Integrated Analysis of the Intestinal Microbiota and Transcriptome of Fenneropenaeus chinensis Response to Low-Salinity Stress
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Shrimp Rearing and Sample Collection
2.2. Microbiota Analysis Using 16S rDNA Genes Sequencing
2.3. Gene Expression Analysis Using RNA Sequencing
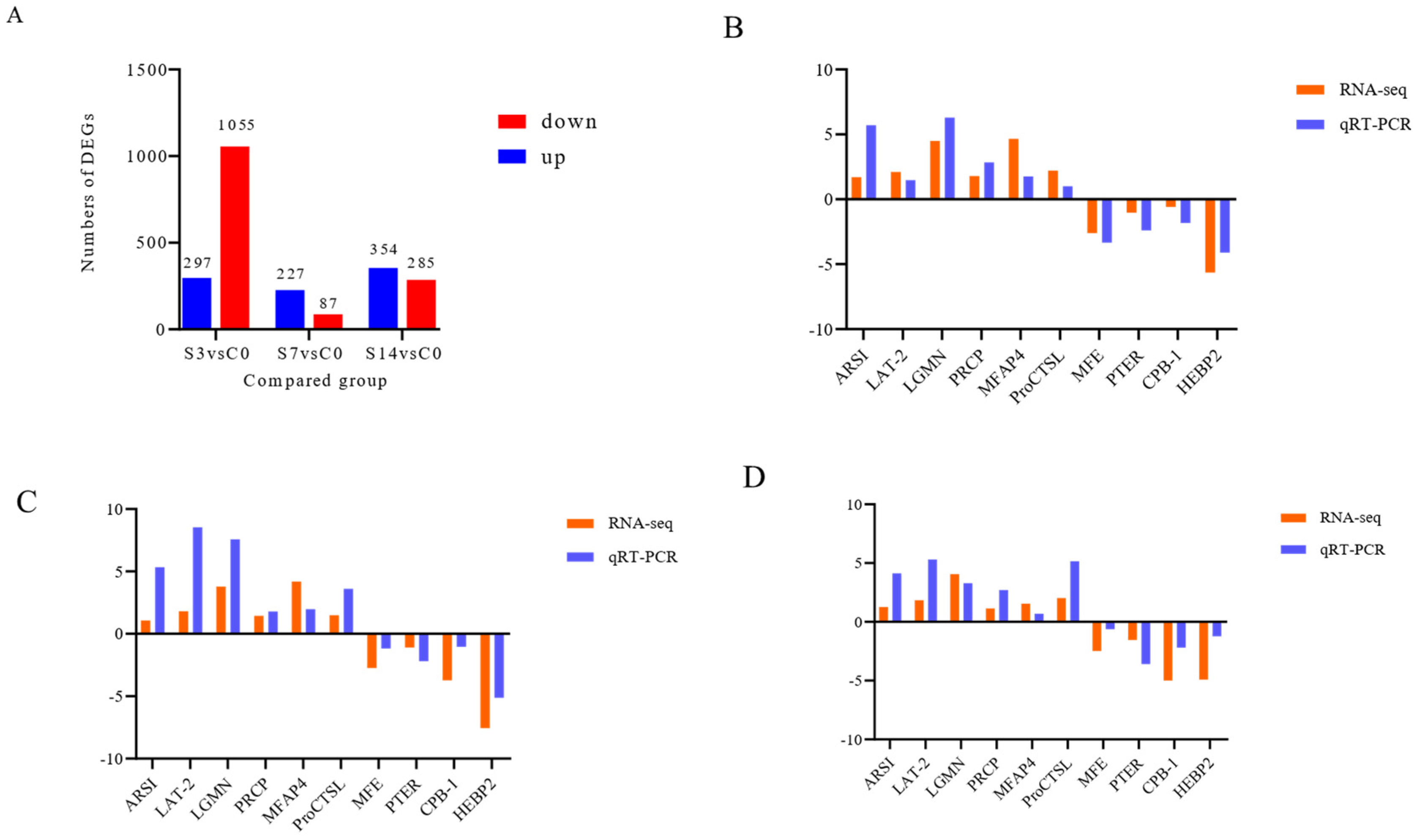
2.4. Verification of Real-Time Quantitative PCR (qPCR)
2.5. Correlations between ASVs and DEGs
3. Results
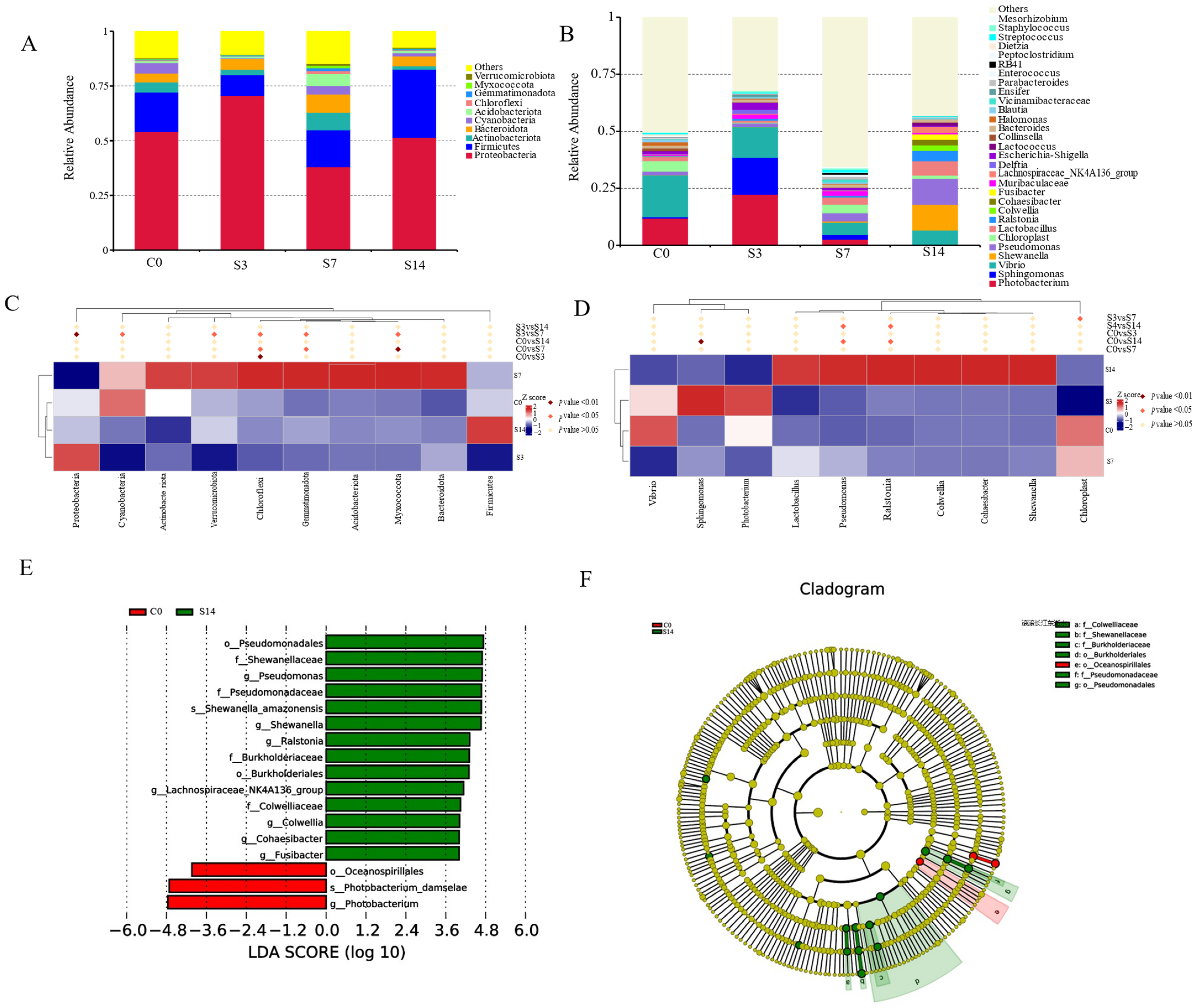
3.1. Intestinal Microbiota Community Analysis
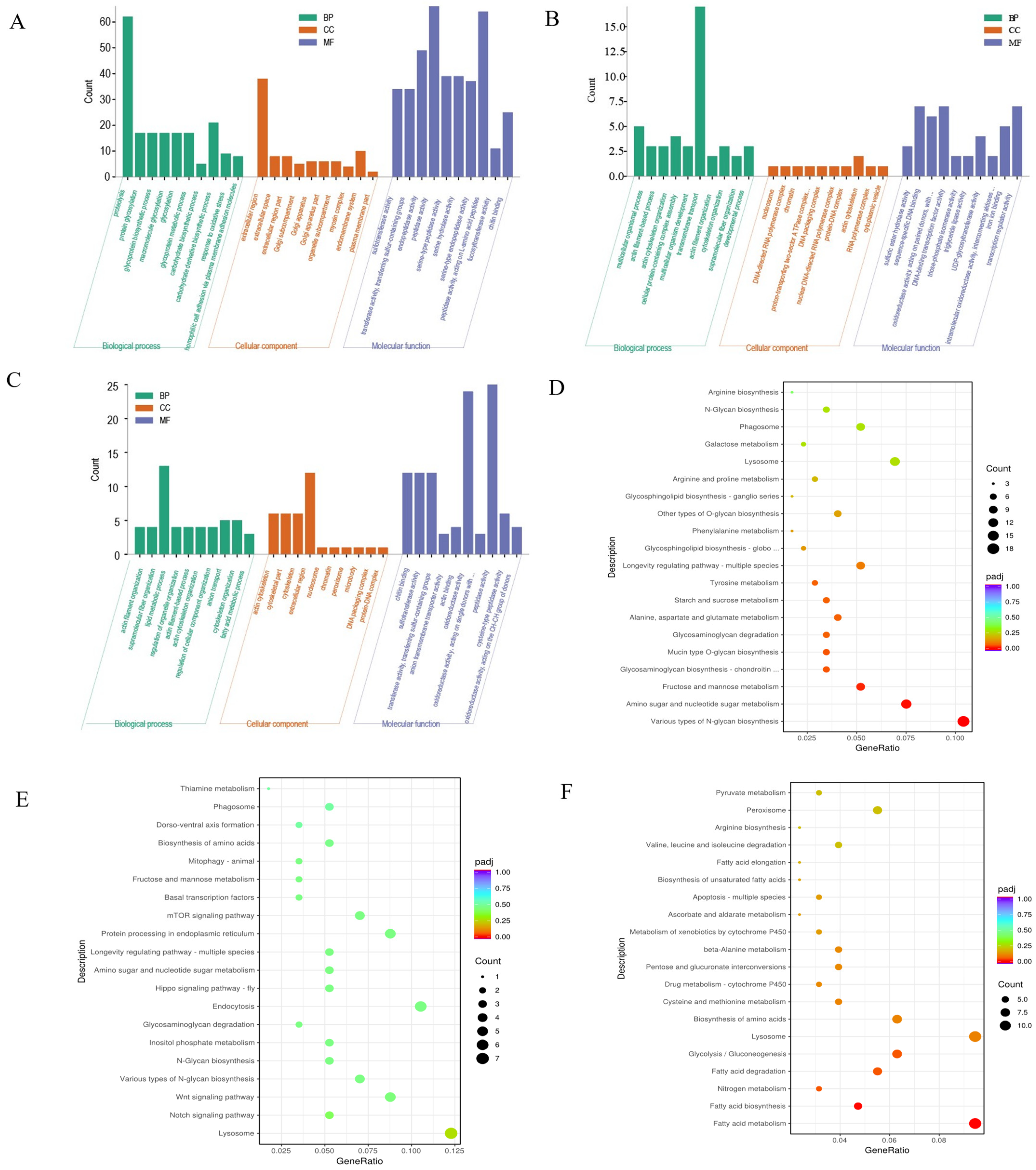
3.2. Intestinal Transcriptome Analysis
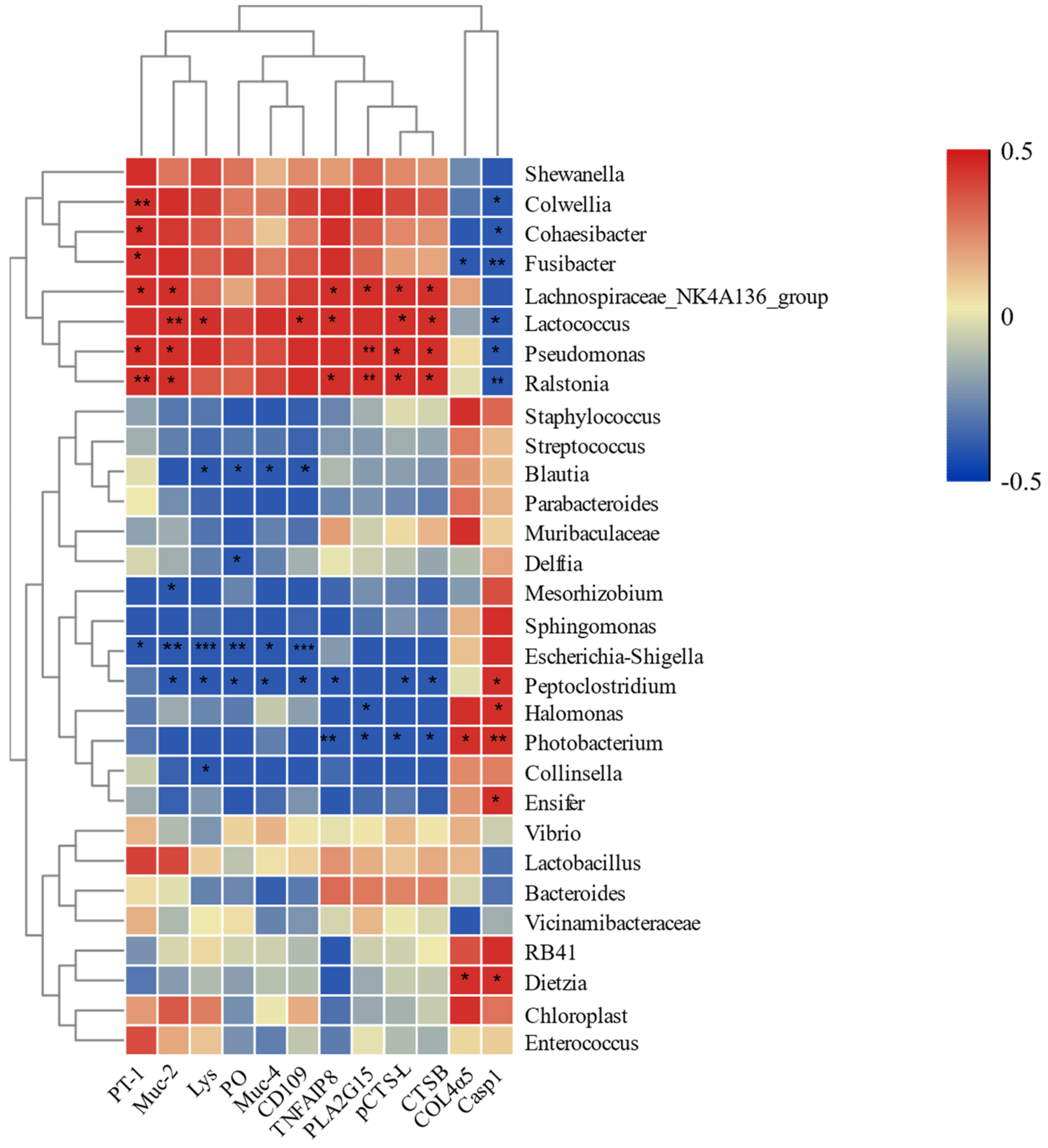
3.3. Correlation between Intestinal Microbiota and Immune-Related DEGs
4. Discussion
4.1. Intestinal Microbial Community Changed under Low-Salinity Stress
4.2. Participation of Immune-Related Genes in Response to Low-Salinity Stress
4.3. Relationship between Intestinal Microbial Community and Expression of Immune-Related Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, S.; Jeon, H.; Im, J.H.; Han, H.S. Evaluation of biofloc-based aquaculture of Pacific white shrimp (Litopenaeus vannamei) using water from a low-salinity artificial reservoir in Korea. Aquac. Res. 2022, 53, 4246–4255. [Google Scholar] [CrossRef]
- Liu, B.; Wang, Q.; Li, J.; Liu, P.; He, Y. A genetic linkage map of marine shrimp Penaeus (Fenneropenaeus) chinensis based on AFLP, SSR, and RAPD markers. Chin. J. Oceanol. Limnol. 2010, 28, 815–825. [Google Scholar] [CrossRef]
- Chen, L.H.; Zeng, W.H.; Rong, Y.Z.; Lou, B. Characterisation of taste-active compositions, umami attributes and aroma compounds in Chinese shrimp. Int. J. Food Sci. Technol. 2021, 56, 6311–6321. [Google Scholar] [CrossRef]
- Yu, H.; Liu, Y.; Liu, C.; Fan, F. Spatiotemporal variation and inequality in China’s economic resilience across cities and urban agglomerations. Sustainability 2018, 10, 4754. [Google Scholar] [CrossRef]
- Liu, B.; Yu, Z.M.; Song, X.X.; Guan, Y.Q.; Jian, X.F.; He, J.G. The effect of acute salinity change on white spot syndrome (WSS) outbreaks in Fenneropenaeus chinensis. Aquaculture 2006, 253, 163–170. [Google Scholar] [CrossRef]
- Jiang, H.F.; Liu, X.L.; Chang, Y.Q.; Liu, M.T.; Wang, G.X. Effects of dietary supplementation of probiotic Shewanella colwelliana WA64, Shewanella olleyana WA65 on the innate immunity and disease resistance of abalone, Haliotis discus hannai Ino. Fish Shellfish Immunol. 2013, 35, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Chaiyapechara, S.; Uengwetwanit, T.; Arayamethakorn, S.; Bunphimpapha, P.; Phromson, M.; Jangsutthivorawat, W.; Tala, S.; Karoonuthaisiri, N.; Rungrassamee, W. Understanding the host-microbe-environment interactions: Intestinal microbiota and transcriptomes of black tiger shrimp Penaeus monodon at different salinity levels. Aquaculture 2022, 546, 737371. [Google Scholar] [CrossRef]
- Roy, L.A.; Davis, D.A.; Saoud, I.P.; Boyd, C.A.; Pine, H.J.; Boyd, C.E. Shrimp culture in inland low salinity waters. Rev. Aquac. 2010, 2, 191–208. [Google Scholar] [CrossRef]
- Prapaiwong, N.; Boyd, C.E. Effluent volume and pollutant loads at an inland, low-salinity, shrimp farm in Alabama. Aquac. Eng. 2012, 48, 1–5. [Google Scholar] [CrossRef]
- Castillo-Soriano, F.A.; Ibarra-Junquera, V.; Escalante-Minakata, P.; Mendoza-Cano, O.; de Jesús Ornelas-Paz, J.; Almanza-Ramírez, J.C.; Meyer-Willerer, A.O. Nitrogen dynamics model in zero water exchange, low salinity intensive ponds of white shrimp, Litopenaeus vannamei, at Colima, Mexico. Lat. Am. J. Aquat. Res. 2013, 41, 68–79. [Google Scholar] [CrossRef]
- Izral, N.M.; Brua, R.B.; Culp, J.M.; Yates, A.G. Developing metabolomics-based bioassessment: Crayfish metabolome sensitivity to food and dissolved oxygen stress. Environ. Sci. Pollut. Res. 2018, 25, 36184–36193. [Google Scholar] [CrossRef] [PubMed]
- Ayi, Q.; Zeng, B.; Yang, K.; Lin, F.; Zhang, X.; Van Bodegom, P.M.; Cornelissen, J.H. Similar growth performance but contrasting biomass allocation of root-flooded terrestrial plant Alternanthera philoxeroides (Mart.) Griseb. in response to nutrient versus dissolved oxygen stress. Front. Plant Sci. 2019, 10, 111. [Google Scholar] [CrossRef]
- Kumar, N.; Kumar, S.; Singh, A.K.; Gite, A.; Patole, P.B.; Thorat, S.T. Exploring mitigating role of zinc nanoparticles on arsenic, ammonia and temperature stress using molecular signature in fish. J. Trace Elem. Med. Biol. 2022, 74, 127076. [Google Scholar] [CrossRef] [PubMed]
- Prymaczok, N.C.; Pasqualino, V.M.; Viau, V.E.; Rodríguez, E.M.; Medesani, D.A. Involvement of the crustacean hyperglycemic hormone (CHH) in the physiological compensation of the freshwater crayfish Cherax quadricarinatus to low temperature and high salinity stress. J. Comp. Physiology. B Biochem. Syst. Environ. Physiol. 2016, 186, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Dawood, M.A.; Koshio, S.; Ishikawa, M.; El-Sabagh, M.; Yokoyama, S.; Wang, W.L.; Zhang, Y.; Olivier, A. Physiological response, blood chemistry profile and mucus secretion of red sea bream (Pagrus major) fed diets supplemented with Lactobacillus rhamnosus under low salinity stress. Fish Physiol. Biochem. 2017, 43, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Pan, J.; Wang, Y.; Qiao, Y.; Han, F.; Xu, C.; Farhadi, A.; Li, E. Growth, health status and gut microbiota of the scalloped spiny lobster (Panulirus homarus) at different salinities. Aquaculture 2023, 562, 738779. [Google Scholar] [CrossRef]
- Adeyemi, J.A.; Deaton, L.E.; Pesacreta, T.C.; Klerks, P.L. Effects of copper on osmoregulation in sheepshead minnow, Cyprinodon variegatus acclimated to different salinities. Aquat. Toxicol. 2012, 109, 111–117. [Google Scholar] [CrossRef]
- Joseph, A.; Philip, R. Acute salinity stress alters the haemolymph metabolic profile of Penaeus monodon and reduces immunocompetence to white spot syndrome virus infection. Aquaculture 2007, 272, 87–97. [Google Scholar] [CrossRef]
- González-Ortegón, E.; Pascual, E.; Drake, P. Respiratory responses to salinity, temperature and hypoxia of six caridean shrimps from different aquatic habitats. J. Exp. Mar. Biol. Ecol. 2013, 445, 108–115. [Google Scholar] [CrossRef]
- Xu, W.B.; Zhang, Y.M.; Li, B.Z.; Lin, C.Y.; Chen, D.Y.; Cheng, Y.X.; Guo, X.L.; Dong, W.R.; Shu, M.A. Effects of low salinity stress on osmoregulation and gill transcriptome in different populations of mud crab Scylla paramamosain. Sci. Total Environ. 2023, 867, 161522. [Google Scholar] [CrossRef]
- Izvekova, I.G.; Solovyev, M.M. Characteristics of the effect of cestodes parasitizing the fish intestine on the activity of the host proteinases. Biol. Bull. 2016, 43, 146–151. [Google Scholar] [CrossRef]
- Kim, P.S.; Shin, N.R.; Lee, J.B.; Kim, M.S.; Whon, T.W.; Hyun, D.W.; Yun, J.H.; Jung, M.J.; Kim, J.Y.; Bae, J.W. Host habitat is the major determinant of the gut microbiome of fish. Microbiome 2021, 9, 166. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Xin, W.; Li, T.; Xue, M.; Ma, Z.; Jiang, N.; Luo, L. Comparative study on the effects of different feeding habits and diets on intestinal microbiota in Acipenser baeri Brandt and Huso huso. BMC Microbiol. 2019, 19, 297. [Google Scholar] [CrossRef] [PubMed]
- Gibiino, G.; De Siena, M.; Sbrancia, M.; Binda, C.; Sambri, V.; Gasbarrini, A.; Fabbri, C. Dietary habits and gut microbiota in healthy adults: Focusing on the right diet. A Systematic Review. Int. J. Mol. Sci. 2021, 22, 6728. [Google Scholar] [CrossRef] [PubMed]
- Cornejo-Granados, F.; Lopez-Zavala, A.A.; Gallardo-Becerra, L.; Mendoza-Vargas, A.; Sánchez, F.; Vichido, R.; Brieba, L.G.; Viana, M.T.; Sotelo-Mundo, R.R.; Ochoa-Leyva, A. Microbiome of Pacific Whiteleg shrimp reveals differential bacterial community composition between Wild, Aquacultured and AHPND/EMS outbreak conditions. Sci. Rep. 2017, 7, 11783. [Google Scholar] [CrossRef] [PubMed]
- Patrice, D.C.; Matthias, V.H.; Charlotte, L.; Clara, D.; Marialetizia, R. Amandine Everard. Microbial regulation of organismal energy homeostasis. Nat. Metab. 2019, 1, 34–46. [Google Scholar]
- Pérez, T.; Balcázar, J.L.; Ruiz-Zarzuela, I.; Halaihel, N.; Vendrell, D.; de Blas, I.; Múzquiz, J.L. Host-microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 2010, 3, 355–360. [Google Scholar] [CrossRef]
- Wang, A.R.; Ran, C.; Ringø, E.; Zhou, Z.G. Progress in fish gastrointestinal microbiota research. Rev. Aquac. 2018, 10, 620–640. [Google Scholar] [CrossRef]
- Qi, X.; Tu, X.; Zha, J.; Huang, A.; Wang, G.; Ling, F. Immunosuppression-induced alterations in fish gut microbiota may increase the susceptibility to pathogens. Fish Shellfish Immunol. 2019, 88, 540–545. [Google Scholar] [CrossRef]
- Whittamore, J.M. Osmoregulation and epithelial water transport: Lessons from the intestine of marine teleost fish. J. Comp. Physiology. B Biochem. Syst. Environ. Physiol. 2012, 182, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Duan, G.; Zhu, Y.; Zhu, D. Gut microbiota and transcriptome response of earthworms (Metaphire guillelmi) to polymyxin B exposure. J. Environ. Sci. 2023, 133, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.X. Study on Behavior and Related Functional Genes in the cAMP Pathway of Fenneropenaeus chinensis under Acute Low Salt and High Density Stress. Master’s Thesis, Shanghai Ocean University, Shanghai, China, 2022. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2, High-resolution sample inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Minchin, P.R. An evaluation of the relative robustness of techniques for ecological ordination. Vegetatio 1987, 69, 89–107. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Huttenhower, G.C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D.L. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zou, S.; Zhai, L.; Wang, Y.; Zhang, F.; An, L.; Yang, G. Pathogen invasion changes the intestinal microbiota composition and induces innate immune responses in the zebrafish intestine. Fish Shellfish Immunol. 2017, 71, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Z.; Huang, J.S.; Zhang, J.D.; Wang, Z.L.; Li, H.J.; Amenyogbe, E.; Chen, G. Effects of hypoxia stress on the intestinal microflora of juvenile of cobia (Rachycentron canadum). Aquaculture 2021, 536, 736419. [Google Scholar] [CrossRef]
- Christiane, H.; Hannes, R.; Andreas, K.; Inken, T.; Astrid, G. Effects of thermal stress on the gut microbiome of Juvenile Milkfish (Chanos chanos). Microorganisms 2020, 9, 5. [Google Scholar]
- Garibay-Valdez, E.; Martínez-Córdova, L.R.; Vargas-Albores, F.; Gollas-Galván, T.; Lago-Leston, A.; Calderón, K.; Martínez-Porchas, M. Biofilm consumption shapes the intestinal microbiota of shrimp (Penaeus vannamei). Aquac. Nutr. 2019, 25, 427–435. [Google Scholar] [CrossRef]
- Cai, M.l.; Li, H.H.; Gu, X.Z.; Tian, H.Y.; Liu, F.; Yang, W.P.; Ren, S.J.; Chu, W.Y.; Hu, Y.; Wang, A.M.; et al. Re-aliment regains feed deprivation-induced microflora dysbiosis and immune stress in the gut of red swamp crayfish (Procambarus clarkii). Aquac. Rep. 2022, 22, 100992. [Google Scholar] [CrossRef]
- Litvak, Y.; Byndloss, M.X.; Tsolis, R.M.; Bäumler, A.J. Dysbiotic Proteobacteria expansion: A microbial signature of epithelial dysfunction. Curr. Opin. Microbiol. 2017, 39, 1–6. [Google Scholar] [CrossRef]
- Shin, N.; Whon, T.W.; Bae, J. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Jia, Z.; Wu, A.; He, M.; Zhang, L.; Wang, C.; Chen, A. Metabolites of stable fly reduce diarrhea in mice by modulating the immune system, antioxidants, and composition of gut microbiota. Microb. Pathog. 2019, 134, 103557. [Google Scholar] [CrossRef]
- Feng, W.W.; Liu, J.; Tan, Y.Z.; Ao, H.; Wang, J.; Peng, C. Polysaccharides from Atractylodes macrocephala Koidz. Ameliorate ulcerative colitis via extensive modification of gut microbiota and host metabolism. Food Res. Int. 2020, 138, 109777. [Google Scholar] [CrossRef]
- Fan, X.J.; Zhang, Y.Y.; Song, Y.; Zhao, Y.Y.; Xu, Y.N.; Guo, F.; Shao, M.W.; Ma, X.J.; Zhang, W.; Wei, F.Y.; et al. Compound Danshen Dripping Pills moderate intestinal flora and the TLR4/MyD88/NF-κB signaling pathway in alleviating cognitive dysfunction in type 2 diabetic KK-Ay mice. Phytomedicine 2023, 111, 154656. [Google Scholar] [CrossRef] [PubMed]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed]
- García de la Banda, I.; Lobo, C.; Chabrillón, M.; León-Rubio, J.M.; Arijo, S.; Pazos, G.; Lucas, L.M.; Moriñigo, M.Á. Influence of dietary administration of a probiotic strain Shewanella putrefaciens on Senegalese sole (Solea senegalensis, Kaup 1858) growth, body composition and resistance to Photobacterium damselae subsp piscicida. Aquac. Res. 2012, 43, 662–669. [Google Scholar] [CrossRef]
- Kumar, J.; D’Souza, S.F. An optical microbial biosensor for detection of methyl parathion using Sphingomonas sp. immobilized on microplate as a reusable biocomponent. Biosens. Bioelectron. 2010, 26, 1292–1296. [Google Scholar] [CrossRef] [PubMed]
- Silveira, D.R.; Milan, C.; Rosa, J.V.D.; Timm, C.D. Fatores de patogenicidade de Vibrio spp. de importância em doenças transmitidas por alimentos. Arq. Inst. Biológico 2016, 83. [Google Scholar] [CrossRef]
- Angthong, P.; Uengwetwanit, T.; Arayamethakorn, S.; Chaitongsakul, P.; Rungrassamee, W. Bacterial analysis in the early developmental stages of the black tiger shrimp (Penaeus monodon). Sci. Rep. 2020, 10, 4896. [Google Scholar] [PubMed]
- Zadeh, S.S. Bacterial Flora Associated with Hatchery-Reared Juvenile Giant Tiger Prawn, Penaeus monodon (fabricius), and Screening of Putative Bacteria as Probiotic Candidate. Ph.D. Dissertation, Universiti Putra Malaysia, Seri Kembangan, Malaysia, 2012. [Google Scholar]
- Kinjo, Y.; Pei, B.; Bufali, S.; Raju, R.; Richardson, S.K.; Imamura, M.; Fujio, M.; Wu, D.; Khurana, A.; Kawahara, K.; et al. Natural Sphingomonas glycolipids vary greatly in their ability toactivate natural killer T cells. J. Biol. Chem. 2008, 15, 654–664. [Google Scholar] [CrossRef]
- Wang, J.; Sandoval, K.; Ding, Y.; Stoeckel, D.; Minard-Smith, A.; Andersen, G.; Dubinsky, E.A.; Atlas, R.; Gardinali, P. Biodegradation of dispersed Macondo crude oil by indigenous gulf of Mexico microbial communities. Sci. Total Environ. 2016, 557, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jiang, Q.; Ji, H.; Ning, J.; Li, C.; Zheng, H. Type 1 diabetes induces cognitive dysfunction in rats associated with alterations of the gut microbiome and metabolomes in serum and hippocampus. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2019, 1865, 165541. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elrahim, R.; Tohamy, M.R.A.; Atia, M.M.; Elashtokhy, M.M.A.; Ali, M.A.S. Bactericidal activity of some plant essential oils against Ralstonia solanacearum infection. Saudi J. Biol. Sci. 2022, 29, 2163–2172. [Google Scholar] [CrossRef] [PubMed]
- Effantin, G.; Hamasaki, R.; Kawasaki, T.; Bacia, M.; Moriscot, C.; Weissenhorn, W.; Yamada, T.; Schoehn, G. Cryo-Electron Microscopy Three-Dimensional Structure of the Jumbo Phage ΦRSL1 Infecting the phytopathogen Ralstonia solanacearum. Structure 2013, 21, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.S.S.; Aravind, R.; Balasubramanian, C.P.; Kumar, S.; Antony, J.; Biju, I.F.; Sangeetha, V.L.; Ambasankar, K.; Vijayan, K.K. Growth, survival, and osmo-ionic regulation in post larval and juvenile Indian white shrimp, Penaeus indicus, reared under three levels of salinity in a semifloc system. Aquaculture 2023, 564, 739042. [Google Scholar] [CrossRef]
- Xu, D.; Liu, W.; Alvarez, A.; Huang, T. Cellular immune responses against viral pathogens in shrimp. Dev. Comp. Immunol. 2014, 47, 287–297. [Google Scholar] [CrossRef]
- Luo, T.; Zhang, X.; Shao, Z.; Xu, X. PmAV, a novel gene involved in virus resistance of shrimp Penaeus monodon. FEBS Lett. 2003, 551, 53–57. [Google Scholar] [CrossRef]
- Sun, M.; Li, S.; Zhang, X.; Xiang, J.; Li, F. Isolation and transcriptome analysis of three subpopulations of shrimp hemocytes reveals the underlying mechanism of their immune functions. Dev. Comp. Immunol. 2020, 108, 103689. [Google Scholar] [CrossRef] [PubMed]
- Melo-Gonzalez, F.; Fenton, T.M.; Forss, C.; Smedley, C.; Goenka, A.; MacDonald, A.S.; Thornton, D.J.; Travis, M.A. Intestinal mucin activates human dendritic cells and IL-8 production in a glycan-specific manner. J. Biol. Chem. 2018, 293, 8543–8553. [Google Scholar] [CrossRef]
- Shan, M.; Gentile, M.; Yeiser, J.R.; Walland, A.C.; Bornstein, V.U.; Chen, K.; He, B.; Cassis, L.; Bigas, A.; Cols, M.; et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science 2013, 342, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.V.; Larsson, J.M.H.; Hansson, G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host--microbial interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 4659–4665. [Google Scholar] [CrossRef] [PubMed]
- Daniel, T.; Masooma, S.; Andrea, S. Irinotecan induces enterocyte cell death and changes to muc2 and muc4 composition during mucositis in a tumour-bearing DA rat model. Cancer Chemother. Pharmacol. 2019, 83, 893–904. [Google Scholar]
- Gao, X.; Zhong, S.; Tong, Y.; Liang, Y.; Feng, G.; Zhou, X.; Zhang, Z.; Huang, G. Alteration and prognostic values of collagen gene expression in patients with gastric cancer under different treatments. Pathol.—Res. Pract. 2020, 216, 152831. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wang, Y.; Xiong, D.; Zhang, J. RNA-seq revealed the signatures of immunity and metabolism in the Litopenaeus vannamei intestine in response to dietary succinate. Fish Shellfish Immunol. 2019, 95, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wang, Y.; Liu, Q.; Zhang, J.; Xiong, D. Changes in the intestine barrier function of Litopenaeus vannamei in response to pH stress. Fish Shellfish Immunol. 2019, 88, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.C.; Li, L.L.; Wang, D.F.; Ren, L.K.; Li, J.S.; Lu, Y.L.; Meng, Y.Q.; Ma, R.; Wang, S.L.; Li, X.P.; et al. Fe/N-doped carbon dots-based nanozyme with super peroxidase activity, high biocompatibility and antibiofilm ability for food preservation. Chem. Eng. J. 2023, 473, 145291. [Google Scholar] [CrossRef]
- Shayman, J.A.; Kelly, R.; Kollmeyer, J.; He, Y.; Abe, A. Group XV phospholipase A2, a lysosomal phospholipase A2. Progress Lipid Res. 2011, 50, 1–13. [Google Scholar] [CrossRef]
- Kuester, D.; Lippert, H.; Roessner, A.; Krueger, S. The cathepsin family and their role in colorectal cancer. Pathol.—Res. Pract. 2008, 204, 491–500. [Google Scholar] [CrossRef]
- Creasy, B.M.; McCoy, K.L. Cytokines regulate cysteine cathepsins during TLR responses. Cell. Immunol. 2011, 267, 56–66. [Google Scholar] [CrossRef]
- Aono, Y.; Suzuki, Y.; Horiguchi, R.; Inoue, Y.; Karayama, M.; Hozumi, H.; Furuhashi, K.; Enomoto, N.; Fujisawa, T.; Nakamura, Y.; et al. CD109 on dendriticcells regulates airway hyperreactivity and eosinophilic airway inflammation. Am. J. Respir. Cell Mol. Biol. 2023, 68, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Tan, W.; Dong, L.J.; Tang, Q.; Yang, F.; Shi, X.X.; Jiang, D.M.; Qian, Y.W. TNFAIP8 influences the motor function in mice after spinal cord injury (SCI) through meditating inflammation dependent on AKT. Biochem. Biophys. Res. Commun. 2020, 528, 234–241. [Google Scholar] [CrossRef]
- Tellam, R.L.; Wijffels, G.; Willadsen, P. Peritrophic matrix proteins. Insect Biochem. Mol. Biol. 1999, 29, 87–101. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, M.Y.; Feng, X.H.; Zeng, Y.X.; Lin, D.J. Expression and prognosis analyses of CASP1 in acute myeloid leukemia. Aging 2021, 13, 14088. [Google Scholar] [CrossRef] [PubMed]
- Romero-Geraldo, R.D.J.; García-Lagunas, N.; Hernández-Saavedra, N.Y. Crassostrea gigas exposure to the dinoflagellate Prorocentrum lima: Histological and gene expression effects on the digestive gland. Mar. Environ. Res. 2016, 120, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Liu, M.; Mao-Ying, Q.; Liu, H.; Wang, Z.; Zhang, M.; Wang, J.; Li, Q.; Liu, S.; Mi, W.; et al. Early single Aspirin-triggered Lipoxin blocked morphine anti-nociception tolerance through inhibiting NALP1 inflammasome: Involvement of PI3k/Akt signaling pathway. Brain Behav. Immun. 2015, 50, 63–77. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, C.; Wang, Q.; Wang, J.; Li, J.; Guan, C.; He, Y.; Gao, H. Integrated Analysis of the Intestinal Microbiota and Transcriptome of Fenneropenaeus chinensis Response to Low-Salinity Stress. Biology 2023, 12, 1502. https://doi.org/10.3390/biology12121502
Tian C, Wang Q, Wang J, Li J, Guan C, He Y, Gao H. Integrated Analysis of the Intestinal Microbiota and Transcriptome of Fenneropenaeus chinensis Response to Low-Salinity Stress. Biology. 2023; 12(12):1502. https://doi.org/10.3390/biology12121502
Chicago/Turabian StyleTian, Caijuan, Qiong Wang, Jiajia Wang, Jitao Li, Chenhui Guan, Yuying He, and Huan Gao. 2023. "Integrated Analysis of the Intestinal Microbiota and Transcriptome of Fenneropenaeus chinensis Response to Low-Salinity Stress" Biology 12, no. 12: 1502. https://doi.org/10.3390/biology12121502
APA StyleTian, C., Wang, Q., Wang, J., Li, J., Guan, C., He, Y., & Gao, H. (2023). Integrated Analysis of the Intestinal Microbiota and Transcriptome of Fenneropenaeus chinensis Response to Low-Salinity Stress. Biology, 12(12), 1502. https://doi.org/10.3390/biology12121502






