Germ Line/Multipotency Genes Show Differential Expression during Embryonic Development of the Annelid Enchytraeus coronatus
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Material and Fixation
2.2. Immunohistochemistry
2.3. Sequence Retrieval, Gene Cloning, and Phylogenetic Analysis
2.4. Whole-Mount in Situ Hybridization
2.5. Data Visualization
3. Results
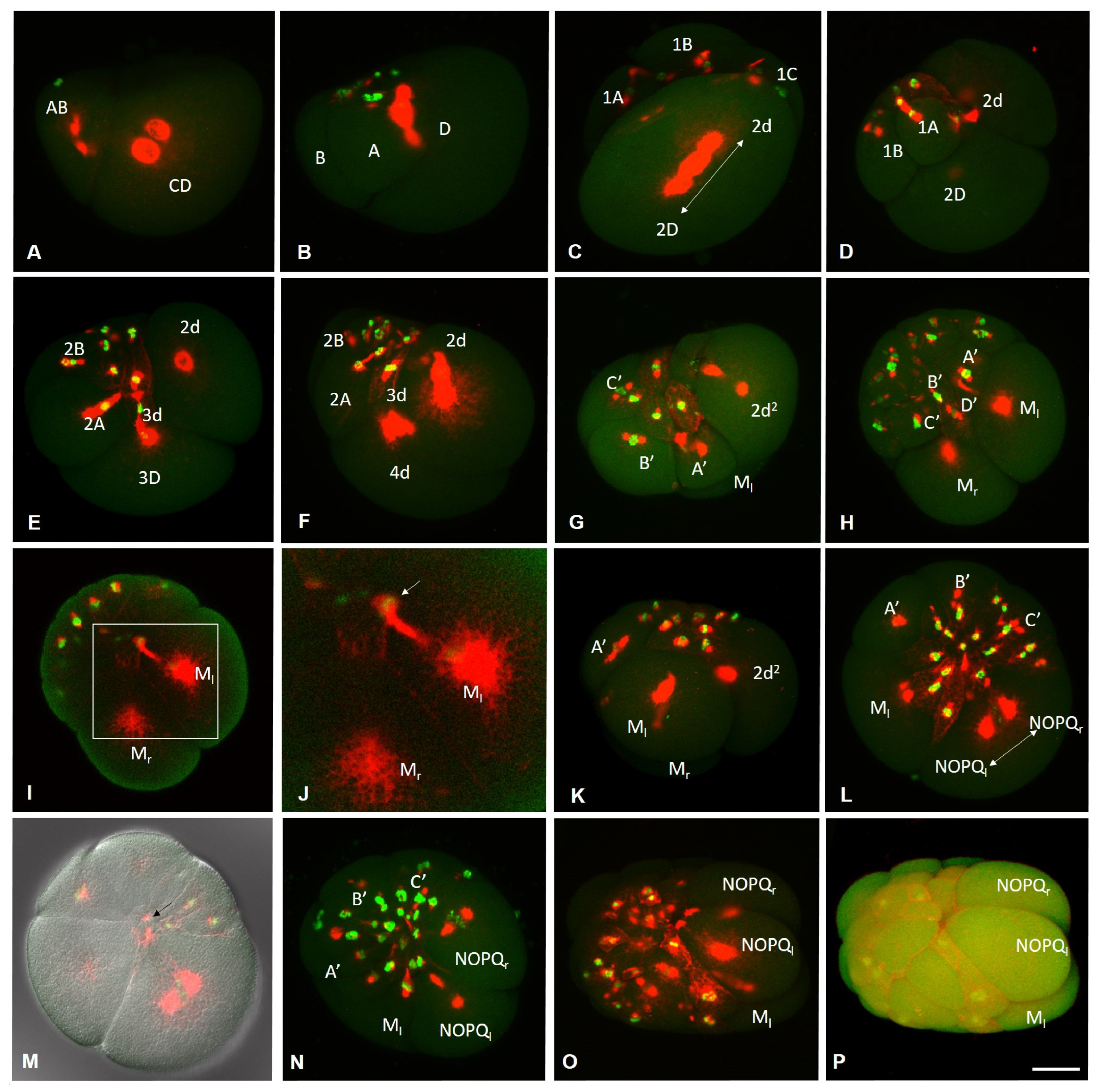
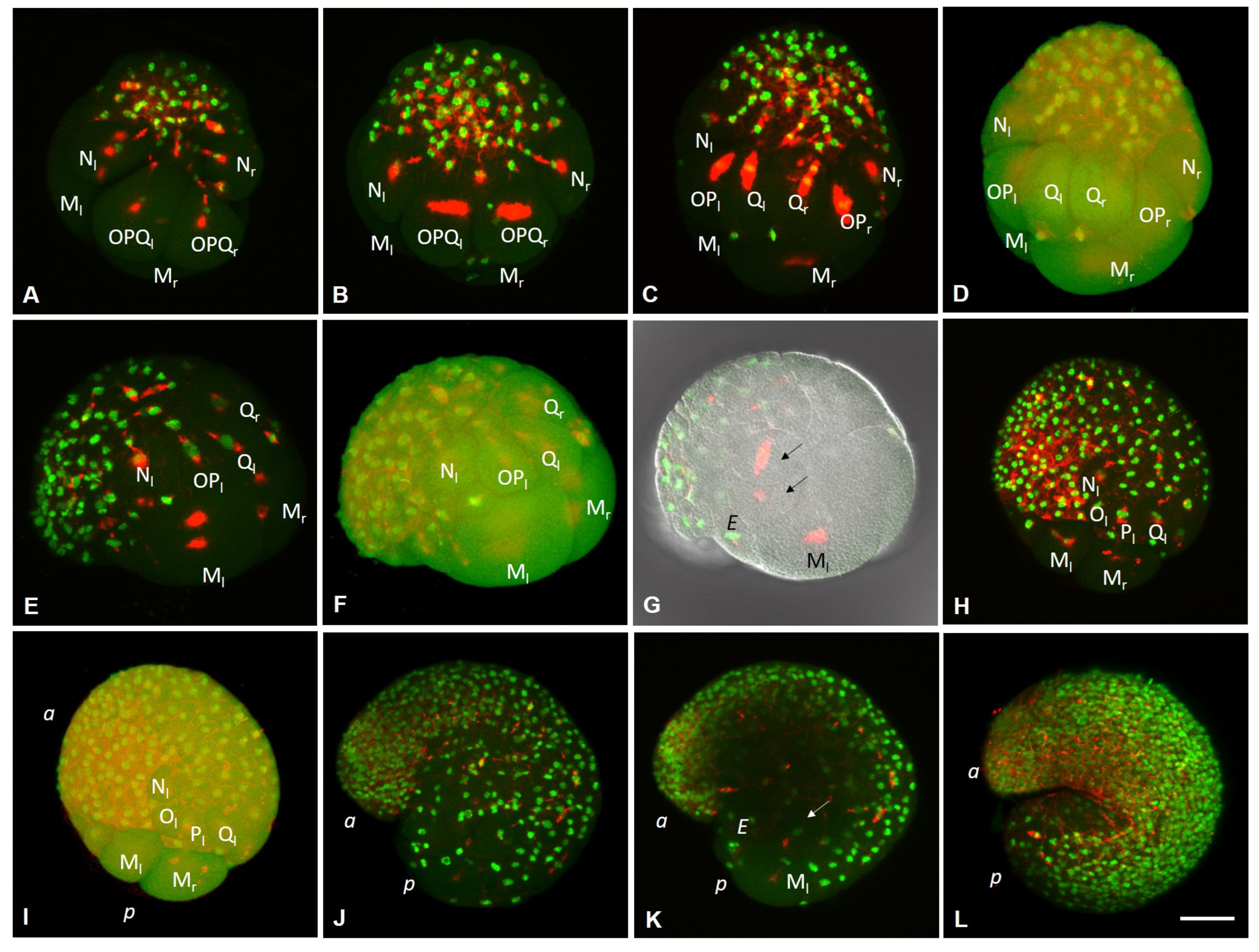
3.1. Life Cycle of the Annelid Enchytraeus Coronatus
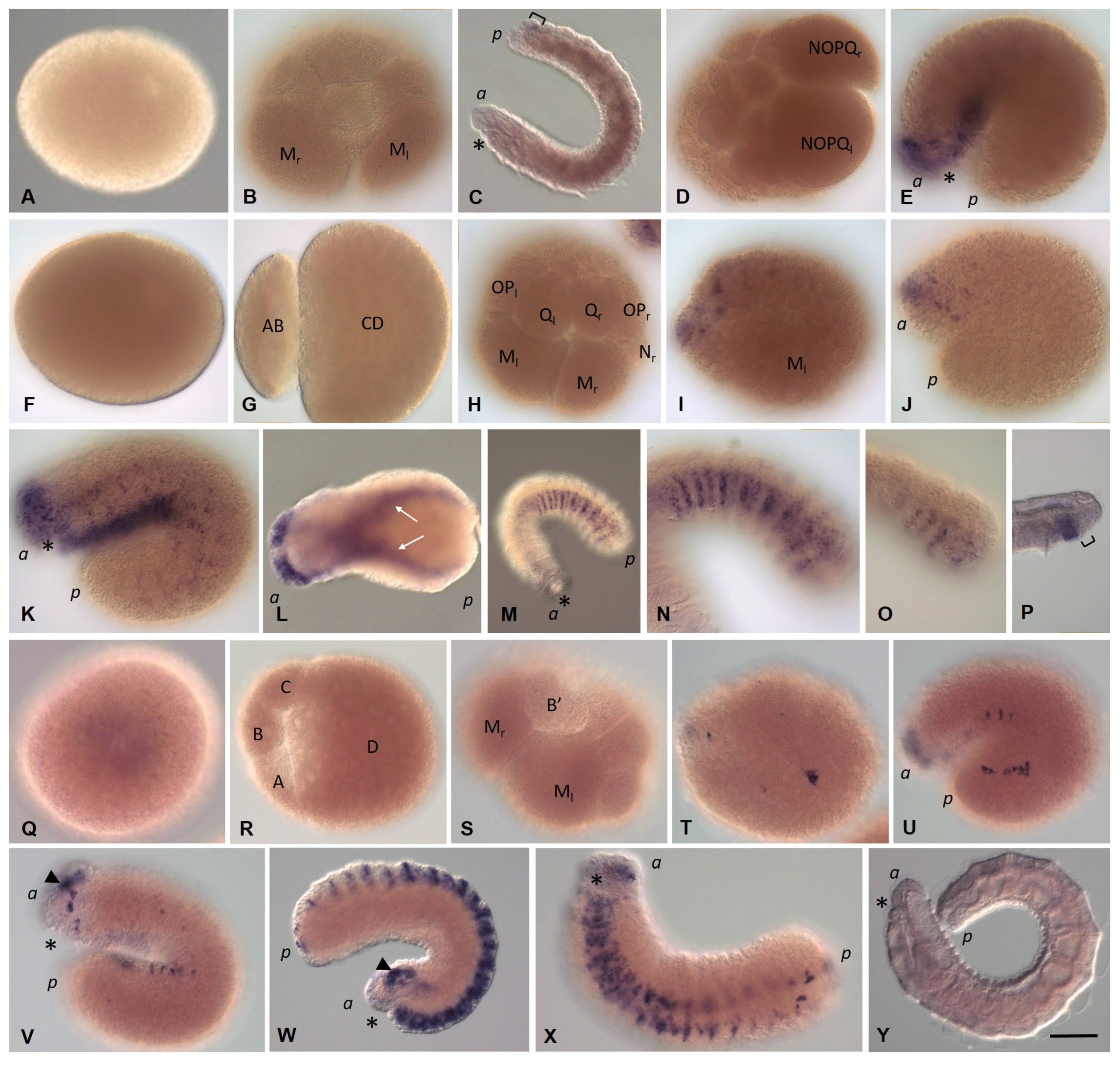
3.2. Development of the Annelid E. coronatus
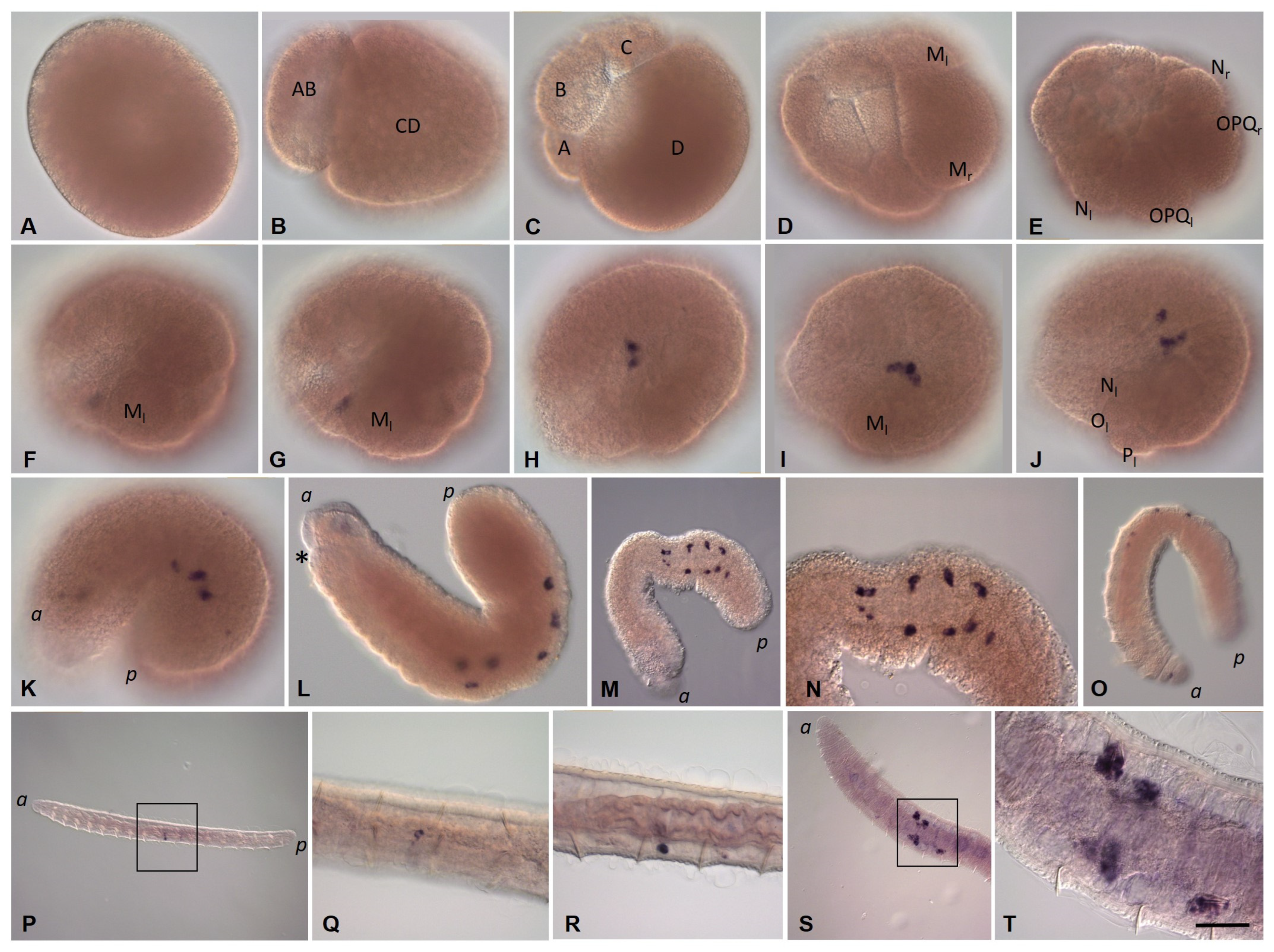
3.3. Expression of Germ Line/Multipotency Genes during Embryonic Development and at Juvenile Stage of E. coronatus
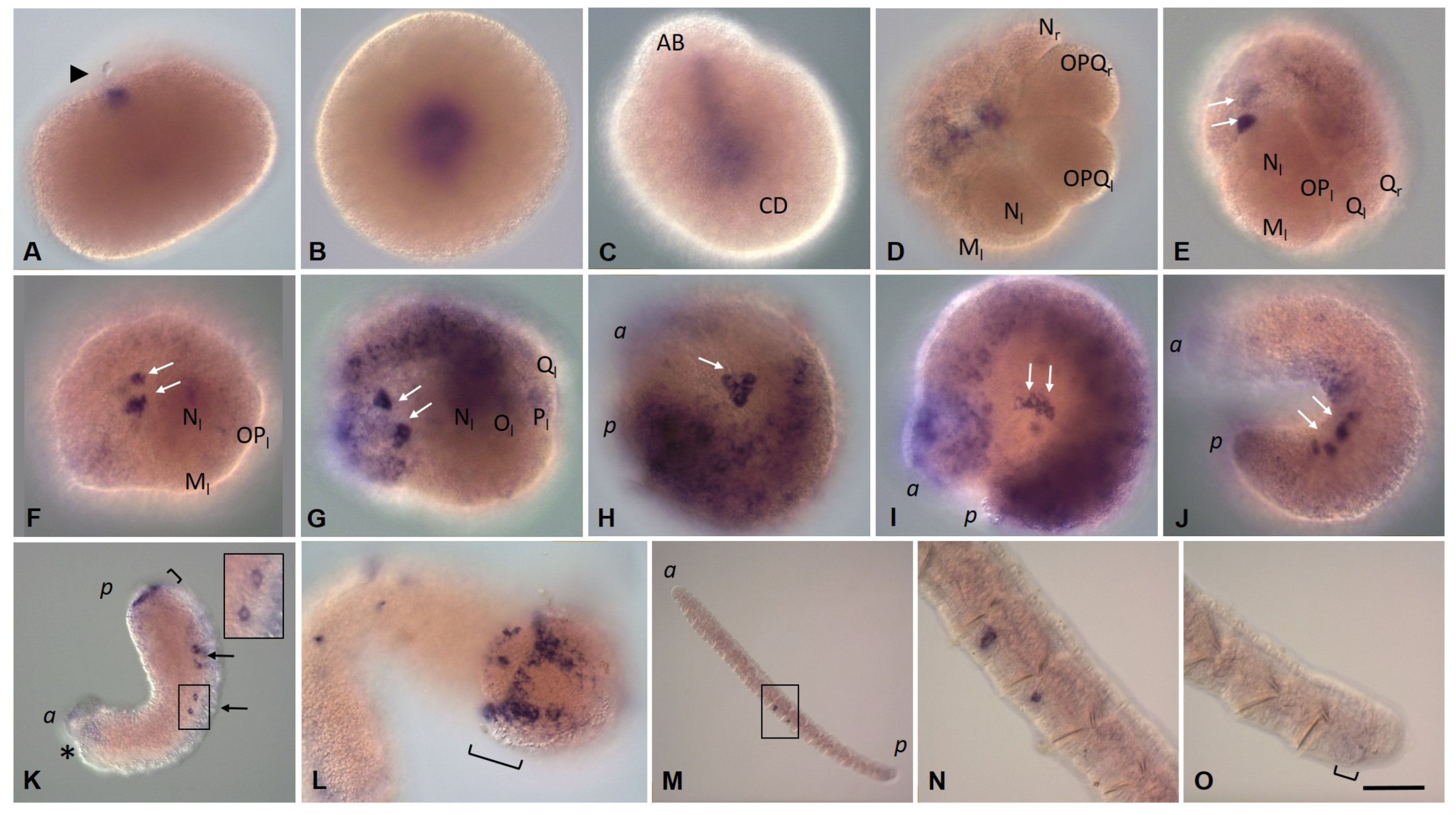
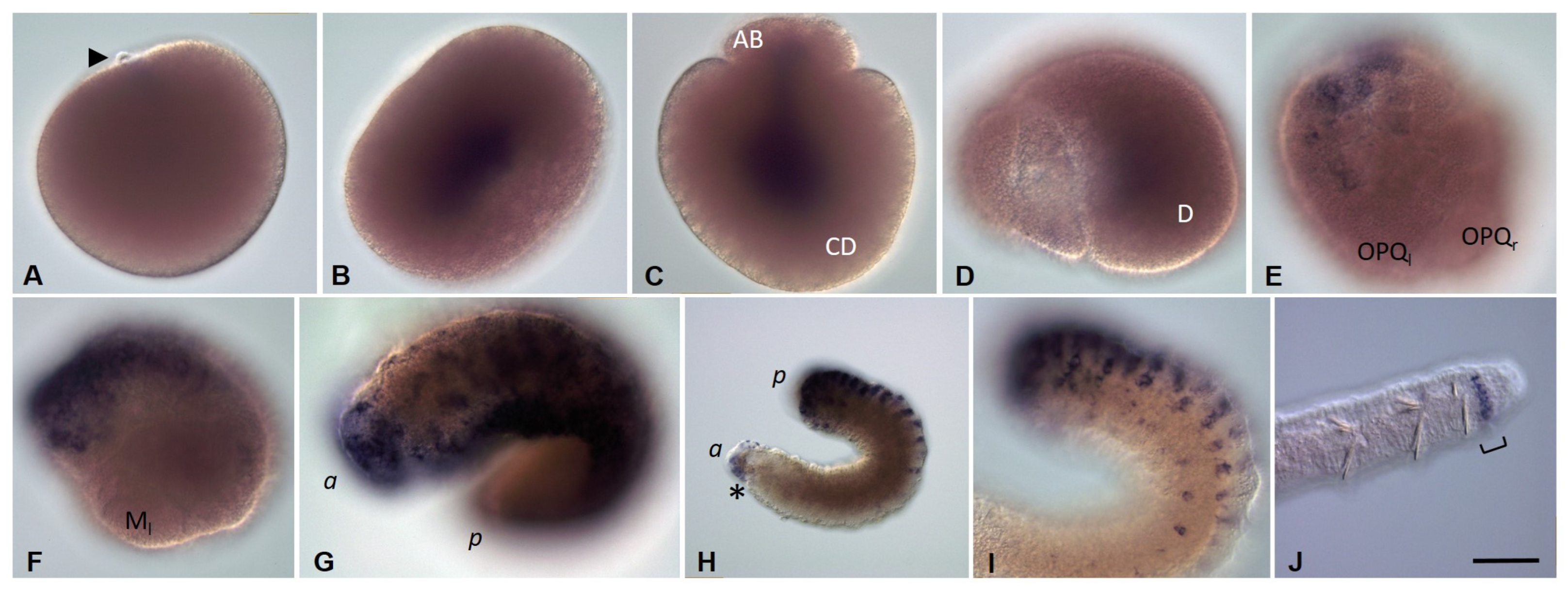
3.3.1. Eco-vasa1 and Eco-vasa2 Embryonic and Juvenile Expression Patterns
3.3.2. Eco-pl10-1 and Eco-pl10-2 Embryonic and Juvenile Expression Patterns
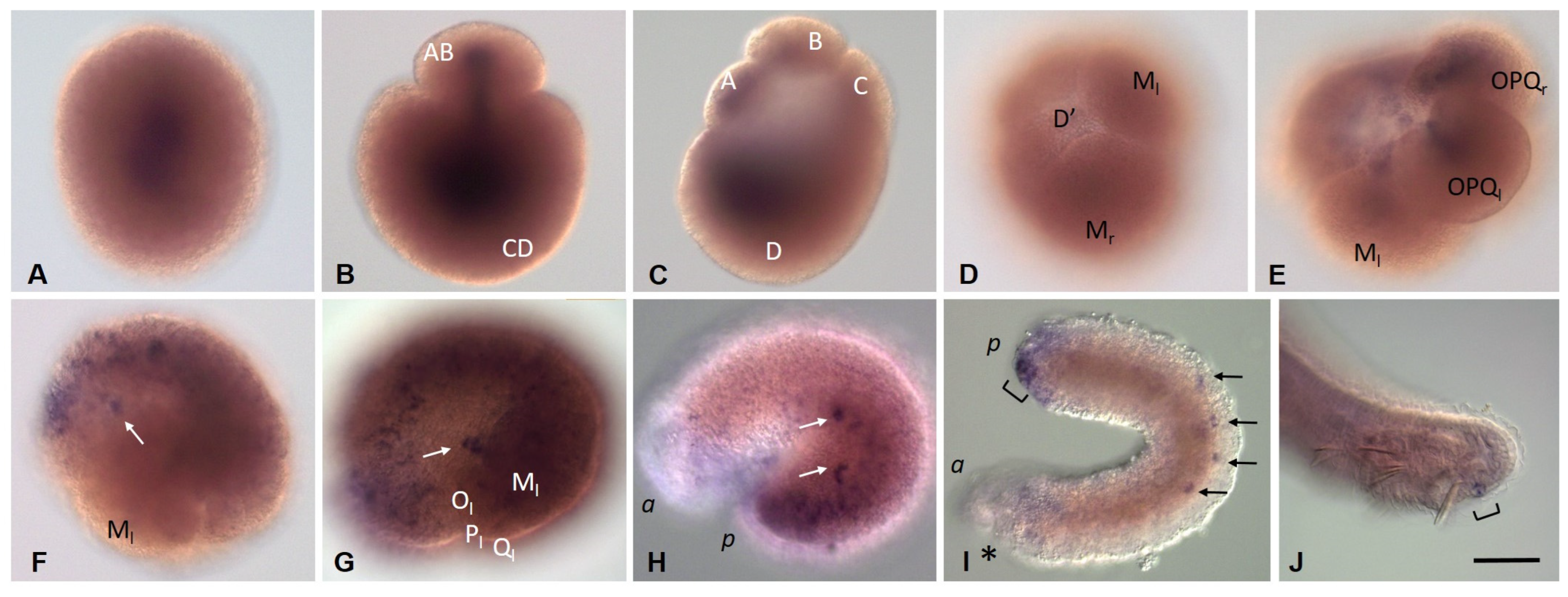
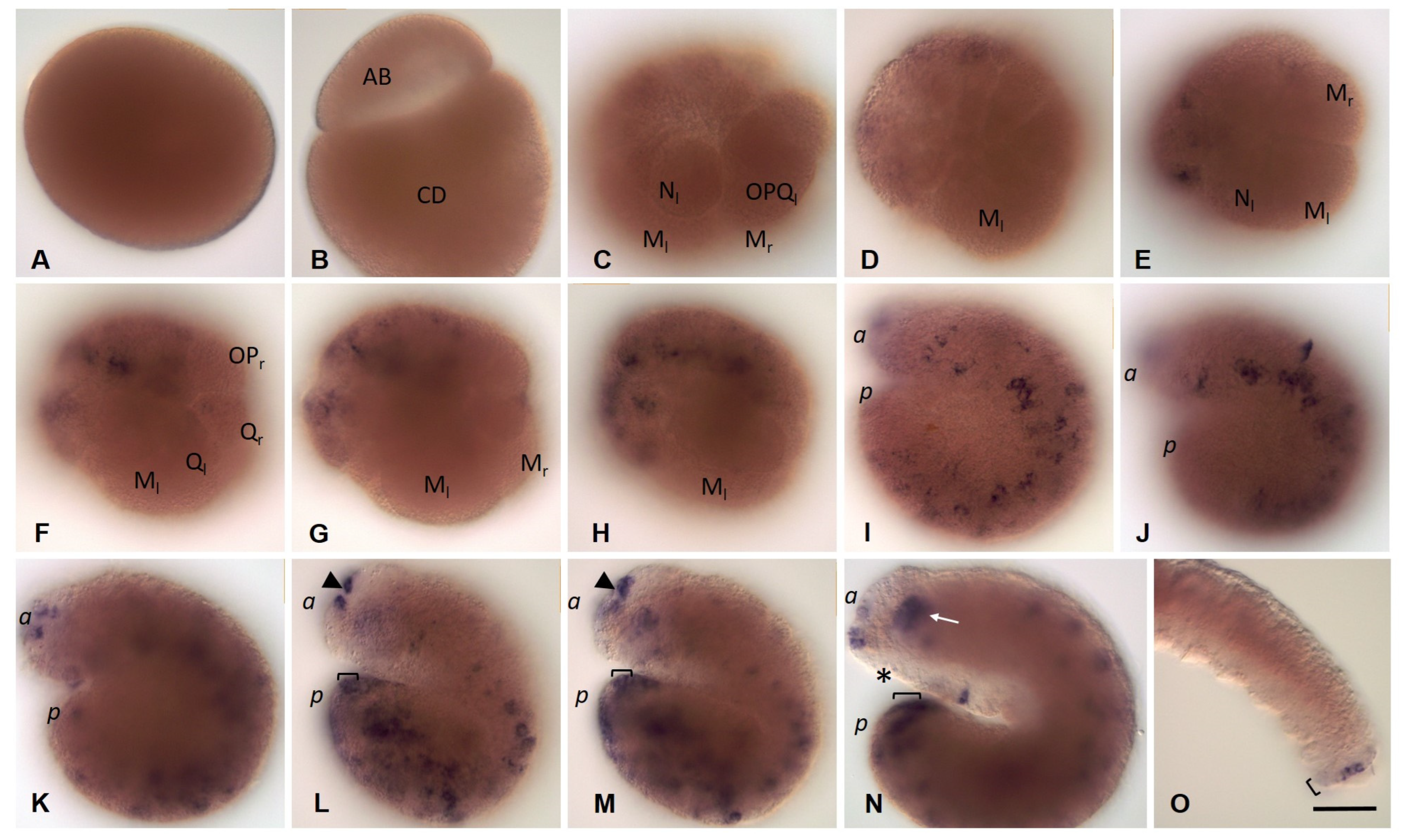
3.3.3. Piwi Homolog Expression during Development of the Annelid E. coronatus
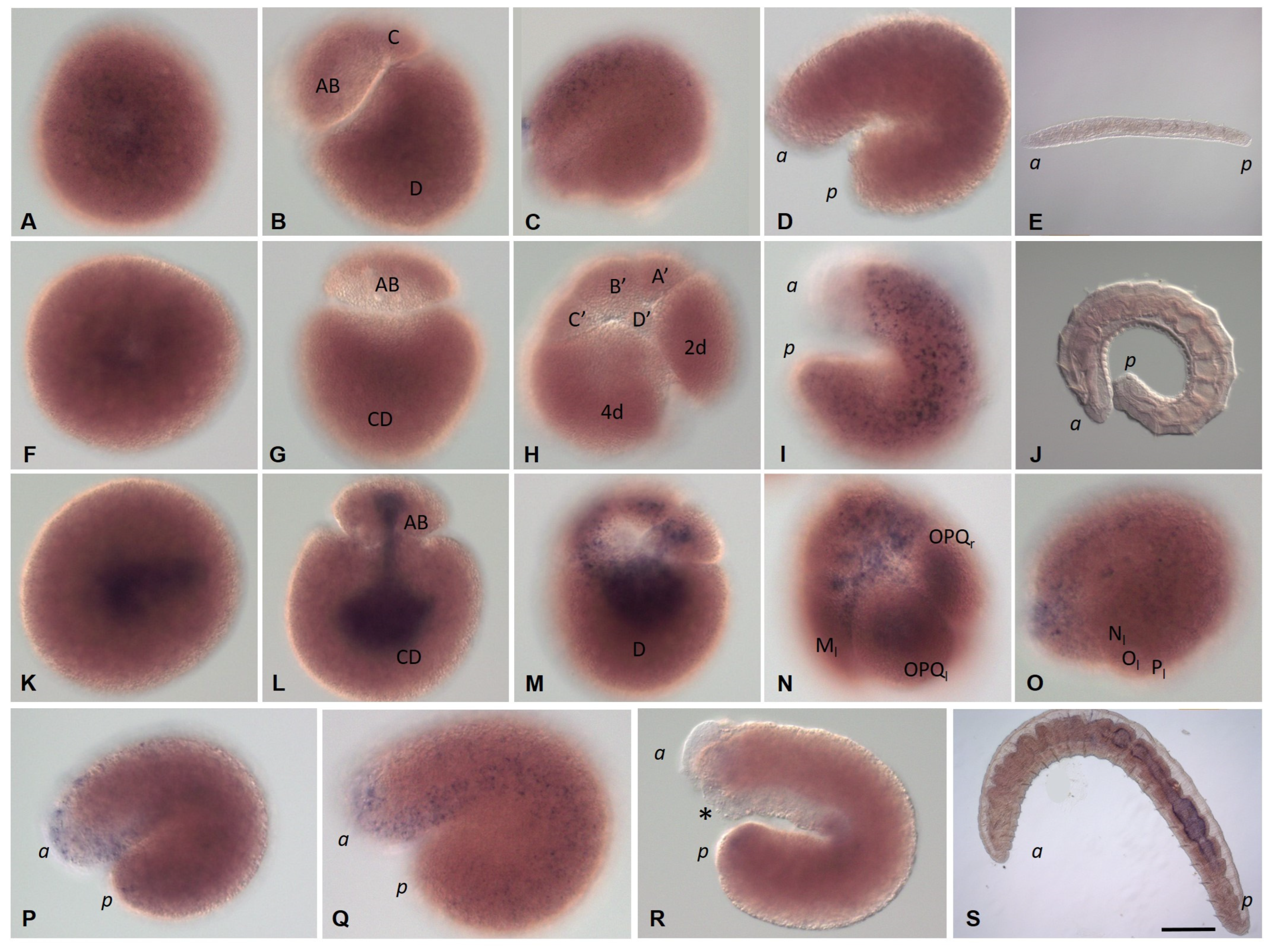
3.3.4. Nanos Homolog Expression during Development of the Annelid E. coronatus
3.3.5. Eco-myc Embryonic and Juvenile Expression Patterns
3.3.6. Pumilio Homolog Expression during Development of the Annelid E. coronatus
3.3.7. Tudor, Boule, and Bruno homolog expression during development of the annelid E. coronatus
4. Discussion
4.1. Early Embryogenesis and Possible Origin of PGCs in E. coronatus
4.2. Germ Line/Multipotency Genes in E. coronatus
4.3. Expression of the Germ Line/Multipotency Genes in E. coronatus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Extavour, C.; Akam, M. Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development 2003, 130, 5869–5884. [Google Scholar] [CrossRef]
- Juliano, C.E.; Swartz, S.Z.; Wessel, G.M. A conserved germline multipotency program. Development 2010, 137, 4113–4126. [Google Scholar] [CrossRef]
- Bosch, T.C.G.; David, C.N. Stem cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. Dev. Biol. 1987, 121, 182–191. [Google Scholar] [CrossRef]
- Varley, A.; Horkan, H.R.; McMahon, E.T.; Krasovec, G.; Frank, U. Pluripotent, germ cell competent adult stem cells underlie cnidarian regenerative ability and clonal growth. Curr. Biol. 2023, 33, 1883–1892.e3. [Google Scholar] [CrossRef]
- Siebert, S.; Farrell, J.A.; Cazet, J.F.; Abeykoon, Y.; Primack, A.S.; Schnitzler, C.E.; Juliano, C.E. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science. 2019, 365, eaav9314. [Google Scholar] [CrossRef] [PubMed]
- Giani, V.C.; Yamaguchi, E.; Boyle, M.J.; Seaver, E.C. Somatic and germline expression of piwi during development and regeneration in the marine polychaete annelid Capitella teleta. Evodevo 2011, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, R.; Sugio, M.; Kutsuna, J.; Tochinai, S.; Takahashi, Y. Early Segregation of Germ and Somatic Lineages during Gonadal Regeneration in the Annelid Enchytraeus Japonensis. Curr. Biol. 2006, 16, 1012–1017. [Google Scholar] [CrossRef]
- Özpolat, B.D.; Bely, A.E. Gonad Establishment during Asexual Reproduction in the Annelid Pristina leidyi. Dev. Biol. 2015, 405, 123–136. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P.; Smirnova, N.P. Vasa, Piwi, and Pl10 Expression during Sexual Maturation and Asexual Reproduction in the Annelid Pristina longiseta. J. Dev. Biol. 2023, 11, 34. [Google Scholar] [CrossRef]
- Sugio, M.; Yoshida-noro, C.; Ozawa, K.; Tochinai, S. Stem cells in asexual reproduction of Enchytraeus japonensis (Oligochaeta, Annelid): Proliferation and migration of neoblasts. Dev. Growth Differ. 2012, 54, 439–450. [Google Scholar] [CrossRef]
- Wagner, D.E.; Ho, J.J.; Reddien, P.W. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell 2012, 10, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Yoshida-noro, C.; Tochinai, S. Stem cell system in asexual and sexual reproduction of Enchytraeus japonensis (Oligochaeta, Annelida). Dev. Growth Differ. 2010, 52, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Nikanorova, D.D.; Kupriashova, E.E.; Kostyuchenko, R.P. Regeneration in Annelids: Cell Sources, Tissue Remodeling, and Differential Gene Expression. Russ. J. Dev. Biol. 2020, 51, 148–161. [Google Scholar] [CrossRef]
- Rosner, A.; Moiseeva, E.; Rinkevich, Y.; Lapidot, Z.; Rinkevich, B. Vasa and the germ line lineage in a colonial urochordate. Dev. Biol. 2009, 331, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Rinkevich, B.; Ballarin, L.; Martinez, P.; Somorjai, I.; Ben-Hamo, O.; Borisenko, I.; Berezikov, E.; Ereskovsky, A.; Gazave, E.; Khnykin, D.; et al. A pan-metazoan concept for adult stem cells: The wobbling Penrose landscape. Biol. Rev. 2022, 97, 299–325. [Google Scholar] [CrossRef]
- Fierro-Constaín, L.; Schenkelaars, Q.; Gazave, E.; Haguenauer, A.; Rocher, C.; Ereskovsky, A.; Borchiellini, C.; Renard, E. The conservation of the germline multipotency program, from sponges to vertebrates: A stepping stone to understanding the somatic and germline origins. Genome Biol. Evol 2017, 9, 474–488. [Google Scholar] [CrossRef] [PubMed]
- Dannenberg, L.C.; Seaver, E.C. Regeneration of the Germline in the Annelid Capitella Teleta. Dev. Biol. 2018, 440, 74–87. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P.; Kozin, V.V. Comparative Aspects of Annelid Regeneration: Towards Understanding the Mechanisms of Regeneration. Genes 2021, 12, 1148. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P. Nanos Is Expressed in Somatic and Germline Tissue during Larval and Post-Larval Development of the Annelid Alitta virens. Genes 2022, 13, 270. [Google Scholar] [CrossRef]
- Kozin, V.V.; Kostyuchenko, R.P. Vasa, PL10, and Piwi gene expression during caudal regeneration of the polychaete annelid Alitta virens. Dev. Genes Evol. 2015, 225, 129–138. [Google Scholar] [CrossRef]
- Xu, C.-M.; Sun, S.-C. Expression of Piwi Genes during the Regeneration of Lineus sanguineus (Nemertea, Pilidiophora, Heteronemertea). Genes 2020, 11, 1484. [Google Scholar] [CrossRef]
- Palakodeti, D.; Smielewska, M.; Lu, Y.-C.; Yeo, G.W.; Graveley, B.R. The PIWI Proteins SMEDWI-2 and SMEDWI-3 are Required for Stem Cell Function and piRNA Expression in Planarians. RNA 2008, 14, 1174–1186. [Google Scholar] [CrossRef] [PubMed]
- Rosner, A.; Paz, G.; Rinkevich, B. Divergent roles of the DEAD-box protein BS-PL10, the urochordate homologue of human DDX3 and DDX3Y proteins, in colony astogeny and ontogeny. Dev. Dyn. 2006, 235, 1508–1521. [Google Scholar] [CrossRef] [PubMed]
- Özpolat, B.D.; Sloane, E.S.; Zattara, E.E.; Bely, A.E. Plasticity and regeneration of gonads in the annelid Pristina leidyi. EvoDevo 2016, 7, 22. [Google Scholar] [CrossRef]
- Rebscher, N.; Zelada-González, F.; Banisch, T.U.; Raible, F.; Arendt, D. Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev. Biol. 2007, 306, 599–611. [Google Scholar] [CrossRef]
- Planques, A.; Malem, J.; Parapar, J.; Vervoort, M.; Gazave, E. Morphological, Cellular and Molecular Characterization of Posterior Regeneration in the Marine Annelid Platynereis dumerilii. Dev. Biol. 2019, 445, 189–210. [Google Scholar] [CrossRef]
- Álvarez-Campos, P.; García-Castro, H.; Emili, E.; Pérez-Posada, A.; Salamanca-Díaz, D.A.; Mason, V.; Metzger, B.; Bely, A.E.; Kenny, N.; Özpolat, B.D.; et al. Annelid adult cell type diversity and their pluripotent cellular origins. bioRxiv 2023. bioRxiv:2023.04.25.537979. [Google Scholar] [CrossRef]
- Forbes, A.; Lehmann, R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 1998, 125, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, K.; Seydoux, G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 1999, 126, 4861–4871. [Google Scholar] [CrossRef]
- Tsuda, M.; Sasaoka, Y.; Kiso, M.; Abe, K.; Haraguchi, S.; Kobayashi, S.; Saga, Y. Conserved role of nanos proteins in germ cell development. Science 2003, 301, 1239–1241. [Google Scholar] [CrossRef]
- Wang, Y.; Zayas, R.M.; Guo, T.; Newmark, P.A. nanos function is essential for development and regeneration of planarian germ cells. Proc. Natl. Acad. Sci. USA 2007, 104, 5901–5906. [Google Scholar] [CrossRef]
- Hansen, C.L.; Pelegri, F. Primordial germ cell specification in vertebrate embryos: Phylogenetic distribution and conserved molecular features of preformation and induction. Front. Cell Dev. Biol. 2021, 9, 730332. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Peters, A.H.F.M.; Newmark, P.A. A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 2006, 11, 159–169. [Google Scholar] [CrossRef]
- Chuma, S.; Hosokawa, M.; Kitamura, K.; Kasai, S.; Fujioka, M.; Hiyoshi, M.; Takamune, K.; Noce, T.; Nakatsuji, N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc. Natl. Acad. Sci. USA 2006, 103, 15894–15899. [Google Scholar] [CrossRef] [PubMed]
- Uyhazi, K.E.; Yang, Y.; Liu, N.; Qi, H.; Huang, X.A.; Mak, W.; Weatherbee, S.D.; de Prisco, N.; Gennarino, V.A.; Song, X.; et al. Pumilio proteins utilize distinct regulatory mechanisms to achieve complementary functions required for pluripotency and embryogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 7851–7862. [Google Scholar] [CrossRef]
- Sekiné, K.; Furusawa, T.; Hatakeyama, M. The boule gene is essential for spermatogenesis of haploid insect male. Dev. Biol. 2015, 399, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Irish, V.; Lehmann, R.; Akam, M. The Drosophila posterior group gene nanos functions by repressing hunchback activity. Nature 1989, 338, 646–648. [Google Scholar] [CrossRef]
- De Keuckelaere, E.; Hulpiau, P.; Saeys, Y.; Berx, G.; van Roy, F. Nanos genes and their role in development and beyond. Cell. Mol. Life Sci. 2018, 75, 1929–1946. [Google Scholar] [CrossRef]
- Skinner, D.E.; Popratiloff, A.; Alrefaei, Y.N.; Mann, V.H.; Rinaldi, G.; Brindley, P.J. Functional analysis of vasa/PL10-like genes in the ovary of Schistosoma mansoni. Mol. Biochem. Parasitol. 2020, 236, 111259. [Google Scholar] [CrossRef]
- Chang, T.-C.; Liu, W.-S. The molecular evolution of PL10 homologs. BMC Evol. Biol. 2010, 10, 127. [Google Scholar] [CrossRef]
- Lasko, P. The DEAD-box helicase Vasa: Evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 810–816. [Google Scholar] [CrossRef]
- Moore, F.L.; Jaruzelska, J.; Fox, M.S.; Urano, J.; Firpo, M.T.; Turek, P.J.; Dorfman, D.M.; Pera, R.A.R. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 538–543. [Google Scholar] [CrossRef]
- Urano, J.; Fox, M.S.; Pera, R.A.R. Interaction of the conserved meiotic regulators, BOULE (BOL) and PUMILIO-2 (PUM2). Mol. Reprod. Dev. 2005, 71, 290–298. [Google Scholar] [CrossRef]
- Fox, M.; Urano, J.; Pera, R.A.R. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics 2005, 85, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Jaruzelska, J.; Kotecki, M.; Kusz, K.; Spik, A.; Firpo, M.; Pera, R.A.R. Conservation of a Pumilio-Nanos complex from Drosophila germ plasm to human germ cells. Dev Genes Evol. 2003, 213, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, T.; Ladd, A.N. The importance of CELF control: Molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip Rev. RNA 2012, 3, 104–121. [Google Scholar] [CrossRef]
- Shi, D.L.; Grifone, R. RNA-Binding Proteins in the Post-transcriptional Control of Skeletal Muscle Development, Regenerationand Disease. Front. Cell Dev. Biol. 2021, 9, 738978. [Google Scholar] [CrossRef] [PubMed]
- Brusca, R.C.; Brusca, G.J. Invertebrates, 2nd ed.; Sinauer Associates: Sunderland, UK, 2003; 936p. [Google Scholar]
- Korn, H. Morphogenese der Tiere. Lieferung 5: H-I. Annelida (einschließlich Echiurida und Sipunculida); VEB Gustav Fischer Verlag: Jena, Germany, 1982; 599 S. [Google Scholar]
- Anderson, D.T. Embryology and Phylogeny in Annelids and Arthropods; Pergamon Press: London, UK, 1973; 495p. [Google Scholar]
- Kostyuchenko, R.P.; Dondua, A.K. Development of the prototroch in embryogenesis of Nereis virens (polychaeta). Russ. J. Dev. Biol. 2006, 37, 69–76. [Google Scholar] [CrossRef]
- Carrillo-Baltodano, A.M.; Seudre, O.; Guynes, K.; Martin-Duran, J.M. Early embryogenesis and organogenesis in the annelid Owenia fusiformis. EvoDevo 2021, 12, 5. [Google Scholar] [CrossRef]
- Kuo, D.-H. The polychaete-to-clitellate transition: An EvoDevo perspective. Dev. Biol. 2017, 427, 230–240. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P.; Kozin, V.V.; Kupriashova, E.E. Regeneration and asexual reproduction in annelids: Cells, genes, and evolution. Biol. Bull. 2016, 43, 185–194. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P.; Kozin, V.V. Morphallaxis versus Epimorphosis? Cellular and Molecular Aspects of Regeneration and Asexual Reproduction in Annelids. Biol. Bull. 2020, 47, 237–246. [Google Scholar] [CrossRef]
- Zattara, E.E.; Bely, A.E. Phylogenetic distribution of regeneration and asexual reproduction in Annelida: Regeneration is ancestral and fission evolves in regenerative clades. Invert. Biol. 2016, 135, 400–414. [Google Scholar] [CrossRef]
- Sugio, M.; Takeuchi, K.; Kutsuna, J.; Tadokoro, R.; Takahashi, Y.; Yoshida-noro, C.; Tochinai, S. Exploration of embryonic origins of germline stem cells and neoblasts in Enchytraeus japonensis (Oligochaeta, Annelida). Gene Expr. Patterns 2008, 8, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Oyama, A.; Shimizu, T. Transient occurrence of vasa-expressing cells in nongenital segments during embryonic development in the oligochaete annelid Tubifex tubifex. Dev. Genes Evol. 2007, 217, 675–690. [Google Scholar] [CrossRef] [PubMed]
- Bely, A.E.; Sikes, J.M. Latent regeneration abilities persist following recent evolutionary loss in asexual annelids. Proc. Natl. Acad. Sci. USA 2010, 107, 1464–1469. [Google Scholar] [CrossRef]
- de Jong, D.M.; Seaver, E.C. Investigation into the Cellular Origins of Posterior Regeneration in the Annelid Capitella Teleta. Regeneration 2018, 5, 61–77. [Google Scholar] [CrossRef]
- Gazave, E.; Behague, J.; Laplane, L.; Guillou, A.; Preau, L.; Demilly, A.; Balavoine, G.; Vervoort, M. Posterior elongation in the annelid Platynereis dumerilii involves stem cells molecularly related to primordial germ cells. Dev. Biol. 2013, 382, 246–267. [Google Scholar] [CrossRef]
- Pelosi, C.; Römbke, J. Enchytraeids as bioindicators of land use and management. Appl. Soil Ecol. 2018, 123, 775–779. [Google Scholar] [CrossRef]
- Rodriguez, P.; Arrate, J.A.; Martinez-Madrid, M. Life history of the oligochaete Enchytraeus coronatus (Annelida, Enchytraeidae) in agar culture. Invert. Biol. 2002, 121, 350–356. [Google Scholar] [CrossRef]
- Bergter, A.; Beck, L.A.; Paululat, A. Embryonic Development of the Oligochaete Enchytraeus Coronatus: An SEM and Histological Study of Embryogenesis from One-Cell Stage to Hatching. J. Morphol. 2004, 261, 26–42. [Google Scholar] [CrossRef]
- Bergter, A.; Paululat, A. Pattern of Body-Wall Muscle Differentiation during Embryonic Development of Enchytraeus Coronatus (Annelida: Oligochaeta; Enchytraeidae). J. Morphol. 2007, 268, 537–549. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P.; Amosov, A.V. Spatial colinear but broken temporal expression of duplicated ParaHox genes in asexually reproducing annelids, Nais communis and Pristina longiseta. Genes 2023, 14, 1501. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Kostyuchenko, R.P.; Kozin, V.V.; Filippova, N.A.; Sorokina, E.V. FoxA expression pattern in two polychaete species, Alitta virens and Platynereis dumerilii: Examination of the conserved key regulator of the gut development from cleavage through larval life, postlarval growth, and regeneration. Dev. Dyn. 2019, 248, 728–743. [Google Scholar] [CrossRef]
- Dondua, A.K.; Kostyuchenko, R.P.; Fedorova, Z.E. Effects of some cytoskeleton inhibitors on ooplasmic segregation in the Nereis virens egg. Int. J. Dev. Biol. 1997, 41, 853–858. [Google Scholar]
- Kostyuchenko, R.P.; Dondua, A.K. Ooplasmic segregation and axis formation in the polychaeteNereis virens embryo. Russ. J. Dev. Biol. 2000, 31, 95–105. [Google Scholar] [CrossRef]
- Shimizu, T. Cytoskeletal mechanisms of ooplasmic segregation in annelid eggs. Int. J. Dev. Biol. 1999, 43, 11–18. [Google Scholar]
- Yoshida, N.; Arai, A.; Aoki, M.; Moriya, M.; Sekiguchi, K.; Shimizu, T. The Cleavage Program in the 2d Cell Lineage of Tubifex Embryos. J. Morphol. 2019, 280, 568–586. [Google Scholar] [CrossRef]
- Iwanoff, P.P. Die Entwiklung der Larvalsegmente bei den Annelide. Z. Morph. Oekol. Tiere. 1928, 10, 62–161. [Google Scholar] [CrossRef]
- Kumar, S.; Tumu, S.C.; Helm, C.; Hausen, H. The Development of Early Pioneer Neurons in the Annelid Malacoceros Fuliginosus. BMC Evol. Biol. 2020, 20, 117. [Google Scholar] [CrossRef]
- Zhang, Q.-J.; Luo, Y.-J.; Wu, H.-R.; Chen, Y.-T.; Yu, J.-K. Expression of Germline Markers in Three Species of Amphioxus Supports a Preformation Mechanism of Germ Cell Development in Cephalochordates. EvoDevo 2013, 4, 17. [Google Scholar] [CrossRef]
- Mohri, K.; Nakamoto, A.; Shimizu, T. The ontogeny of nanos homologue expression in the oligochaete annelid Tubifex tubifex. Gene Expr. Patterns 2016, 20, 32–41. [Google Scholar] [CrossRef]
- Dill, K.K.; Seaver, E. Vasa and nanos are coexpressed in somatic and germ line tissue from early embryonic cleavage stages through adulthood in the polychaete Capitella sp. I. Dev. Genes Evol. 2008, 218, 453–463. [Google Scholar] [CrossRef]
- Kang, D.; Pilon, M.; Weisblat, D.A. Maternal and zygotic expression of a nanos-class gene in the leech Helobdella robusta: Primordial germ cells arise from segmental mesoderm. Dev. Biol. 2002, 245, 28–41. [Google Scholar] [CrossRef]
- Cho, S.-J.; Vallès, Y.; Weisblat, D.A. Differential Expression of Conserved Germ Line Markers and Delayed Segregation of Male and Female Primordial Germ Cells in a Hermaphrodite, the Leech Helobdella. Mol. Biol. Evol. 2014, 31, 341–354. [Google Scholar] [CrossRef][Green Version]
- Rabinowitz, J.S.; Chan, X.Y.; Kingsley, E.P.; Lambert, J.D. Nanos is required in somatic blast cell lineages in the posterior of a mollusc embryo. Curr. Biol. 2008, 18, 331–336. [Google Scholar] [CrossRef][Green Version]
- Kranz, A.M.; Tollenaere, A.; Norris, B.J.; Degnan, B.M.; Degnan, S.M. Identifying the germline in an equally cleaving mollusc: Vasa and Nanos expression during embryonic and larval development of the vetigastropod Haliotis asinina. J. Exp. Zool. (Mol. Dev. Evol.) 2010, 314, 267–279. [Google Scholar] [CrossRef]
- Joiner, M.L.; Wu, C.F. Nervous system function for the testis RNA-binding protein BOULE in Drosophila. J. Neurogenet. 2004, 18, 341–363. [Google Scholar] [CrossRef]
- Hoopfer, E.D.; Penton, A.; Watts, R.J.; Luo, L. Genomic analysis of Drosophila neuronal remodeling: A role for the RNA-binding protein Boule as a negative regulator of axon pruning. J. Neurosci. 2008, 28, 6092–6103. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, E.; Clausen, D.S.; Null, R.W.; Metzger, B.M.; Willis, A.D.; Özpolat, B.D. Segment number threshold determines juvenile onset of germline cluster expansion in Platynereis dumerilii. J. Exp. Zool. (Mol. Dev. Evol.) 2021, 338, 225–240. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostyuchenko, R.P.; Nikanorova, D.D.; Amosov, A.V. Germ Line/Multipotency Genes Show Differential Expression during Embryonic Development of the Annelid Enchytraeus coronatus. Biology 2023, 12, 1508. https://doi.org/10.3390/biology12121508
Kostyuchenko RP, Nikanorova DD, Amosov AV. Germ Line/Multipotency Genes Show Differential Expression during Embryonic Development of the Annelid Enchytraeus coronatus. Biology. 2023; 12(12):1508. https://doi.org/10.3390/biology12121508
Chicago/Turabian StyleKostyuchenko, Roman P., Daria D. Nikanorova, and Artem V. Amosov. 2023. "Germ Line/Multipotency Genes Show Differential Expression during Embryonic Development of the Annelid Enchytraeus coronatus" Biology 12, no. 12: 1508. https://doi.org/10.3390/biology12121508
APA StyleKostyuchenko, R. P., Nikanorova, D. D., & Amosov, A. V. (2023). Germ Line/Multipotency Genes Show Differential Expression during Embryonic Development of the Annelid Enchytraeus coronatus. Biology, 12(12), 1508. https://doi.org/10.3390/biology12121508





