potato Spindle Tuber Viroid Modulates Its Replication through a Direct Interaction with a Splicing Regulator. J. Virol. 2018, 92, e01004-18. [Google Scholar] [CrossRef]
- Gas, M.-E.; Hernandez, C.; Flores, R.; Daròs, J.-A. Processing of Nuclear Viroids In Vivo: An Interplay between RNA Conformations. PLOS Pathog. 2007, 3, e182. [Google Scholar] [CrossRef]
- Gas, M.-E.; Molina-Serrano, D.; Hernández, C.; Flores, R.; Daròs, J.-A. Monomeric Linear RNA of Citrus Exocortis Viroid Resulting from Processing In Vivo Has 5′-Phosphomonoester and 3′-Hydroxyl Termini: Implications for the RNase and RNA Ligase Involved in Replication. J. Virol. 2008, 82, 10321–10325. [Google Scholar] [CrossRef]
- Nohales, M.; Flores, R.; Daròs, J.-A. Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc. Natl. Acad. Sci. USA 2012, 109, 13805–13810. [Google Scholar] [CrossRef]
- Daròs, J.-A.; Elena, S.; Flores, R. Viroids: An Ariadne’s thread into the RNA labyrinth. EMBO Rep. 2006, 7, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Gomez, G.; Pallás, V. Noncoding RNA Mediated Traffic of Foreign mRNA into Chloroplasts Reveals a Novel Signaling Mechanism in Plants. PLoS ONE 2010, 5, e12269. [Google Scholar] [CrossRef] [PubMed]
- Gomez, G.; Pallas, V. Studies on Subcellular Compartmentalization of Plant Pathogenic Noncoding RNAs Give New Insights into the Intracellular RNA-Traffic Mechanisms. Plant Physiol. 2012, 159, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Navarro, J.A.; Vera, A.; Flores, R. A Chloroplastic RNA Polymerase Resistant to Tagetitoxin Is Involved in Replication of Avocado Sunblotch Viroid. Virology 2000, 268, 218–225. [Google Scholar] [CrossRef]
- Rodio, M.-E.; Delgado, S.; De Stradis, A.; Gómez, M.-D.; Flores, R.; Di Serio, F. A Viroid RNA with a Specific Structural Motif Inhibits Chloroplast Development. Plant Cell 2007, 19, 3610–3626. [Google Scholar] [CrossRef] [PubMed]
- Delgado, S.; de Alba, A.E.M.; Hernández, C.; Flores, R. A Short Double-Stranded RNA Motif of Peach Latent Mosaic Viroid Contains the Initiation and the Self-Cleavage Sites of Both Polarity Strands. J. Virol. 2005, 79, 12934–12943. [Google Scholar] [CrossRef]
- Motard, J.; Bolduc, F.; Thompson, D.; Perreault, J.-P. The peach latent mosaic viroid replication initiation site is located at a universal position that appears to be defined by a conserved sequence. Virology 2008, 373, 362–375. [Google Scholar] [CrossRef]
- López-Carrasco, A.; Gago-Zachert, S.; Mileti, G.; Minoia, S.; Flores, R.; Delgado, S. The transcription initiation sites of eggplant latent viroid strands map within distinct motifs in their in vivo RNA conformations. RNA Biol. 2015, 13, 83–97. [Google Scholar] [CrossRef]
- Hutchins, C.J.; Rathjen, P.D.; Forster, A.C.; Symons, R.H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986, 14, 3627–3640. [Google Scholar] [CrossRef]
- Prody, G.A.; Bakos, J.T.; Buzayan, J.M.; Schneider, I.R.; Bruening, G. Autolytic Processing of Dimeric Plant Virus Satellite RNA. Science 1986, 231, 1577–1580. [Google Scholar] [CrossRef]
- Webb, C.-H.T.; Riccitelli, N.J.; Ruminski, D.J.; Lupták, A. Widespread Occurrence of Self-Cleaving Ribozymes. Science 2009, 326, 953. [Google Scholar] [CrossRef] [PubMed]
- de la Peña, M.; Garcia-Robles, I. Intronic hammerhead ribozymes are ultraconserved in the human genome. EMBO Rep. 2010, 11, 711–716. [Google Scholar] [CrossRef]
- de la Peña, M.; García-Robles, I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA 2010, 16, 1943–1950. [Google Scholar] [CrossRef] [PubMed]
- Forster, A.C.; Symons, R.H. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell 1987, 50, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Pley, H.W.; Flaherty, K.M.; McKay, D.B. Three-dimensional structure of a hammerhead ribozyme. Nature 1994, 372, 68–74. [Google Scholar] [CrossRef]
- Khvorova, A.; Lescoute, A.; Westhof, E.; Jayasena, S.D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Mol. Biol. 2003, 10, 708–712. [Google Scholar] [CrossRef]
- De La Peña, M.; Gago, S.; Flores, R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003, 22, 5561–5570. [Google Scholar] [CrossRef]
- Murray, J.B.; Seyhan, A.A.; Walter, N.G.; Burke, J.M.; Scott, W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998, 5, 587–595. [Google Scholar] [CrossRef]
- O’Rear, J.L.; Wang, S.; Feig, A.L.; Beigelman, L.; Uhlenbeck, O.C.; Herschlag, D. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA 2001, 7, 537–545. [Google Scholar] [CrossRef]
- Roychowdhury-Saha, M.; Burke, D.H. Distinct reaction pathway promoted by non-divalent-metal cations in a tertiary stabilized hammerhead ribozyme. RNA 2007, 13, 841–848. [Google Scholar] [CrossRef]
- Lee, T.-S.; López, C.S.; Giambaşu, G.M.; Martick, M.; Scott, W.G.; York, D.M. Role of Mg2+ in Hammerhead Ribozyme Catalysis from Molecular Simulation. J. Am. Chem. Soc. 2008, 130, 3053–3064. [Google Scholar] [CrossRef]
- Mir, A.; Golden, B.L. Two Active Site Divalent Ions in the Crystal Structure of the Hammerhead Ribozyme Bound to a Transition State Analogue. Biochemistry 2016, 55, 633–636. [Google Scholar] [CrossRef]
- Canny, M.D.; Jucker, F.M.; Pardi, A. Efficient Ligation of the Schistosoma Hammerhead Ribozyme. Biochemistry 2007, 46, 3826–3834. [Google Scholar] [CrossRef]
- Przybilski, R.; Hammann, C. Idiosyncratic cleavage and ligation activity of individual hammerhead ribozymes and core sequence variants thereof. Biol. Chem. 2007, 388, 737–741. [Google Scholar] [CrossRef]
- Daròs, J.-A.; Flores, R. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 2002, 21, 749–759. [Google Scholar] [CrossRef]
- Nohales, M.; Molina-Serrano, D.; Flores, R.; Daròs, J.-A. Involvement of the Chloroplastic Isoform of tRNA Ligase in the Replication of Viroids Belonging to the Family Avsunviroidae. J. Virol. 2012, 86, 8269–8276. [Google Scholar] [CrossRef]
- Englert, M.; Latz, A.; Becker, D.; Gimple, O.; Beier, H.; Akama, K. Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie 2007, 89, 1351–1365. [Google Scholar] [CrossRef]
- Molina-Serrano, D.; Suay, L.; Salvador, M.L.; Flores, R.; Daròs, J.-A. Processing of RNAs of the Family Avsunviroidae in Chlamydomonas reinhardtii Chloroplasts. J. Virol. 2007, 81, 4363–4366. [Google Scholar] [CrossRef]
- Martínez, F.; Marqués, J.; Salvador, M.L.; Daròs, J.-A. Mutational analysis of eggplant latent viroid RNA processing in Chlamydomonas reinhardtii chloroplast. J. Gen. Virol. 2009, 90, 3057–3065. [Google Scholar] [CrossRef]
- Daròs, J.-A.; Aragonés, V.; Cordero, T. A viroid-derived system to produce large amounts of recombinant RNA in Escherichia coli. Sci. Rep. 2018, 8, 1904. [Google Scholar] [CrossRef]
- Daròs, J.-A.; Aragonés, V.; Cordero, M.-T. Recombinant RNA Production 2014. Available online: https://patents.google.com/patent/US20170088871A1/en (accessed on 16 January 2023).
- Cordero, T.; Ortolá, B.; Daròs, J.-A. Mutational Analysis of Eggplant Latent Viroid RNA Circularization by the Eggplant tRNA Ligase in Escherichia coli. Front. Microbiol. 2018, 9, 635. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Kwon, M.-O.; Hammond, R.; Owens, R. Cell-to-cell movement of potato spindle tuber viroid. Plant J. 1997, 12, 931–936. [Google Scholar] [CrossRef]
- Palukaitis, P. Potato spindle tuber viroid: Investigation of the long-distance, intra-plant transport route. Virology 1987, 158, 239–241. [Google Scholar] [CrossRef]
- Zhu, Y.; Green, L.; Woo, Y.-M.; Owens, R.; Ding, B. Cellular Basis of Potato Spindle Tuber Viroid Systemic Movement. Virology 2001, 279, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Bani-Hashemian, S.M.; Pensabene-Bellavia, G.; Duran-Vila, N.; Serra, P. Phloem restriction of viroids in three citrus hosts is overcome by grafting with Etrog citron: Potential involvement of a translocatable factor. J. Gen. Virol. 2015, 96, 2405–2410. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Tao, X.; Stombaugh, J.; Leontis, N.; Ding, B. Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. EMBO J. 2007, 26, 3836–3846. [Google Scholar] [CrossRef] [PubMed]
- Gómez, G.; Pallás, V. Identification of an In Vitro Ribonucleoprotein Complex Between a Viroid RNA and a Phloem Protein from Cucumber Plants. Mol. Plant-Microbe Interact. 2001, 14, 910–913. [Google Scholar] [CrossRef] [PubMed]
- Gómez, G.; Pallás, V. A Long-Distance Translocatable Phloem Protein from Cucumber Forms a Ribonucleoprotein Complex In Vivo with Hop Stunt Viroid RNA. J. Virol. 2004, 78, 10104–10110. [Google Scholar] [CrossRef]
- Gómez, G.; Torres, H.; Pallás, V. Identification of translocatable RNA-binding phloem proteins from melon, potential components of the long-distance RNA transport system. Plant J. 2005, 41, 107–116. [Google Scholar] [CrossRef]
- Morozov, S.Y.; Makarova, S.S.; Erokhina, T.N.; Kopertekh, L.; Schiemann, J.; Owens, R.A.; Solovyev, A.G. Plant 4/1 protein: Potential player in intracellular, cell-to-cell and long-distance signaling. Front. Plant Sci. 2014, 5, 26. [Google Scholar] [CrossRef]
- Solovyev, A.G.; Makarova, S.S.; Remizowa, M.V.; Lim, H.-S.; Hammond, J.; Owens, R.A.; Kopertekh, L.; Schiemann, J.; Morozov, S.Y. Possible role of the Nt-4/1 protein in macromolecular transportin vascular tissue. Plant Signal. Behav. 2013, 8, e25784. [Google Scholar] [CrossRef] [PubMed]
- Adkar-Purushothama, C.R.; Brosseau, C.; Giguère, T.; Sano, T.; Moffett, P.; Perreault, J.-P. Small RNA Derived from the Virulence Modulating Region of the Potato spindle tuber viroid Silences callose synthase Genes of Tomato Plants. Plant Cell 2015, 27, 2178–2194. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, V.A.; Meins, F. Movement of plant viruses is delayed in a beta-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 2000, 21, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Bucher, G.L.; Tarina, C.; Heinlein, M.; Di Serio, F.; Meins, F.; Iglesias, V.A. Local expression of enzymatically active class I β-1, 3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 2001, 28, 361–369. [Google Scholar] [CrossRef]
- Li, W.; Zhao, Y.; Liu, C.; Yao, G.; Wu, S.; Hou, C.; Zhang, M.; Wang, D. Callose deposition at plasmodesmata is a critical factor in restricting the cell-to-cell movement of Soybean mosaic virus. Plant Cell Rep. 2012, 31, 905–916. [Google Scholar] [CrossRef]
- Qi, Y.; Pélissier, T.; Itaya, A.; Hunt, E.; Wassenegger, M.; Ding, B. Direct Role of a Viroid RNA Motif in Mediating Directional RNA Trafficking across a Specific Cellular Boundary. Plant Cell 2004, 16, 1741–1752. [Google Scholar] [CrossRef]
- Takeda, R.; Petrov, A.I.; Leontis, N.B.; Ding, B. A Three-Dimensional RNA Motif in Potato spindle tuber viroid Mediates Trafficking from Palisade Mesophyll to Spongy Mesophyll in Nicotiana benthamiana. Plant Cell 2011, 23, 258–272. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Y. Studies on Viroid Shed Light on the Role of RNA Three-Dimensional Structural Motifs in RNA Trafficking in Plants. Front. Plant Sci. 2022, 13, 836267. [Google Scholar] [CrossRef]
- Papaefthimiou, I.; Hamilton, A.; Denti, M.; Baulcombe, D.; Tsagris, M.; Tabler, M. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 2001, 29, 2395–2400. [Google Scholar] [CrossRef]
- Itaya, A.; Folimonov, A.; Matsuda, Y.; Nelson, R.S.; Ding, B. Potato spindle tuber viroid as Inducer of RNA Silencing in Infected Tomato. Mol. Plant-Microbe Interact. 2007, 14, 1332–1334. [Google Scholar] [CrossRef]
- de Alba, A.E.M.; Flores, R.; Hernández, C. Two Chloroplastic Viroids Induce the Accumulation of Small RNAs Associated with Posttranscriptional Gene Silencing. J. Virol. 2002, 76, 13094–13096. [Google Scholar] [CrossRef] [PubMed]
- Markarian, N.; Li, H.W.; Ding, S.W.; Semancik, J.S. RNA silencing as related to viroid induced symptom expression. Arch. Virol. 2004, 149, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Navarro, B.; Pantaleo, V.; Gisel, A.; Moxon, S.; Dalmay, T.; Bisztray, G.; Di Serio, F.; Burgyán, J. Deep Sequencing of Viroid-Derived Small RNAs from Grapevine Provides New Insights on the Role of RNA Silencing in Plant-Viroid Interaction. PLoS ONE 2009, 4, e7686. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Donaire, L.; Llave, C.; Pallas, V.; Gomez, G. High-throughput sequencing of Hop stunt viroid-derived small RNAs from cucumber leaves and phloem. Mol. Plant Pathol. 2010, 11, 347–359. [Google Scholar] [CrossRef]
- Minoia, S.; Carbonell, A.; Di Serio, F.; Gisel, A.; Carrington, J.C.; Navarro, B.; Flores, R. Specific Argonautes Selectively Bind Small RNAs Derived from Potato Spindle Tuber Viroid and Attenuate Viroid Accumulation In Vivo. J. Virol. 2014, 88, 11933–11945. [Google Scholar] [CrossRef]
- Carbonell, A.; de Alba, Á.-E.M.; Flores, R.; Gago, S. Double-stranded RNA interferes in a sequence-specific manner with the infection of representative members of the two viroid families. Virology 2008, 371, 44–53. [Google Scholar] [CrossRef]
- Schwind, N.; Zwiebel, M.; Itaya, A.; Ding, B.; Wang, M.-B.; Krczal, G.; Wassenegger, M. RNAi-mediated resistance to Potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol. Plant Pathol. 2009, 10, 459–469. [Google Scholar] [CrossRef]
- Adkar-Purushothama, C.R.; Kasai, A.; Sugawara, K.; Yamamoto, H.; Yamazaki, Y.; He, Y.-H.; Takada, N.; Goto, H.; Shindo, S.; Harada, T.; et al. RNAi mediated inhibition of viroid infection in transgenic plants expressing viroid-specific small RNAs derived from various functional domains. Sci. Rep. 2015, 5, 17949. [Google Scholar] [CrossRef]
- Carbonell, A.; Daròs, J.-A. Artificial microRNAs and synthetic trans-acting small interfering RNAs interfere with viroid infection. Mol. Plant Pathol. 2017, 18, 746–753. [Google Scholar] [CrossRef]
- Gómez, G.; Pallás, V. Mature monomeric forms of Hop stunt viroid resist RNA silencing in transgenic plants. Plant J. 2007, 51, 1041–1049. [Google Scholar] [CrossRef]
- Di Serio, F.; de Alba, A.-E.M.; Navarro, B.; Gisel, A.; Flores, R. RNA-Dependent RNA Polymerase 6 Delays Accumulation and Precludes Meristem Invasion of a Viroid That Replicates in the Nucleus. J. Virol. 2010, 84, 2477–2489. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, Z.; Zhou, C.; Li, S. RNA-dependent RNA polymerase 1 delays the accumulation of viroids in infected plants. Mol. Plant Pathol. 2021, 22, 1195–1208. [Google Scholar] [CrossRef]
- Serra, P.; Hashemian, S.M.B.; Fagoaga, C.; Romero, J.; Ruiz-Ruiz, S.; Gorris, M.T.; Bertolini, E.; Duran-Vila, N. Virus-Viroid Interactions: Citrus Tristeza Virus Enhances the Accumulation of Citrus Dwarfing Viroid in Mexican Lime via Virus-Encoded Silencing Suppressors. J. Virol. 2014, 88, 1394–1397. [Google Scholar] [CrossRef] [PubMed]
- Campos, L.; Granell, P.; Tárraga, S.; López-Gresa, P.; Conejero, V.; Bellés, J.M.; Rodrigo, I.; Lisón, P. Salicylic acid and gentisic acid induce RNA silencing-related genes and plant resistance to RNA pathogens. Plant Physiol. Biochem. 2014, 77, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Itaya, A.; Zhong, X.; Bundschuh, R.; Qi, Y.; Wang, Y.; Takeda, R.; Harris, A.R.; Molina, C.; Nelson, R.S.; Ding, B. A Structured Viroid RNA Serves as a Substrate for Dicer-Like Cleavage To Produce Biologically Active Small RNAs but Is Resistant to RNA-Induced Silencing Complex-Mediated Degradation. J. Virol. 2007, 81, 2980–2994. [Google Scholar] [CrossRef]
- Dalakouras, A.; Dadami, E.; Bassler, A.; Zwiebel, M.; Krczal, G.; Wassenegger, M. Replicating Potato spindle tuber viroid mediates de novo methylation of an intronic viroid sequence but no cleavage of the corresponding pre-mRNA. RNA Biol. 2015, 12, 268–275. [Google Scholar] [CrossRef]
- Wang, M.-B.; Bian, X.-Y.; Wu, L.-M.; Liu, L.-X.; Smith, N.A.; Isenegger, D.; Wu, R.-M.; Masuta, C.; Vance, V.B.; Watson, J.M.; et al. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA 2004, 101, 3275–3280. [Google Scholar] [CrossRef]
- Bally, J.; Fishilevich, E.; Bowling, A.J.; Pence, H.E.; Narva, K.E.; Waterhouse, P.M. Improved insect-proofing: Expressing double-stranded RNA in chloroplasts. Pest Manag. Sci. 2018, 74, 1751–1758. [Google Scholar] [CrossRef]
- Flores, R.; Hernández, C.; de Alba, A.E.M.; Daròs, J.-A.; Di Serio, F. Viroids and Viroid-Host Interactions. Annu. Rev. Phytopathol. 2005, 43, 117–139. [Google Scholar] [CrossRef]
- Shimura, H.; Pantaleo, V.; Ishihara, T.; Myojo, N.; Inaba, J.-I.; Sueda, K.; Burgyán, J.; Masuta, C. A Viral Satellite RNA Induces Yellow Symptoms on Tobacco by Targeting a Gene Involved in Chlorophyll Biosynthesis using the RNA Silencing Machinery. PLOS Pathog. 2011, 7, e1002021. [Google Scholar] [CrossRef]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Di Serio, F. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012, 70, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Malfitano, M.; Di Serio, F.; Covelli, L.; Ragozzino, A.; Hernández, C.; Flores, R. Peach latent mosaic viroid variants inducing peach calico (extreme chlorosis) contain a characteristic insertion that is responsible for this symptomatology. Virology 2003, 313, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Aviña-Padilla, K.; Rivera-Bustamante, R.; Kovalskaya, N.Y.; Hammond, R.W. Pospiviroid Infection of Tomato Regulates the Expression of Genes Involved in Flower and Fruit Development. Viruses 2018, 10, 516. [Google Scholar] [CrossRef] [PubMed]
- Adkar-Purushothama, C.R.; Iyer, P.S.; Perreault, J.-P. Potato spindle tuber viroid infection triggers degradation of chloride channel protein CLC-b-like and Ribosomal protein S3a-like mRNAs in tomato plants. Sci. Rep. 2017, 7, 8341. [Google Scholar] [CrossRef] [PubMed]
- Adkar-Purushothama, C.R.; Sano, T.; Perreault, J.-P. Viroid-derived small RNA induces early flowering in tomato plants by RNA silencing. Mol. Plant Pathol. 2018, 19, 2446–2458. [Google Scholar] [CrossRef]
- Thibaut, O.; Claude, B. Innate Immunity Activation and RNAi Interplay in Citrus Exocortis Viroid—Tomato Pathosystem. Viruses 2018, 10, 587. [Google Scholar] [CrossRef]
- Bao, S.; Owens, R.A.; Sun, Q.; Song, H.; Liu, Y.; Eamens, A.L.; Feng, H.; Tian, H.; Wang, M.-B.; Zhang, R. Silencing of transcription factor encoding gene StTCP23 by small RNAs derived from the virulence modulating region of potato spindle tuber viroid is associated with symptom development in potato. PLOS Pathog. 2019, 15, e1008110. [Google Scholar] [CrossRef]
- Gómez, G.; Martínez, G.; Pallás, V. Interplay between viroid-induced pathogenesis and RNA silencing pathways. Trends Plant Sci. 2009, 14, 264–269. [Google Scholar] [CrossRef]
- Di Serio, F.; Gisel, A.; Navarro, B.; Delgado, S.; Martínez De Alba, Á.-E.; Donvito, G.; Flores, R. Deep Sequencing of the Small RNAs Derived from Two Symptomatic Variants of a Chloroplastic Viroid: Implications for Their Genesis and for Pathogenesis. PLoS ONE 2009, 4, e7539. [Google Scholar] [CrossRef]
- Jiang, D.-M.; Wang, M.; Li, S.-F.; Zhang, Z.-X. High-Throughput Sequencing Analysis of Small RNAs Derived from Coleus Blumei Viroids. Viruses 2019, 11, 619. [Google Scholar] [CrossRef]
- Torchetti, E.M.; Pegoraro, M.; Navarro, B.; Catoni, M.; Di Serio, F.; Noris, E. A nuclear-replicating viroid antagonizes infectivity and accumulation of a geminivirus by upregulating methylation-related genes and inducing hypermethylation of viral DNA. Sci. Rep. 2016, 6, 35101. [Google Scholar] [CrossRef] [PubMed]
- Wassenegger, M.; Heimes, S.; Riedel, L.; Sänger, H.L. RNA-directed de novo methylation of genomic sequences in plants. Cell 1994, 76, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Vogt, U.; Pélissier, T.; Pütz, A.; Razvi, F.; Fischer, R.; Wassenegger, M. Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J. 2004, 38, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Dalakouras, A.; Dadami, E.; Wassenegger, M.; Krczal, G. RNA-directed DNA methylation efficiency depends on trigger and target sequence identity. Plant J. 2016, 87, 202–214. [Google Scholar] [CrossRef]
- Wassenegger, M.; Dalakouras, A. Viroids as a Tool to Study RNA-Directed DNA Methylation in Plants. Cells 2021, 10, 1187. [Google Scholar] [CrossRef]
- Lv, D.-Q.; Liu, S.-W.; Zhao, J.-H.; Zhou, B.; Wang, S.-P.; Guo, H.-S.; Fang, Y.-Y. Replication of a pathogenic non-coding RNA increases DNA methylation in plants associated with a bromodomain-containing viroid-binding protein. Sci. Rep. 2016, 6, 35751. [Google Scholar] [CrossRef]
- Gómez, G.; Marquez-Molins, J.; Martinez, G.; Pallas, V. Plant epigenome alterations: An emergent player in viroid-host interactions. Virus Res. 2022, 318, 198844. [Google Scholar] [CrossRef]
- Castellano, M.; Pallas, V.; Gomez, G. A pathogenic long noncoding RNA redesigns the epigenetic landscape of the infected cells by subverting host Histone Deacetylase 6 activity. New Phytol. 2016, 211, 1311–1322. [Google Scholar] [CrossRef]
- Castellano, M.; Martinez, G.; Marques, M.C.; Moreno-Romero, J.; Köhler, C.; Pallas, V.; Gomez, G. Changes in the DNA methylation pattern of the host male gametophyte of viroid-infected cucumber plants. J. Exp. Bot. 2016, 67, 5857–5868. [Google Scholar] [CrossRef]
- Castellano, M.; Martinez, G.; Pallás, V.; Gómez, G. Alterations in host DNA methylation in response to constitutive expression of Hop stunt viroid RNA in Nicotiana benthamiana plants. Plant Pathol. 2015, 64, 1247–1257. [Google Scholar] [CrossRef]
- Martinez, G.; Castellano, M.; Tortosa, M.; Pallas, V.; Gomez, G. A pathogenic non-coding RNA induces changes in dynamic DNA methylation of ribosomal RNA genes in host plants. Nucleic Acids Res. 2014, 42, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, Y.; Ding, B.; Fei, Z. Comprehensive Transcriptome Analyses Reveal that Potato Spindle Tuber Viroid Triggers Genome-Wide Changes in Alternative Splicing, Inducible trans-Acting Activity of Phased Secondary Small Interfering RNAs, and Immune Responses. J. Virol. 2017, 91, e00247-17. [Google Scholar] [CrossRef] [PubMed]
- Štajner, N.; Radišek, S.; Mishra, A.K.; Nath, V.S.; Matoušek, J.; Jakše, J. Evaluation of Disease Severity and Global Transcriptome Response Induced by Citrus bark cracking viroid, Hop latent viroid, and Their Co-Infection in Hop (Humulus lupulus L.). Int. J. Mol. Sci. 2019, 20, 3154. [Google Scholar] [CrossRef] [PubMed]
- Herranz, M.C.; Niehl, A.; Rosales, M.; Fiore, N.; Zamorano, A.; Granell, A.; Pallas, V. A remarkable synergistic effect at the transcriptomic level in peach fruits doubly infected by prunus necrotic ringspot virus and peach latent mosaic viroid. Virol. J. 2013, 10, 164. [Google Scholar] [CrossRef] [PubMed]
- Lavagi-Craddock, I.; Dang, T.; Comstock, S.; Osman, F.; Bodaghi, S.; Vidalakis, G. Transcriptome Analysis of Citrus Dwarfing Viroid Induced Dwarfing Phenotype of Sweet Orange on Trifoliate Orange Rootstock. Microorganisms 2022, 10, 1144. [Google Scholar] [CrossRef] [PubMed]
- Lisón, P.; Tárraga, S.; López-Gresa, P.; Saurí, A.; Torres, C.; Campos, L.; Bellés, J.M.; Conejero, V.; Rodrigo, I. A noncoding plant pathogen provokes both transcriptional and posttranscriptional alterations in tomato. Proteomics 2013, 13, 833–844. [Google Scholar] [CrossRef]
- Cottilli, P.; Belda-Palazón, B.; Adkar-Purushothama, C.R.; Perreault, J.-P.; Schleiff, E.; Rodrigo, I.; Ferrando, A.; Lisón, P. Citrus exocortis viroid causes ribosomal stress in tomato plants. Nucleic Acids Res. 2019, 47, 8649–8661. [Google Scholar] [CrossRef]
- Prol, F.V.; Márquez-Molins, J.; Rodrigo, I.; López-Gresa, M.; Bellés, J.; Gómez, G.; Pallás, V.; Lisón, P. Symptom Severity, Infection Progression and Plant Responses in Solanum Plants Caused by Three Pospiviroids Vary with the Inoculation Procedure. Int. J. Mol. Sci. 2021, 22, 6189. [Google Scholar] [CrossRef]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-strandedRNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef]
- Marquez-Molins, J.; Juarez-Gonzalez, V.T.; Gomez, G.; Pallas, V.; Martinez, G. Occurrence of RNA post-transcriptional modifications in plant viruses and viroids and their correlation with structural and functional features. Virus Res. 2023, 323, 198958. [Google Scholar] [CrossRef]
- Márquez-Molins, J.; Villalba-Bermell, P.; Corell-Sierra, J.; Pallás, V.; Gómez, G.; Gustavo Gomez, C.G. Multiomic Analisys Reveals That Viroid Infection Induces a Temporal Reprograming of Plant-Defence Mechanisms at Multiple Regulatory Levels; Cold Spring Harbor Laboratory: Laurel Hollow, NY, USA, 2022. [Google Scholar]
- Li, S.; Wu, Z.-G.; Zhou, Y.; Dong, Z.-F.; Fei, X.; Zhou, C.-Y.; Li, S.-F. Changes in metabolism modulate induced by viroid infection in the orchid Dendrobium officinale. Virus Res. 2022, 308, 198626. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Navarro, B.; Delgado, S.; Serra, P.; Di Serio, F. Viroid pathogenesis: A critical appraisal of the role of RNA silencing in triggering the initial molecular lesion. FEMS Microbiol. Rev. 2020, 44, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Xu, C.; Kotta-Loizou, I.; Jiang, J.; Lv, R.; Kong, L.; Li, S.; Hong, N.; Wang, G.; Coutts, R.H.A.; et al. Novel Viroid-Like RNAs Naturally Infect a Filamentous Fungus. Adv. Sci. 2022, e2204308. [Google Scholar] [CrossRef] [PubMed]
- Minoia, S.; Navarro, B.; Covelli, L.; Barone, M.; García-Becedas, M.T.; Ragozzino, A.; Alioto, D.; Flores, R.; Di Serio, F. Viroid-like RNAs from cherry trees affected by leaf scorch disease: Further data supporting their association with mycoviral double-stranded RNAs. Arch. Virol. 2014, 159, 589–593. [Google Scholar] [CrossRef]
- Wei, S.; Bian, R.; Andika, I.B.; Niu, E.; Liu, Q.; Kondo, H.; Yang, L.; Zhou, H.; Pang, T.; Lian, Z.; et al. Symptomatic plant viroid infections in phytopathogenic fungi. Proc. Natl. Acad. Sci. USA 2019, 116, 13042–13050. [Google Scholar] [CrossRef]
- Navarro, B.; Li, S.; Gisel, A.; Chiumenti, M.; Minutolo, M.; Alioto, D.; Di Serio, F. A Novel Self-Cleaving Viroid-Like RNA Identified in RNA Preparations from a Citrus Tree Is Not Directly Associated with the Plant. Viruses 2022, 14, 2265. [Google Scholar] [CrossRef]
- Tian, M.; Wei, S.; Bian, R.; Luo, J.; Khan, H.A.; Tai, H.; Kondo, H.; Hadidi, A.; Andika, I.B.; Sun, L. Natural Cross-Kingdom Spread of Apple Scar Skin Viroid from Apple Trees to Fungi. Cells 2022, 11, 3686. [Google Scholar] [CrossRef]
- Serra, P.; Carbonell, A.; Navarro, B.; Gago-Zachert, S.; Li, S.; Di Serio, F.; Flores, R. Symptomatic plant viroid infections in phytopathogenic fungi: A request for a critical reassessment. Proc. Natl. Acad. Sci. USA 2020, 117, 10126–10128. [Google Scholar] [CrossRef]
- Daròs, J.-A. Eggplant latent viroid: A friendly experimental system in the familyAvsunviroidae. Mol. Plant Pathol. 2016, 17, 1170–1177. [Google Scholar] [CrossRef]
- Matsushita, Y. Chrysanthemum Stunt Viroid. Jpn. Agric. Res. Q. 2013, 47, 237–242. [Google Scholar] [CrossRef]
- Vernière, C.; Perrier, X.; Dubois, C.; Dubois, A.; Botella, L.; Chabrier, C.; Bové, J.M.; Vila, N.D. Interactions Between Citrus Viroids Affect Symptom Expression and Field Performance of Clementine Trees Grafted on Trifoliate Orange. Phytopathology 2006, 96, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Kanematsu, S.; Koganezawa, H.; Tsuchizaki, T.; Yoshida, K. Detection of a Viroid Associated with Apple Fruit Crinkle Disease. Ann. Phytopath. Soc. Jpn. 1993, 59, 520–527. [Google Scholar] [CrossRef]
- Kim, H.-R.; Lee, S.-H.; Lee, D.-H.; Kim, J.-S.; Park, J.-W. Transmission of Apple scar skin viroid by Grafting, Using Contaminated Pruning Equipment, and Planting Infected Seeds. Plant Pathol. J. 2006, 22, 63–67. [Google Scholar] [CrossRef]
- Hagemann, M.H.; Born, U.; Sprich, E.; Seigner, L.; Oechsner, H.; Hülsemann, B.; Steinbrenner, J.; Winterhagen, P.; Lehmair, E. Degradation of hop latent viroid during anaerobic digestion of infected hop harvest residues. Eur. J. Plant Pathol. 2021, 161, 579–591. [Google Scholar] [CrossRef]
- Jakse, J.; Radisek, S.; Pokorn, T.; Matousek, J.; Javornik, B. Deep-sequencing revealed Citrus bark cracking viroid (CBCVd) as a highly aggressive pathogen on hop. Plant Pathol. 2015, 64, 831–842. [Google Scholar] [CrossRef]
- Kerins, G.; Blackburn, J.; Nixon, T.; Daly, M.; Conyers, C.; Pietravalle, S.; Noble, R.; Henry, C.M. Composting to sanitize plant-based waste infected with organisms of plant health importance. Plant Pathol. 2018, 67, 411–417. [Google Scholar] [CrossRef]
- Kryczyński, S.; Paduch-Cichal, E.; Skrzeczkowski, L.J. Transmission of Three Viroids Through Seed and Pollen of Tomato Plants. J. Phytopathol. 1988, 121, 51–57. [Google Scholar] [CrossRef]
- Antignus, Y.; Lachman, O.; Pearlsman, M. Spread of Tomato apical stunt viroid (TASVd) in Greenhouse Tomato Crops Is Associated with Seed Transmission and Bumble Bee Activity. Plant Dis. 2007, 91, 47–50. [Google Scholar] [CrossRef]
- Desjardins, P.R.; Drake, R.J.; Atkins, L.; Bergh, B.O. Pollen Transmission of Avocado Sunblotch Virus Experimentally Demonstrated. Calif Agric 1979, 33, 14–15. [Google Scholar]
- Schnell, R.J.; Tondo, C.L.; Kuhn, D.N.; Winterstein, M.C.; Ayala-Silva, T.; Moore, J.M. Spatial Analysis of Avocado Sunblotch Disease in an Avocado Germplasm Collection. J. Phytopathol. 2011, 159, 773–781. [Google Scholar] [CrossRef]
- Fukuta, S.; Kuwayama, S.; Hirano, T.; Hattori, H.; Nakamura, Y.; Ohishi, K. Contact transmission of Chrysanthemum stunt viroid through root. Annu. Rep. Kansai Plant Prot. Soc. 2012, 54, 47–51. [Google Scholar] [CrossRef]
- Mehle, N.; Gutiérrez-Aguirre, I.; Prezelj, N.; Delić, D.; Vidic, U.; Ravnikar, M. Survival and Transmission of Potato Virus Y, Pepino Mosaic Virus, and Potato Spindle Tuber Viroid in Water. Appl. Environ. Microbiol. 2014, 80, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Vachev, T.; Ivanova, D.; Minkov, I.; Tsagris, M.; Gozmanova, M. Trafficking of the Potato spindle tuber viroid between tomato and Orobanche ramosa. Virology 2010, 399, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Leichtfried, T.; Reisenzein, H.; Steinkellner, S.; Gottsberger, R.A. Transmission studies of the newly described apple chlorotic fruit spot viroid using a combined RT-qPCR and droplet digital PCR approach. Arch. Virol. 2020, 165, 2665–2671. [Google Scholar] [CrossRef]
- Schumann, G.L.; Tingey, W.M.; Thurston, H.D. Evaluation of six insect pests for transmission of potato spindle tuber viroid. Am. J. Potato Res. 1980, 57, 205–211. [Google Scholar] [CrossRef]
- Desvignes, J. Peach latent mosaic and its relation to peach mosaic and peach yellow mosaic virus diseases. Acta Hortic. 1986, 193, 51–58. [Google Scholar] [CrossRef]
- Syller, J.; Marczewski, W.; Pawłowicz, J. Transmission by aphids of potato spindle tuber viroid encapsidated by potato leafroll luteovirus particles. Eur. J. Plant Pathol. 1997, 103, 285–289. [Google Scholar] [CrossRef]
- Cohen, O.; Batuman, O.; Moskowits, Y.; Rozov, A.; Gootwine, E.; Mawassi, M.; Bar-Joseph, M. Goat Horns: Platforms for Viroid Transmission to Fruit Trees? Phytoparasitica 2005, 33, 141–148. [Google Scholar] [CrossRef]
- Matsushita, Y.; Aoki, K.; Sumitomo, K. Selection and inheritance of resistance to Chrysanthemum stunt viroid. Crop Prot. 2012, 35, 1–4. [Google Scholar] [CrossRef]
- Naoi, T.; Hataya, T. Tolerance Even to Lethal Strain of Potato Spindle Tuber Viroid Found in Wild Tomato Species Can Be Introduced by Crossing. Plants 2021, 10, 575. [Google Scholar] [CrossRef]
- Niblett, C.; Dickson, E.; Fernow, K.; Horst, R.; Zaitlin, M. Cross protection among four viroids. Virology 1978, 91, 198–203. [Google Scholar] [CrossRef] [PubMed]
- De la Peña, M.; Flores, R. Chrysanthemum Chlorotic Mottle Viroid RNA: Dissection of the Pathogenicity Determinant and Comparative Fitness of Symptomatic and Non-symptomatic Variants. J. Mol. Biol. 2002, 321, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A. RNAi tools for controlling viroid diseases. Virus Res. 2022, 313, 198729. [Google Scholar] [CrossRef]
- Ma, J.; Mudiyanselage, S.D.D.; Wang, Y. Emerging value of the viroid model in molecular biology and beyond. Virus Res. 2022, 313, 198730. [Google Scholar] [CrossRef] [PubMed]
- Cordero, T.; Aragonés, V.; Daròs, J.-A. Large-scale Production of Recombinant RNAs on a Circular Scaffold Using a Viroid-derived System in Escherichia coli. J. Vis. Exp. 2018, 141, e58472. [Google Scholar] [CrossRef] [PubMed]
- Ortolá, B.; Cordero, T.; Hu, X.; Daròs, J.-A. Intron-assisted, viroid-based production of insecticidal circular double-stranded RNA in Escherichia coli. RNA Biol. 2021, 18, 1846–1857. [Google Scholar] [CrossRef]
- Ortolá, B.; Daròs, J.-A. Production of Recombinant RNA in Escherichia coli Using Eggplant Latent Viroid as a Scaffold. In Viroids: Methods and Protocols; Rao Ayala, L.N., Lavagi-Craddock, I., Georgios, V., Eds.; Springer: New York, NY, USA, 2021; ISBN 978-1-0716-1463-1. [Google Scholar]
- Marquez-Molins, J.; Hernandez-Azurdia, A.G.; Urrutia-Perez, M.; Pallas, V.; Gomez, G. A circular RNA vector for targeted plant gene silencing based on an asymptomatic viroid. Plant J. 2022, 112, 284–293. [Google Scholar] [CrossRef]
| Family Pospiviroidae | ||
|---|---|---|
| Genus Pospiviroid | PSTVd | Potato spindle tuber viroid |
| CEVd | Citrus exocortis viroid | |
| CSVd | Chrysanthemum stunt viroid | |
| CLVd | Columnea latent viroid | |
| IrVd-1 | Iresine viroid 1 | |
| PCFVd | Pepper chat fruit viroid | |
| TASVd | Tomato apical stunt viroid | |
| TCDVd | Tomato chlorotic dwarf viroid | |
| TPMVd | Tomato planta macho viroid | |
| Genus Hostuviroid | HSVd 1 | Hop stunt viroid |
| DLVd | Dahlia latent virus | |
| Genus Apscaviroid | ASSVd | Apple scar skin viroid |
| ADFVd | Apple dimple fruit viroid | |
| AGVd | Australian grapevine viroid | |
| CBLVd 1 | Citrus bent leaf viroid | |
| CDVd 1 | Citrus dwarfing viroid | |
| CVd-V 1 | Citrus viroid V | |
| CVd-VI 1 | Citrus viroid VI | |
| GYSVd-1 | Grapevine yellow speckle viroid 1 | |
| GYSVd-2 | Grapevine yellow speckle viroid 2 | |
| PBCVd | Pear blister canker viroid | |
| Genus Cocadviroid | CCCVd | Coconut cadang-cadang viroid |
| CTiVd | Coconut tinangaja viroid | |
| CBCVd 1 | Citrus bark cracking viroid | |
| HLVd | Hop latent viroid | |
| Genus Coleviroid | CbVd-1 | Coleus blumei viroid 1 |
| CbVd-2 | Coleus blumei viroid 2 | |
| CbVd-3 | Coleus blumei viroid 3 | |
| Family Avsunviroidae | ||
| Genus Avsunviroid | ASBVd | Avocado sunblotch viroid |
| Genus Pelamoviroid | PLMVd | Peach latent mosaic viroid |
| CChMVd | Chrysanthemum chlorotic mottle viroid | |
| AHVd | Apple hammerhead viroid | |
| Genus Elaviroid | ELVd | Eggplant latent viroid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortolá, B.; Daròs, J.-A. Viroids: Non-Coding Circular RNAs Able to Autonomously Replicate and Infect Higher Plants. Biology 2023, 12, 172. https://doi.org/10.3390/biology12020172
Ortolá B, Daròs J-A. Viroids: Non-Coding Circular RNAs Able to Autonomously Replicate and Infect Higher Plants. Biology. 2023; 12(2):172. https://doi.org/10.3390/biology12020172
Chicago/Turabian StyleOrtolá, Beltrán, and José-Antonio Daròs. 2023. "Viroids: Non-Coding Circular RNAs Able to Autonomously Replicate and Infect Higher Plants" Biology 12, no. 2: 172. https://doi.org/10.3390/biology12020172
APA StyleOrtolá, B., & Daròs, J.-A. (2023). Viroids: Non-Coding Circular RNAs Able to Autonomously Replicate and Infect Higher Plants. Biology, 12(2), 172. https://doi.org/10.3390/biology12020172







